Introduction
5-Fluorouracil (5-FU) is a key chemotherapeutic
agent in colorectal cancer (CRC) treatment, primarily acting by
inhibiting thymidylate synthase, which disrupts DNA synthesis, and
by incorporating toxic metabolites into RNA to impair cell
functions (1). Its efficacy is
enhanced in combination regimens such as leucovorin, 5-FU and
oxaliplatin) and FOLFIRI (leucovorin, 5-FU, and Irinotecan), often
alongside leucovorin, stabilizing its cytotoxic effects (2). Despite its effectiveness, 5-FU
treatment is frequently limited by resistance mechanisms and
adverse effects, such as myelosuppression and gastrointestinal
toxicity, highlighting the importance of biomarkers and
personalized strategies to predict treatment response and mitigate
side effects (3).
Advances in pharmacogenomics and combinatorial
approaches continue to refine the use of 5-FU, offering improved
outcomes for patients with CRC (4).
The efficacy and safety of 5-FU are notably influenced by
interindividual variability in drug metabolism, largely attributed
to genetic polymorphisms in the dihydropyrimidine dehydrogenase
(DPYD) gene, which encodes dihydropyrimidine dehydrogenase
(DPD), the key enzyme responsible for 5-FU catabolism (5). Deficient or decreased DPD activity,
caused by DPYD polymorphisms such as DPYD 85T>C
and DPYD 1896T>C, leads to the accumulation of toxic 5-FU
metabolites, resulting in severe and sometimes fatal toxicity,
including neutropenia, diarrhea and mucositis (6,7).
The three DPYD variants DPYD 85T>C,
DPYD 1627A>G, and DPYD 1896T>C are relatively
more prevalent in East and Southeast Asian populations compared
with European risk alleles such as DPYD*2A
(c.1905+1G>A) and DPYD 13 (c.1679T>G) (8). Previous pharmacogenetic studies,
including analyses in Thai cohorts and population databases, have
consistently demonstrated higher allele frequencies for these
variants in Asian populations (9,10).
Each variant has potential functional relevance: DPYD
85T>C is associated with decreased DPD enzymatic activity and
variable risk of fluoropyrimidine-related toxicity; DPYD
1627A>G represents a missense substitution with conflicting
reports on its functional consequences but occurs at a relatively
high frequency in Asian populations and DPYD 1896T>C has
been observed in patients with severe 5-FU-induced toxicity,
suggesting its role as a putative risk allele (6,7,11).
Finally, these variants are not currently incorporated into
international dosing guidelines such as those issued by the
Clinical Pharmacogenetics Implementation Consortium (4,12).
However, evidence (9,10) indicates that they may hold clinical
value in non-European populations, thereby justifying further
investigation.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
~22 nucleotides in length. miRNAs regulate gene expression at the
post-transcriptional level by mRNA degradation or translational
inhibition (13). The key roles of
miRNAs involve several biological processes such as cell
proliferation, differentiation and apoptosis (14). The aberrant expression of miRNAs has
been investigated in various cancer types and related to drug
resistance (15-17).
Circulating miRNAs may be a non-invasive biomarker for diagnosing
and predicting disease progression (18). Ferracin et al (19) found that miR-21-5p expression is
high in the plasma of patients with CRC patients. A previous study
also showed that miR-145, miR-106a and miR-17-3p were significantly
differentially expressed between patients with pre- and
post-operative stage II/III CRC. High levels of miR-17-3p and
miR-106a are associated with shorter disease-free survival,
suggesting they may serve as serum-miRNA-based biomarkers for
prognosis and predicting disease recurrence in patients with stage
II/III CRC (20). In addition,
recent studies have identified altered expression of other miRNAs
in patients with CRC patients, including upregulation of miR-374a
in both plasma and tumor tissues (21), as well as specific miRNA signatures
in saliva and lymphatic samples, which may serve as non-invasive
diagnostic biomarkers and help identify patients at high risk of
lymph node metastasis (22,23). Offer et al (24) demonstrated that miR-27a and miR-27b
may be pharmacological modulators of hepatic DPD enzyme function.
DPD is an important enzyme in the uracil catabolic pathway
converting the anti-cancer drug 5-FU to the inactive metabolite
5-dihydrofluorouracil. Deficiency of DPD resulting from inadequate
expression or deleterious variants in DPYD is associated
with severe toxic responses to 5-FU (24).
Moreover, patients who are heterozygous for miR-27a
SNV (rs895819) have an increased risk of fluoropyrimidine toxicity,
which was investigated in both DPYD wild-type and
DPYD variant carriers (25).
This suggests that miR-27a rs895819 may serve as a biomarker for
fluoropyrimidine-associated toxicity prediction. Previous studies
have predominantly focused on common DPYD variants observed
in European populations, such as DPYD 2A (c.1905+1G>A),
DPYD 13 (c.1679T>G), and HapB3 (c.1236G>A),
which are well-established markers of fluoropyrimidine toxicity
(8,26). To the best of our knowledge, few
studies (24,25) have concurrently assessed germline
DPYD polymorphisms alongside miRNA expression profiles to
investigate their combined impact on 5-FU toxicity. The present
study aimed to determine the miRNA expression profiles in the
DPYD variant (DPYD 85T>C and DPYD
1896T>C) in patients with CRC to facilitate use of miRNAs as
potential targets for predicting the toxicity of 5-FU and the
development of personalized patient profiles and therapeutic
interventions.
Materials and methods
Subjects
A total of 48 patients with CRC were recruited
between October 2020 and October 2023 at the Division of Oncology,
Ramathibodi Hospital, Mahidol University, Bangkok, Thailand. The
mean age was 64.7±11.9 years, ranging from 18-90 years. Among the
48 patients 28 (58.3%) were male and 20 (41.7% were female.
Inclusion criteria were as follows: CRC confirmed histologically or
cytologically; age ≥18 years; no previous treatment with 5-FU;
Eastern Cooperative Oncology Group (ECOG) (27) performance status of 0-2; life
expectancy >3 months; white blood cell,
4.5-11.0x109/l; hemoglobin, 13.2-16.6 g/dl for male
patients and 11.6-15.0 g/dl for female patients; neutrophil count
<1.5x109/l; platelet count <8x1010/l
and serum creatinine <1.5 mg/dl. Exclusion criteria were
pregnancy and any laboratory evidence of renal or hepatic
abnormality.
The present study was carried out in compliance with
the Declaration of Helsinki and approved by the Ethics Committee of
Ramathibodi Hospital, Mahidol University, Thailand (approval no.
MURA2020/1613 Ref.2419). The study procedure was explained to the
patients before the study andall patients signed the consent form
to participate in the study.
Molecular analysis
EDTA blood samples were collected to perform DNA
extraction using MagNA Pure Compact System (Roche Diagnostics
GmbH). TaqMan® real-time (RT)PCR ViiA7™
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was
used to detect three DPYD variants: DPYD 85T>C
(rs1801265, cat. no. C_9491497_10), DPYD 1627 A>G
(rs1801159, cat. no. C_1823316_20) and DPYD 1896T>C
(rs17376848, cat. no. C_25471727_20). All reagents were obtained
from Applied Biosystems, Thermo Fisher Scientific, Inc., and used
according to the manufacturer's instructions.
Toxicity assessment
The Common Terminology Criteria for Adverse Events
(CTCAE) v5.0(28) was used to
evaluate toxicity at first and second cycles of treatment. The
hematological toxicity included leukopenia, neutropenia,
thrombocytopenia and anemia. Grade 1-4 was regarded as
toxicity.
miRNA extraction and cDNA
synthesis
Plasma samples from patients with CRC were used for
miRNA extraction. Prior to extraction, plasma was centrifuged at
1,107 x g for 15 min at room temperature. Following the
manufacturer's protocol, 100 µl plasma was processed using the
miRNeasy® Serum/Plasma kit (cat. no. 217184; Qiagen
GmbH) for miRNA isolation. The RNA concentration was measured using
a NanoDrop 2000/2000c Spectrophotometer (Thermo Fisher Scientific,
Inc.). To synthesize cDNA, 200 ng RNA was used with the miRCURY
LNA™ RT kit (cat. no. 339340; Qiagen GmbH). The reaction
mixture included 5X miRCURY SYBR Green RT reaction buffer, 10X
miRCURY RT enzyme mix, Uni Sp6 RNA spike-in and the RNA template.
The reverse transcription reaction was performed in a thermal
cycler at 42˚C for 60 min, followed by enzyme inactivation at 95˚C
for 5 min and a final hold at 4˚C. cDNA was then stored at -20˚C
until further use.
miRNA array
The miRNA expression profiles were analyzed using
RT-PCR with a customized miRNA array panel (cat. no. 217184; Qiagen
GmbH) coated with specific primers for 43 miRNAs (sequences not
provided) involved in the 5-FU metabolic pathway. miR target
sequences are listed in Table SI.
The reactions were performed using the miRCURY LNA™ SYBR
Green PCR kit (cat. no. 339345, Qiagen GmbH). cDNA was diluted to
1:80 before mixing in the reaction containing 2X miRCURY SYBRGreen
mix and nuclease-free water. The 96-well plate of the miRNA array
was subjected to RT-PCR (Bio-Rad Laboratories, Inc.; cat. no. CFX
96). Thermocycling conditions were as follows: Initial heat
activation at 95˚C for 2 min, followed by 50 cycles of denaturation
at 95˚C for 10 sec and annealing/extension at 56˚C for 60 sec. The
relative miRNA expression was determined using the
2-ΔΔCq method (29). The
analysis of miRNA profiles was performed using the Qiagen web
portal at GeneGlobe (geneglobe.qiagen.com). Heatmaps were generated using
the GeneGlobe platform (geneglobe.qiagen.com/th, which applies an unsupervised
clustering algorithm. Unsupervised clustering groups both samples
and genes based on the similarity of their expression patterns,
rather than relying on predefined group labels. This approach
minimizes bias introduced by prior assumptions. This method was
selected because it is widely regarded as a standard for gene
expression analysis and provides an unbiased visualization of
expression profiles (30,31).
Statistical analysis
Hardy-Weinberg equilibrium of DPYD was
assessed using the χ2 test. Data were assessed for
normality of distribution. Descriptive statistics for patients were
presented as mean ± standard deviation (SD) for variables with a
normal distribution. The association between DPYD variants
status and the hematological toxicity was evaluated by Fisher's
exact test. All tests were performed using SPSS software version
21.0 (IBM Corp.). P-values were calculated using unpaired Student's
t-test for each miRNA. The test was performed as a parametric,
unpaired, two-sample t-test assuming equal variance with a
two-tailed distribution. The present study was designed as an
exploratory screen to identify candidate miRNAs for further
validation. Therefore, no multiplicity adjustment was performed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient demographics and clinical
data
A total of 48 patients were included in the present
study. The mean age was 64.7±11.9 years, with a male predominance
(58.3%, n=28 vs. 41.7%, n=20). Most patients had a performance
status, assessed by ECOG score, of 0 or 1, with a wide distribution
of primary tumor locations, including the rectum, sigmoid colon and
other sites across the colon. Metastases were most commonly
observed at sites other than the liver and lung (22.9%, n=11), with
the liver being the second most frequent site (18.8%, n=9). Tumors
were predominantly moderately differentiated (Table I).
 | Table IPatient characteristics (n=48). |
Table I
Patient characteristics (n=48).
| Characteristic | Value |
|---|
| Mean age,
years | 64.7±11.9 |
| Sex (%) | |
|
Male | 28 (58.3) |
|
Female | 20 (41.7) |
| ECOG score (%) | |
|
0 | 21 (43.8) |
|
1 | 22 (45.8) |
|
2 | 5 (10.4) |
| Site of cancer
(%) | |
|
Ascending
colon | 5 (10.4) |
|
Transverse
colon | 3 (6.3) |
|
Descending
colon | 4 (8.3) |
|
Sigmoid
colon | 14 (29.2) |
|
Rectosigmoid | 5 (10.4) |
|
Rectum | 16 (33.3) |
|
Colon,
unspecified | 1 (2.1) |
| Site of metastasis
(%) | |
|
Liver | 9 (18.8) |
|
Lung | 3 (6.3) |
|
Liver and
lung | 2 (4.2) |
|
Other | 11 (22.9) |
|
No | 23 (47.9) |
| Histopathology
(%) | |
|
Well
differentiated | 11 (22.9) |
|
Moderately
differentiated | 36 (75.0) |
|
Poorly
differentiated | 1 (2.1) |
Genotypic and allelic frequencies of
DPYD variants
A total of three single nucleotide polymorphisms
(SNPs) in the DPYD gene were analyzed: 85T>C (rs1801265),
1627A>G (rs1801159), and 1896T>C (rs17376848). All three
variants were present in the study cohort, with genotype
distributions consistent with Hardy-Weinberg equilibrium.
For the DPYD 85T>C polymorphism, the
majority of individuals were homozygous wild-type (72.9%), while
27.1% were heterozygous; no homozygous variant carriers were
observed. The corresponding allele frequencies were 0.86 for the
wild-type allele and 0.14 for the variant allele.
For the DPYD 1627A>G variant, 70.8% of
subjects carried the homozygous wild-type genotype, 25.0% were
heterozygous, and 4.2% were homozygous for the variant allele,
yielding allele frequencies of 0.83 (wild-type) and 0.17
(variant).
Similarly, the DPYD 1896T>C polymorphism
demonstrated 72.9% homozygous wild-type, 27.1% heterozygous, and no
homozygous variant genotypes, corresponding to allele frequencies
of 0.86 and 0.14 for the wild-type and variant alleles,
respectively (Table II).
 | Table IIDihydropyrimidine dehydrogenase
genotype and allele frequency. |
Table II
Dihydropyrimidine dehydrogenase
genotype and allele frequency.
| | Genotype frequency
(%) | Allele
frequency |
|---|
| SNP | rs ID no. | wt/wt | wt/vt | vt/vt | wt | vt |
|---|
| 85T>C | 1801265 | 35 (72.9) | 13 (27.1) | 0 (0.0) | 0.86 | 0.14 |
| 1627A>G | 1801159 | 34 (70.8) | 12 (25.0) | 2 (4.2) | 0.83 | 0.17 |
| 1896 T>C | 17376848 | 35 (72.9) | 13 (27.1) | 0 (0.0) | 0.86 | 0.14 |
Association of DPYD variants with
hematological toxicity
Hematological toxicity, including anemia,
leucopenia, neutropenia and thrombocytopenia, were assessed for
their association with DPYD variants across two chemotherapy
cycles (Table III). DPYD
85T>C showed a trend toward increased anemia, particularly
during the second chemotherapy cycle. Although the difference was
not significant, this suggests a possible cumulative effect of the
variant on drug metabolism over time. By contrast, DPYD
1627A>G and DPYD 1896T>C were not associated with
increased anemia in either cycle.
 | Table IIIDihydropyrimidine dehydrogenase
variants and hematological toxicity. |
Table III
Dihydropyrimidine dehydrogenase
variants and hematological toxicity.
| | First cycle | Second cycle |
|---|
| Toxicity | SNP | Genotype | n | Grade 0 (%) | Grade 1-4 (%) | P-value | Grade 0 (%) | Grade 1-4 (%) | P-value |
|---|
| Anemia | 85T>C | T/T | 35 | 19 (54.3) | 16 (45.7) | 0.371 | 21 (60.0) | 14 (40.0) | 0.070 |
| | | T/C | 13 | 5 (38.5) | 8 (61.5) | | 4 (30.8) | 9 (69.2) | |
| | 1627A>G | A/A | 34 | 18 (52.9) | 16 (47.1) | 0.492 | 17 (50.0) | 17 (50.0) | 0.626 |
| | | A/G or G/G | 14 | 6 (42.9) | 8 (57.1) | | 8 (57.1) | 6 (42.9) | |
| | 1896T>C | T/T | 35 | 17 (48.6) | 18 (51.4) | 0.727 | 18 (51.4) | 17 (48.6) | 0.873 |
| | | T/C | 13 | 7 (53.8) | 6 (46.2) | | 7 (53.8) | 6 (46.2) | |
| Leucopenia | 85T>C | T/T | 35 | 28 (80.0) | 7 (20.0) | 0.446 | 27 (77.1) | 8 (22.9) | 0.064 |
| | | T/C | 13 | 12 (92.3) | 1 (7.7) | | 13 (100.0) | 0 (0.0) | |
| | 1627A>G | A/A | 34 | 28 (82.4) | 6 (17.6) | >0.999 | 28 (82.4) | 6 (17.6) | >0.999 |
| | | A/G or G/G | 14 | 12 (85.7) | 2 (14.3) | | 12 (85.7) | 2 (14.3) | |
| | 1896T>C | T/T | 35 | 27 (77.1) | 8 (22.9) | 0.64 | 29 (82.9) | 6 (17.1) | >0.999 |
| | | T/C | 13 | 13 (100.0) | 0 (0.0) | | 11 (84.6) | 2 (15.4) | |
| Neutropenia | 85T>C | T/T | 35 | 27 (77.1) | 8 (22.9) | 0.724 | 25 (71.4) | 10 (28.6) | 0.498 |
| | | T/C | 13 | 11 (84.6) | 2 (15.4) | | 11 (84.6) | 2 (15.4) | |
| | 1627A>G | A/A | 34 | 26 (76.5) | 8 (23.5) | 0.724 | 25 (73.5) | 9 (26.5) | >0.999 |
| | | A/G or G/G | 14 | 12 (85.7) | 2 (14.3) | | 11 (78.6) | 3 (21.4) | |
| | 1896T>C | T/T | 35 | 26 (74.3) | 9 (25.7) | 0.280 | 26 (74.3) | 9 (25.7) | >0.999 |
| | | T/C | 13 | 12 (92.3) | 1 (7.7) | | 10 (76.9) | 3 (23.1) | |
|
Thrombocytopenia | 85T>C | T/T | 35 | 33 (94.3) | 2 (5.7) | 0.583 | 28 (80.0) | 7 (20.0) | 0.446 |
| | | T/C | 13 | 12 (92.3) | 1 (7.7) | | 12 (92.3) | 1 (7.7) | |
| | 1627A>G | A/G | 34 | 32 (94.1) | 2 (5.9) | >0.999 | 30 (88.2) | 4 (11.8) | 0.205 |
| | | A/G or G/G | 14 | 13 (92.9) | 1 (7.1) | | 10 (71.4) | 4 (28.6) | |
| | 1896T>C | A/A | 35 | 33 (94.3) | 2 (5.7) | 0.583 | 28 (80.0) | 7 (20.0) | 0.446 |
| | | A/G | 13 | 12 (92.3) | 1 (7.7) | | 12 (92.3) | 1 (7.7) | |
For leukopenia, a lower incidence was observed among
DPYD 85T>C variant carriers during the second cycle. This
was not statistically significant. No consistent associations were
found between DPYD 1627A>G or DPYD 1896T>C
variants and any of the evaluated toxicities, including neutropenia
and thrombocytopenia.
No significant associations were found between
DPYD variants and thrombocytopenia. The incidence of grade
1-4 thrombocytopenia was similar across DPYD 85T>C,
DPYD 1627A>G, and DPYD 1896T>C variants for
both cycles. A slightly higher prevalence of thrombocytopenia was
observed among A/G or G/G carriers of 1627A>G in the second
cycle (28.6%) compared with A/A carriers (11.8%), but this
difference was not statistically significant.
miRNA expression profiles associated
with DPYD 85T>C
miRNAs were extracted from plasma samples of nine
patients with CRC (five wild-type and four with the variant
DPYD 85T>C) using the miRNeasy® Serum/Plasma
kit. miRNA array was used to analyze 43 miRNAs related to the
DPYD drug-metabolizing gene. miRNAs were arranged by
unsupervised clustering, which organizes data according to
expression similarity (Fig. 1A).
This approach revealed natural groupings of WT and VT samples,
reflecting their underlying expression profiles. The results
demonstrated differential expression of miRNAs when comparing the
variant DPYD 85T>C with wild-type patients with CRC
(Fig. 1A). The scatter plot
demonstrated up- and downregulated miRNAs in variant vs. wild-type
patients, indicating a potential link between miRNA expression and
the DPYD gene polymorphism (Fig.
1B). There were nine up- and 11 downregulated miRNAs in the
variant DPYD 85T>C compared with the wild-type group
(Table IV).
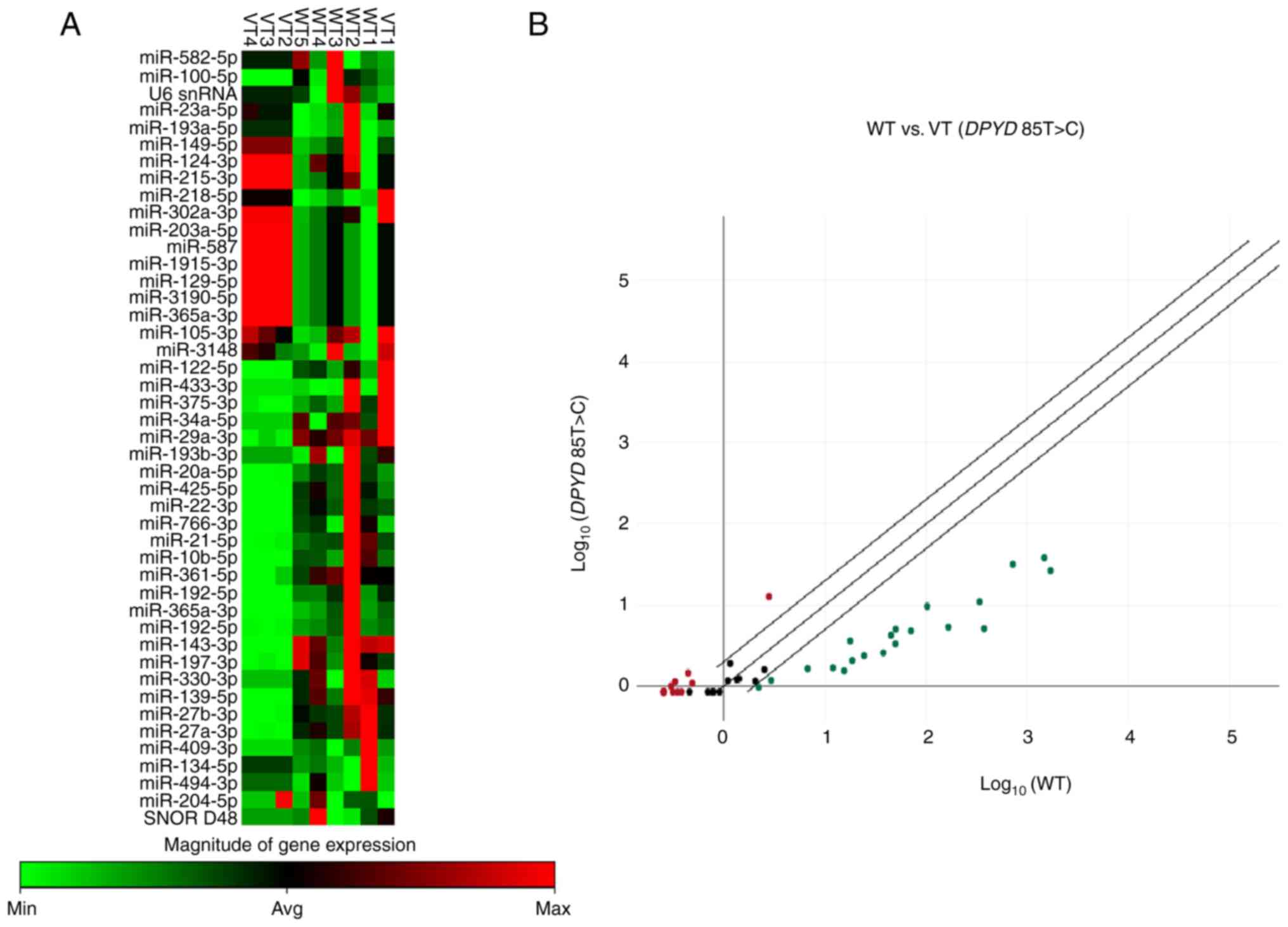 | Figure 1miR expression profiles in patients
with VT (DPYD 85T>C) and WT colorectal cancer. A total of
43 miRs were detected in the plasma of patients with VT (n=4) and
WT colorectal cancer (n=5) using miR array. (A) Heat map of
differential expression of miRs in VT compared with WT colorectal
cancer. Red and green show up- and downregulation, respectively.
(B) Scatter plot showing miR profiles between VT and WT colorectal
cancer. Each dot represents the fold-change in expression of miRNA.
Red, green, and black dots represent up- and downregulated and
unchanged miRNAs, respectively. miR, microRNA; VT, Variant;
DPYD, dihydropyrimidine dehydrogenase gene; WT, Wild type;
min, minimum; avg, average; max, maximum; sn, small nuclear. |
 | Table IVUp- and downregulated miRs in
Dihydropyrimidine dehydrogenase 85T>C compared with patients
with wild-type colorectal cancer. |
Table IV
Up- and downregulated miRs in
Dihydropyrimidine dehydrogenase 85T>C compared with patients
with wild-type colorectal cancer.
| A, Upregulated
miRs |
|---|
| No. | miR | Fold-change | P-value |
|---|
| 1 | miR-218-5p | 3.44 | 0.001 |
| 2 | miR-203a-5p | 3.35 | 0.003 |
| 3 | miR-215-3p | 3.35 | 0.003 |
| 4 | miR-587 | 3.35 | 0.003 |
| 5 | miR-1915-3p | 3.35 | 0.003 |
| 6 | miR-129-5p | 3.35 | 0.003 |
| 7 | miR-3190-5p | 3.35 | 0.003 |
| 8 | miR-365a-3p | 3.35 | 0.003 |
| 9 | miR-302a-3p | 3.33 | 0.001 |
| B, Downregulated
miRs |
| No. | miR | Fold-change | P-value |
| 1 | miR-425-5p | -72.63 | 0.015 |
| 2 | miR-20a-5p | -63.79 | 0.039 |
| 3 | miR-21-5p | -38.30 | 0.042 |
| 4 | miR-27b-3p | -30.85 | 0.020 |
| 5 | miR-27a-3p | -30.45 | 0.015 |
| 6 | miR-22-3p | -22.59 | 0.031 |
| 7 | miR-197-3p | -14.96 | 0.017 |
| 8 | miR-361-5p | -10.53 | 0.019 |
| 9 | miR-766-3p | -10.01 | 0.045 |
| 10 | miR-139-5p | -8.92 | 0.045 |
| 11 | miR-10b-5p | -7.11 | 0.044 |
miRNA expression profiles associated
with DPYD 1896T>C
To assess the miRNA expression profiles in CRC
patients with the DPYD 1896T>C variant compared with
wild-type, miRNA was extracted from plasma samples of six patients
with CRC (three wild-type and three with the DPYD 1896T>C
variant). The results revealed differential miRNA expression
between the variant DPYD 1896T>C and wild-type patients
(Fig. 2A). The scatter plot
demonstrated both up- and downregulated miRNAs in the variant
compared with the wild-type group, suggesting a potential
association between these miRNAs and the DPYD 1896T>C
variant (Fig. 2B). There were five
up-and nine downregulated miRNAs in the variant group compared with
wild-type patients (Table V).
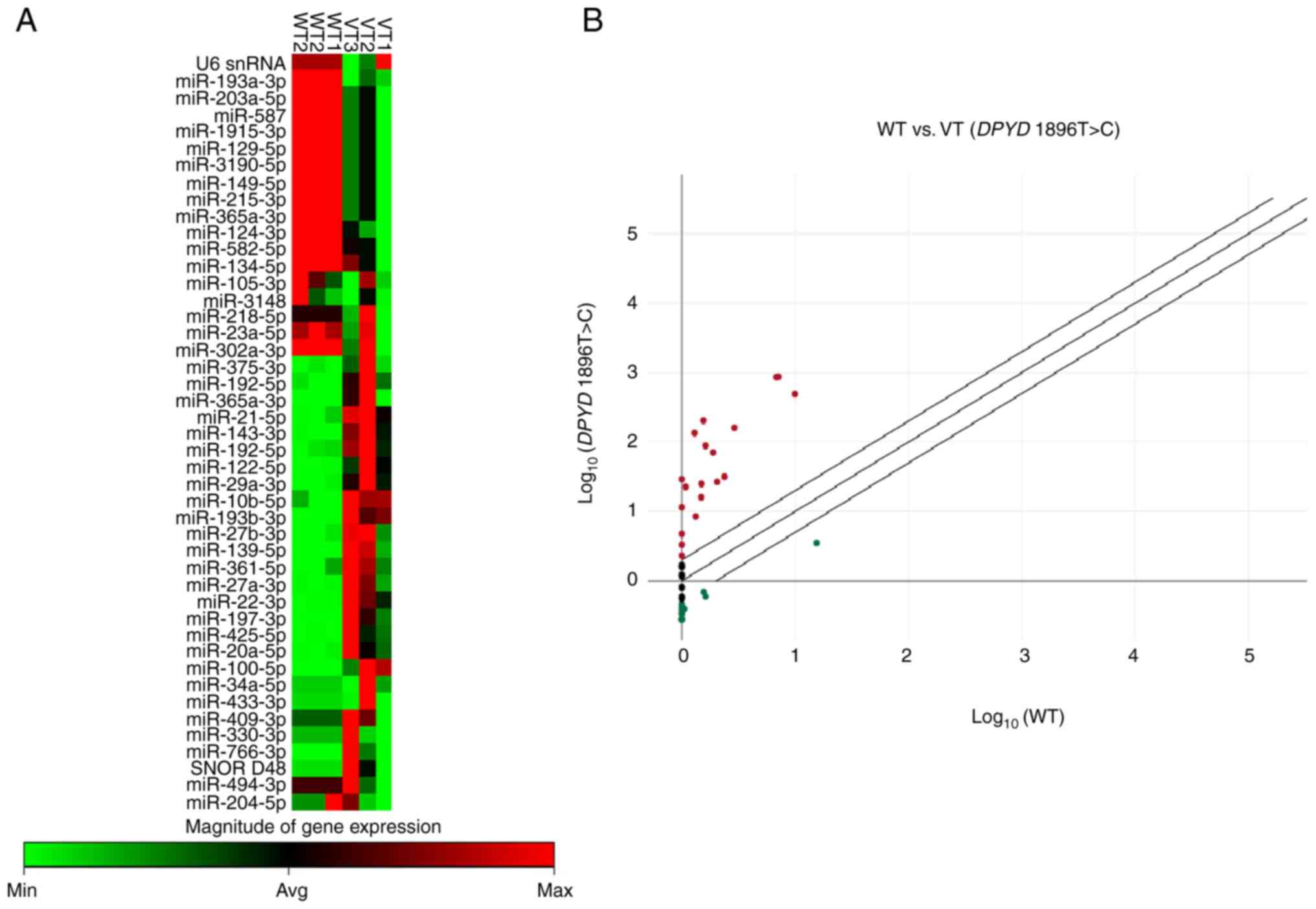 | Figure 2miRNA expression profiles in patients
with VT (DPYD 1896T>C) and WT colorectal cancer. A total
of 43 miRs were detected in the plasma of patients with (n=3) and
WT colorectal cancer (n=3). (A) Heat map of differential expression
of miRs in VT compared with WT colorectal cancer. Red and green
show up- and downregulation, respectively. (B) Scatter plot showing
miR profiles between VT and WT colorectal cancer. Each dot
represents the fold-change in expression of miRNA. Red, green, and
black dots represent up- and downregulated and unchanged miRNAs,
respectively. miR, microRNA; VT, Variant; DPYD,
dihydropyrimidine dehydrogenase gene; WT, Wild type; min, minimum;
avg, average; max, maximum; snRNA, small nuclear RNA. |
 | Table VUp- and downregulated miRs in
dihydropyrimidine dehydrogenase 1896T>C compared with patients
with wild-type colorectal cancer. |
Table V
Up- and downregulated miRs in
dihydropyrimidine dehydrogenase 1896T>C compared with patients
with wild-type colorectal cancer.
| A, Upregulated
miRs |
|---|
| No. | miR | Fold-change | P-value |
|---|
| 1 | miR-21-5p | 122.32 | 0.028 |
| 2 | miR-22-3p | 49.31 | 0.049 |
| 3 | miR-143-3p | 20.91 | 0.043 |
| 4 | miR-10b-5p | 6.42 | <0.001 |
| 5 | miR-193b-3p | 2.31 | 0.016 |
| B, Downregulated
miRs |
| No. | miR | Fold-change | P-value |
| 1 | miR-587 | -3.56 | 0.032 |
| 2 | miR-1915-3p | -3.56 | 0.032 |
| 3 | miR-129-5p | -3.56 | 0.032 |
| 4 | miR-3190-5p | -3.56 | 0.032 |
| 5 | miR-149-5p | -3.56 | 0.032 |
| 6 | miR-215-3p | -3.56 | 0.032 |
| 7 | miR-365a-3p | -3.56 | 0.032 |
| 8 | miR-203a-5p | -3.56 | 0.032 |
| 9 | miR-193a-3p | -2.37 | 0.010 |
miRNA profiling was performed on a randomly selected
subset of 16 patients. Among these, only two individuals had the
DPYD 1627A>G variant. Due to the very limited number of
variant carriers, statistical analysis of the association between
this SNP and miRNA expression as not conducted.
Discussion
DPYD encodes the enzyme DPD, which is
responsible for the catabolism of pyrimidine-based compounds,
including the chemotherapy drug 5-FU. Deficiency or reduced
activity of DPYD leads to the accumulation of 5-FU,
resulting in severe hematological toxicity, including neutropenia,
thrombocytopenia, anemia and leukopenia. Patients with partial DPD
deficiency have a 3.4-fold increased risk of developing grade IV
neutropenia compared with those with normal DPD activity (32,33).
Analysis of the DPYD gene in patients with grade IV
neutropenia revealed that 50% of the individuals tested were either
heterozygous or homozygous for the IVS14+1G>A mutation (34). In addition, the DPYD*5 gene
mutation leads to decreased DPD enzyme activity and impaired 5-FU
metabolism, which is associated with the accumulation of 5-FU and
increased chemotherapeutic toxicity in gastric and colon carcinoma
(35).
The present study assessed the association between
DPYD 85T>C, 1627A>G and 1896T>C and hematological
toxicity across two cycles of 5-FU-based chemotherapy in patients
with CRC. Recent analyses (8,36) have
demonstrated the relevance of DPYD variants in Asian
cohorts. For example, Chan et al (8) conducted a systematic review of
DPYD genotypes in non-European individuals with severe
fluoropyrimidine toxicity, identifying high-frequency variants
[c.1627A>G (DPYD*5) and c.85T>C
(DPYD*9A)] in East and Southeast Asian patients that are not
commonly included in European-focused genotyping panels. Similar to
present study, the frequency of DPYD 85T>C variant was 14% in
Thai patients with CRC. Among these, DPYD 85T>C showed a
trend toward increased anemia, especially during the second
chemotherapy cycle, with 69.2% of T/C carriers developing anemia
compared to 40.0% of T/T carriers (P=0.070). Although not
statistically significant, this trend suggests a possible
cumulative effect of the variant on drug metabolism and hematologic
toxicity. Previous studies (11,34,36)
have reported similar associations between DPYD variants and
hematological adverse effects. Patients with DPYD 85T>C
variant have a significantly increased risk of hematological
toxicity (11,35,37).
Detailleur et al (38) reported a high prevalence of the
DPYD 85T>C variant among patients who experienced severe
toxicity following treatment with 5-FU-based chemotherapy in a
retrospective study. Different DPYD polymorphisms confer
varying levels of residual DPD enzyme activity. For example, the
85T>C (DPYD*9A) variant is associated with a modest
reduction in DPD activity rather than complete loss, which may
explain the milder and variable toxicity profiles seen in certain
carriers (6,9).
The meta-analysis by Leung and Chan (10) revealed that the DPYD
1627A>G variant has a high allele frequency (>20%) in
patients from China, Korea, Japan and Thailand, while the
DPYD 1896T>C variant shows an allele frequency >14% in
Korean and Thai cohorts. The statistical power to detect
associations for both polymorphisms is >75%. Similar to present
study, the allele frequency was 17% in DPYD 1627A>G and
14% in DPYD 1896T>C. The DPYD c.1627A>G variant
is a SNP that results in a missense mutation, causing an
isoleucine-to-valine substitution at codon 543 (p.I543V) of the DPD
enzyme. Several studies have reported that it may influence DPD
enzymatic activity, particularly in compound heterozygous
individuals or in the presence of additional risk alleles (6,10).
The present study found that DPYD 1627A>G
and 1896T>C variants were not significantly associated with
anemia, neutropenia, leukopenia or thrombocytopenia in either
treatment cycle. He et al (9) reported that DPYD enzyme activity does
not differ significantly among carriers of the 85T>C (DPYD
9A), 1627A>G (DPYD 5) or 1896T>C variants.
miRNAs are small, non-coding RNAs that regulate gene
expression by binding to the 3' untranslated region (UTR) of target
mRNAs, leading to mRNA degradation or inhibition of translation
(39). In drug metabolism, miRNAs
have been found to regulate a variety of enzymes involved in drug
processing, including cytochrome P450 and
UDP-glucuronosyltransferases (40).
Sun et al (41) reported
that miR-21, miR-215, miR-218, miR-326 and miR-328 are involved in
the regulation of 5-FU metabolic pathways, and their differential
expression is significantly associated with clinical outcomes and
survival in patients with CRC receiving fluoropyrimidine-based
adjuvant chemotherapy.
The present study compared the miRNA profiles of
wild-type and DPYD 85T>C and 1896T>C variants in
patients with CRC and revealed distinct miRNA profiles between
wild-type and variant patients, with different patterns of up- and
downregulation observed for each variant. Several miRNAs were
upregulated in the DPYD 85T>C variant, but downregulated
in the 1896T>C variant, including miR-587, miR-1915-3p,
miR-129-5p, miR-3190-5p, miR-215-3p, miR-365a-3p and miR-203a-5p.
By contrast, some miRNAs were upregulated in the 1896T>C variant
but downregulated in the 85T>C variant, such as miR-21-5p,
miR-22-3p and miR-10b-5p. This suggested that the regulation of
miRNAs varies across different DPYD variants, and the
expression of miRNAs may be influenced by genetic variants,
potentially modulating their target genes and cellular functions in
CRC, which may affect disease progression, treatment response and
toxicity. miR-21-5p was highly expressed in patients with the
DPYD 1896T>C variant, showing a 122-fold increase
compared with wild-type patients, suggesting its potential as a
diagnostic biomarker for DPYD 1896T>C. This finding
aligns with a previous study, which reported elevated miR-21-5p
levels in the serum of patients with CRC, with fluctuations
observed after surgery and recurrence (42). Moreover, miR-21-5p levels are
associated with TNM staging and lymph node metastasis, suggesting
that miR-21-5p may serve as an oncogene in CRC progression and a
valuable diagnostic biomarker (42). However, these observations warrant
further validation in a larger cohort to confirm their clinical
relevance.
Downregulation of miR-22-3p was observed in the
85T>C variant, which is consistent with previous studies
(43,44) showing that miR-22-3p is
downregulated in CRC and exerts antitumor effects. miR-22-3p
decreases the proliferative, invasive and migratory capacity of CRC
cells (43). Another study found
that low miR-22 in CRC tissue and metastatic cell lines correlated
with metastasis, advanced stage, and relapse, whereas ectopic
miR-22 inhibited CRC growth and metastasis (44). In the present study, miR-10b-5p was
also downregulated in DPYD 85T>C compared with wild-type
patients. The aforementioned study showed that miR-10b-5p is a
target of circular RNAs, such as circ_0021977. Additionally, p21
and p53 are potential target genes of miR-10b-5p (45). The circ_0021977/miR-10b-5p/p21/p53
axis suppresses CRC cell proliferation, migration and invasion,
suggesting its role in CRC progression (45). miR-587 contributes to drug
resistance by downregulating Protein Phosphatase 2, Regulatory
Subunit A, Beta (PPP2R1B), a subunit of the Protein Phosphatase 2
(PP2A) complex, which results in increased AKT activation and
enhanced 5-FU resistance through elevated X-linked Inhibitor of
Apoptosis Protein (XIAP) expression. Targeting the
miR-587/PPP2R1B/phosphorylated AKT/XIAP axis could offer
therapeutic strategies to overcome drug resistance in CRC (46). In the present study, miR-587 was
upregulated in patients with the 85T>C variant, which may be
associated with 5-FU resistance. This preliminary finding suggests
a possible role of miR-587 in CRC treatment and its relevance in
drug resistance, warranting further investigation in larger
cohorts. The present data also suggested that miR-1915-3p may be
upregulated in the 85T>C variant. This aligns with previous
report that exosomal delivery of miR-1915-3p can enhance the
chemotherapeutic efficacy of oxaliplatin in CRC cells by
suppressing epithelial-mesenchymal transition-promoting oncogenes,
such as 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3) and
USP2 (Ubiquitin-Specific Protease 2) (46). In addition, miR-129-5p was
downregulated in the 1896T>C variant. This is consistent with
previous research indicating that miR-129-5p is downregulated in
colon cancer tissue and miR-129-5p mimics suppress the
proliferation of colon cancer cells (48). However, further studies with larger
sample sizes are needed to confirm these observations.
The present study demonstrated an upregulation of
miR-3190-5p in the 85T>C variant while it was downregulated in
the 1896T>C variant compared with wild-type. To the best of our
knowledge, only one previous study has reported this miRNA in CRC
(48). miR-3190-5p regulates the
expression of ABCC4 (ATP-binding cassette sub-family C member 4) by
binding the 3' UTR of the ABCC4 gene, and this regulatory
effect is disrupted by the rs3742106 polymorphism (49). Additionally, miR-3190-5p increases
the intracellular concentration of 5-FU, thereby enhancing the
sensitivity of CRC cells to 5-FU (49), suggesting the miR-3190-5p and
DPYD variant may serve as biomarkers for the personalized
use of 5-FU in CRC treatment.
The present study demonstrated upregulation of
miR-215-3p in the 85T>C variant and a downregulation in the
1896T>C variant compared with the wild-type. Previous studies
(50-52)
have indicated that the levels of miR-215-3p are associated with
the sensitivity of CRC cells to 5-FU, with alterations in
miR-215-3p affecting 5-FU sensitivity. Specifically, miR-215-3p has
been shown to enhance the apoptosis of CRC cells treated with 5-FU
(50). Mechanistically, miR-215-3p
regulates C-X-C chemokine receptor type 1 (CXCR1) expression in
human CRC HCT116) cells, and alterations in CXCR1 affect 5-FU
sensitivity, influencing CRC cell response to the drug (50,51).
The genetic variant-dependent regulation of miR-215-3p may provide
insight into personalized treatment strategies, where miR-215-3p
may serve as a biomarker for predicting response to 5-FU
chemotherapy in patients with CRC. Moreover, p53 is a key regulator
of the DNA damage response and apoptosis, both of which are
critical in determining cell sensitivity to 5-FU (52). Studies have shown that p53 status
influences miRNA expression profiles and may modulate response to
fluoropyrimidine-based chemotherapy (53,54).
Thus, variability in p53 function may interact with DPYD
variants and impact downstream miRNA expression and treatment
outcomes.
miR-365a-3p and miR-203a-5p were upregulated in the
85T>C variant and downregulated in the 1896T>C variant
compared with the wild-type. miR-365a-3p inhibits CRC progression,
at least in part by suppressing ADAM10 expression and the
associated JAK/STAT signaling pathway (55). miR-203a-5p regulates tumorigenesis
by downregulating suppressor of cytokine signaling 3 expression
(56), highlighting the signaling
axis as a potential therapeutic target in CRC.
Increasing evidence (24,25)
suggests that miRNAs may regulate DPYD expression and, by
extension, impact the metabolism of fluoropyrimidines. Offer et
al (24) identified expression
of miR-27a and miR-27b as potential pharmacological modulators of
hepatic DPD enzyme function (24).
DPD serves a crucial role in the uracil catabolic pathway by
converting the chemotherapy drug 5-FU into its inactive metabolite
5-dihydrofluorouracil. A deficiency in DPD, resulting from
insufficient expression or harmful variants in the DPYD
gene, is associated with severe toxic reactions to 5-FU (24). Additionally, patients who are
heterozygous for the miR-27a SNP (rs895819) have an increased risk
of fluoropyrimidine toxicity. This has been studied in both
individuals with wild-type and variant DPYD variant
(25). These findings suggest that
miR-27a rs895819 could serve as a potential biomarker for
predicting fluoropyrimidine-associated toxicity. Hirota et
al (57) demonstrated that
DPYD is a target of several miRNAs, including miR-27a,
miR-27b, miR-134 and miR-582-5p (57). Overexpression of these miRNAs leads
to a significant reduction in reporter activity in a plasmid
containing the 3' UTR of DPYD mRNA in a luciferase assay
(57). Additionally, the
overexpression of these miRNAs also results in a notable decrease
in DPD protein levels in pancreatic carcinoma MIAPaca-2 cells,
suggesting these miRNAs regulate DPD protein expression at the
post-transcriptional level (57).
In the present study, miR-27a-3p and miR-27b-3p were significantly
downregulated in patients with CRC with the variant compared with
wild-type patients, indicating the absence of DPYD deficiency in
the DPYD 85T>C variant.
Hou et al (58) demonstrated similar miRNA expression
in human colon cancer cells (HT29) in response to 5-FU treatment
and nutrient starvation using miRNA microarray analysis (58). Bioinformatic predictions, pathway
and gene network analyses revealed four downregulated miRNAs,
including hsa-miR-302a-3p, and 27 upregulated miRNAs, which may
regulate autophagy in CRC cells during 5-FU-based chemotherapy
(58). The present study showed
that miR-302a-3p was upregulated in DPYD 85T>C compared
with wild-type CRC. The present study had limitations. The sample
size for miRNA analysis was relatively small, particularly in the
variant groups (n=4 for 85T>C and n=3 for 1896T>C), which may
limit statistical power and generalizability. The small sample size
may contribute to potential bias and increased variability and
limit the ability to detect subtle but biologically meaningful
differences. The present study focused on only three SNPs of
DPYD associated with 5-FU-related toxicities; other SNPs of
DPYD variants should be considered for further study.
Additionally, the lack of longitudinal data on toxicity progression
and survival outcomes limits the ability to establish causal links.
Future studies with larger, multi-center cohorts and functional
validation assays are warranted. These should include measurements
of DPD enzyme activity and 5-FU pharmacokinetics to clarify the
mechanistic association between the identified SNP variants, miRNA
expression profiles and clinical responses to 5-FU in CRC.
In conclusion, as miRNAs are regulators of drug
metabolism and toxicity, future research should focus on their
potential role in modulating the impact of DPYD
polymorphisms. Understanding the association between miRNA
expression and DPYD activity may facilitate personalized treatment
strategies, where miRNA profiling could be used alongside
DPYD genotyping to predict toxicity and optimize drug
dosing. Additionally, miRNA-based therapy may be explored as a
potential method to modulate DPYD activity in patients with low
enzyme activity, decreasing the risk of toxicity while maintaining
therapeutic efficacy.
Supplementary Material
miR target sequences used in the miR
array.
Acknowledgements
The authors would like to thank Professor Sam
Ormond (Clinical Research Center, Faculty of Medicine, Thammasat
University, Pathum Thani, Thailand) for English editorial
assistance.
Funding
Funding: The present study was supported by the Health Systems
Research Institute under Genomics Thailand Strategic Fund (grant
no. 65-084) and Thailand Science Research and Innovation
Fundamental Fund, fiscal year 2025.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PP designed the study, performed experiments,
analyzed data and wrote and revised the manuscript. PC, ES, TR, SA
and SS designed the study. PJ performed experiments. CS and CA
conceived and designed the study and analyzed data. CA wrote and
edited the manuscript. PP and CA confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was carried out in compliance
with the Declaration of Helsinki and received approval from the
Ethics Committee of Ramathibodi Hospital, Mahidol University,
Bangkok, Thailand (approval no. MURA2020/1613 Ref.2419). Written
informed consent was provided by all participants before the start
of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Longley DB, Harkin DP and Johnston PG:
5-Fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Van Cutsem E, Cervantes A, Adam R, Sobrero
A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson
A, Bodoky G, et al: ESMO consensus guidelines for the management of
patients with metastatic colorectal cancer. Ann Oncol.
27:1386–1422. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Goto T, Shinmura K, Yokomizo K, Sakuraba
K, Kitamura Y, Shirahata A, Saito M, Kigawa G, Nemoto H, Sanada Y
and Hibi K: Expression levels of thymidylate synthase,
dihydropyrimidine dehydrogenase, and thymidine phosphorylase in
patients with colorectal cancer. Anticancer Res. 32:1757–1762.
2012.PubMed/NCBI
|
|
4
|
Amstutz U, Henricks LM, Offer SM,
Barbarino J, Schellens JHM, Swen JJ, Klein TE, McLeod HL, Caudle
KE, Diasio RB and Schwab M: Clinical pharmacogenetics
implementation consortium (CPIC) guideline for dihydropyrimidine
dehydrogenase genotype and fluoropyrimidine dosing: 2017 Update.
Clin Pharmacol Ther. 103:210–216. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Deac AL, Burz CC, Bocşe HF, Bocşan IC and
Buzoianu AD: A review on the importance of genotyping and
phenotyping in fluoropyrimidine treatment. Med Pharm Rep.
93:223–230. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Offer SM, Wegner NJ, Fossum C, Wang K and
Diasio RB: Phenotypic profiling of DPYD variations relevant to
5-fluorouracil sensitivity using real-time cellular analysis and in
vitro measurement of enzyme activity. Cancer Res. 73:1958–1968.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Falvella FS, Cheli S, Martinetti A,
Mazzali C, Iacovelli R, Maggi C, Gariboldi M, Pierotti MA, Di
Bartolomeo M, Sottotetti E, et al: DPD and UGT1A1 deficiency in
colorectal cancer patients receiving triplet chemotherapy with
fluoropyrimidines, oxaliplatin and irinotecan. Br J Clin Pharmacol.
80:581–588. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chan TH, Zhang JE and Pirmohamed M: DPYD
genetic polymorphisms in non-European patients with severe
fluoropyrimidine-related toxicity: A systematic review. Br J
Cancer. 131:498–514. 2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
He YF, Wei W, Zhang X, Li YH, Li S, Wang
FH, Lin XB, Li ZM, Zhang DS, Huang HQ, et al: Analysis of the DPYD
gene implicated in 5-fluorouracil catabolism in Chinese cancer
patients. J Clin Pharm Ther. 33:307–314. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Leung HWC and Chan ALF: Association and
prediction of severe 5-fluorouracil toxicity with dihydropyrimidine
dehydrogenase gene polymorphisms: A meta-analysis. Biomed Rep.
3:879–883. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Atasilp C, Vanwong N, Yodwongjane P,
Chansriwong P, Sirachainan E, Reungwetwattana T, Jinda P,
Aiempradit S, Sirilerttrakul S, Chamnanphon M, et al: Influence of
DPYD gene polymorphisms on 5-fluorouracil toxicities in Thai
colorectal cancer patients. Cancer Chemother Pharmacol.
95(2)2024.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sarhangi N, Rouhollah F, Niknam N, Sharifi
F, Nikfar S, Larijani B, Patrinos GP and Hasanzad M:
Pharmacogenetic DPYD allele variant frequencies: A comprehensive
analysis across an ancestrally diverse Iranian population. Daru.
32:715–727. 2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sarkar FH, Li Y, Wang Z, Kong D and Ali S:
Implication of microRNAs in drug resistance for designing novel
cancer therapy. Drug Resist Updat. 13:57–66. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Magee P, Shi L and Garofalo M: Role of
microRNAs in chemoresistance. Ann Transl Med. 3(332)2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11(25)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Condrat CE, Thompson DC, Barbu MG, Bugnar
OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM and Voinea SC: miRNAs
as biomarkers in disease: latest findings regarding their role in
diagnosis and prognosis. Cells. 9(276)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ferracin M, Lupini L, Salamon I, Saccenti
E, Zanzi MV, Rocchi A, Da Ros L, Zagatti B, Musa G, Bassi C, et al:
Absolute quantification of cell-free microRNAs in cancer patients.
Oncotarget. 6:14545–14555. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li J, Liu Y, Wang C, Deng T, Liang H, Wang
Y, Huang D, Fan Q, Wang X, Ning T, et al: Serum miRNA expression
profile as a prognostic biomarker of stage II/III colorectal
adenocarcinoma. Sci Rep. 5(12921)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bader El Din NG, El-Shenawy R, Moustafa
RI, Khairy A and Farouk S: Association between the expression level
of miRNA-374a and TGF-β1 in patients with colorectal cancer. World
Acad Sci J. 6(68)2024.
|
|
22
|
Okamoto K, Nozawa H, Ozawa T, Yamamoto Y,
Yokoyama Y, Emoto S, Murono K, Sasaki K, Fujishiro M and Ishihara
S: Comparative microRNA signatures based on liquid biopsy to
identify lymph node metastasis in T1 colorectal cancer patients
undergoing upfront surgery or endoscopic resection. Cell Death
Discov. 11(67)2025.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Schwab S and Nonaka T: Circulating miRNAs
as liquid biopsy biomarkers for diagnosis in patients with
colorectal cancer: A systematic review and meta-analysis. Front
Genet. 16(1574586)2025.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Offer SM, Butterfield GL, Jerde CR, Fossum
CC, Wegner NJ and Diasio RB: microRNAs miR-27a and miR-27b directly
regulate liver dihydropyrimidine dehydrogenase expression through
two conserved binding sites. Mol Cancer Ther. 13:742–751.
2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Medwid S, Wigle TJ, Ross C and Kim RB:
Genetic variation in miR-27a Is associated with
fluoropyrimidine-associated toxicity in patients with
dihydropyrimidine dehydrogenase variants after genotype-guided dose
reduction. Int J Mol Sci. 24(13284)2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Meulendijks D, Henricks LM, Sonke GS,
Deenen MJ, Froehlich TK, Amstutz U, Largiadèr CR, Jennings BA,
Marinaki AM, Sanderson JD, et al: Clinical relevance of DPYD
variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as
predictors of severe fluoropyrimidine-associated toxicity: A
systematic review and meta-analysis of individual patient data.
Lancet Oncol. 16:1639–1650. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the eastern cooperative oncology group. Am J Clin
Oncol. 5:649–655. 1982.PubMed/NCBI
|
|
28
|
Freites-Martinez A, Santana N,
Arias-Santiago S and Viera A: Using the common terminology criteria
for adverse events (CTCAE-version 5.0) to evaluate the severity of
adverse events of anticancer therapies. Actas Dermosifiliogr (Engl
Ed). 112:90–92. 2021.PubMed/NCBI View Article : Google Scholar : (In English,
Spanish).
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wilkinson L and Friendly M: The history of
the cluster heat map. Am Statist. 63:179–184. 2009.
|
|
32
|
Mounier-Boutoille H, Boisdron-Celle M,
Cauchin E, Galmiche JP, Morel A, Gamelin E and Matysiak-Budnik T:
Lethal outcome of 5-fluorouracil infusion in a patient with a total
DPD deficiency and a double DPYD and UTG1A1 gene mutation. Br J
Clin Pharmacol. 70:280–283. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chai J, Dong W, Xie C, Wang L, Han DL,
Wang S, Guo HL and Zhang ZL: MicroRNA-494 sensitizes colon cancer
cells to fluorouracil through regulation of DPYD. IUBMB Life.
67:191–201. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Van Kuilenburg ABP, Meinsma R, Zoetekouw L
and Van Gennip AH: Increased risk of grade IV neutropenia after
administration of 5-fluorouracil due to a dihydropyrimidine
dehydrogenase deficiency: High prevalence of the IVS14+1g>a
mutation. Int J Cancer. 101:253–258. 2002.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang H, Li YM, Zhang H and Jin X: DPYD*5
gene mutation contributes to the reduced DPYD enzyme activity and
chemotherapeutic toxicity of 5-FU: Results from genotyping study on
75 gastric carcinoma and colon carcinoma patients. Med Oncol.
24:251–258. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Raha R, Bhoyar RC, Biswal RP,
Venkatakrishnan R, Rai P, Umashankar E, Kulkarni PM, Sivasubbu S,
Scaria V and Jolly B: Opportunistic analysis of clinically
actionable DPYD gene variants in a germline testing cohort in
India. Pharmacogenomics. 1–6. 2025.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
37
|
Varma A, Jayanthi M, Dubashi B, Shewade DG
and Sundaram R: Genetic influence of DPYD*9A polymorphism on plasma
levels of 5-fluorouracil and subsequent toxicity after oral
administration of capecitabine in colorectal cancer patients of
South Indian origin. Drug Metab Pers Ther. 35:2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Detailleur S, Segelov E, Re MD and Prenen
H: Dihydropyrimidine dehydrogenase deficiency in patients with
severe toxicity after 5-fluorouracil: A retrospective single-center
study. Ann Gastroenterol. 34:68–72. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Dluzen DF and Lazarus P: MicroRNA
regulation of the major drug-metabolizing enzymes and related
transcription factors. Drug Metab Rev. 47:320–334. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sun X, Chen J, Chen X, Gao Q, Chen W, Zou
X, Zhang F, Gao S, Qiu S, Yue X, et al: A systematic review of
clinical validated and potential miRNA markers related to the
efficacy of fluoropyrimidine drugs. Dis Markers.
2022(1360954)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jin XH, Lu S and Wang AF: Expression and
clinical significance of miR-4516 and miR-21-5p in serum of
patients with colorectal cancer. BMC Cancer. 20(241)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jin RR, Zeng C and Chen Y: MiR-22-3p
regulates the proliferation, migration and invasion of colorectal
cancer cells by directly targeting KDM3A through the Hippo pathway.
Histol Histopathol. 37:1241–1252. 2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xia SS, Zhang GJ, Liu ZL, Tian HP, He Y,
Meng CY, Li LF, Wang ZW and Zhou T: MicroRNA-22 suppresses the
growth, migration and invasion of colorectal cancer cells through a
Sp1 negative feedback loop. Oncotarget. 8:36266–36278.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lu C, Jiang W, Hui B, Rong D, Fu K, Dong
C, Tang W and Cao H: The circ_0021977/miR-10b-5p/P21 and P53
regulatory axis suppresses proliferation, migration, and invasion
in colorectal cancer. J Cell Physiol. 235:2273–2285.
2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang Y, Talmon G and Wang J: MicroRNA-587
antagonizes 5-FU-induced apoptosis and confers drug resistance by
regulating PPP2R1B expression in colorectal cancer. Cell Death Dis.
6(e1845)2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Xiao Z, Liu Y, Li Q, Liu Q, Liu Y, Luo Y
and Wei S: EVs delivery of miR-1915-3p improves the
chemotherapeutic efficacy of oxaliplatin in colorectal cancer.
Cancer Chemother Pharmacol. 88:1021–1031. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wu Q, Meng WY, Jie Y and Zhao H: LncRNA
MALAT1 induces colon cancer development by regulating
miR-129-5p/HMGB1 axis. J Cell Physiol. 233:6750–6757.
2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen Q, Meng F, Wang L, Mao Y, Zhou H, Hua
D, Zhang H and Wang W: A polymorphism in ABCC4 is related to
efficacy of 5-FU/capecitabine-based chemotherapy in colorectal
cancer patients. Sci Rep. 7(7059)2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li XW, Qiu SJ and Zhang X: Overexpression
of miR-215-3p sensitizes colorectal cancer to 5-fluorouracil
induced apoptosis through regulating CXCR1. Eur Rev Med Pharmacol
Sci. 22:7240–7250. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jafarzadeh A, Seyedmoalemi S, Dashti A,
Nemati M, Jafarzadeh S, Aminizadeh N, Vosough M, Rajabi A,
Afrasiabi A and Mirzaei H: Interplays between non-coding RNAs and
chemokines in digestive system cancers. Biomed Pharmacother.
152(113237)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Tang X, Shi X, Wang N, Peng W and Cheng Z:
MicroRNA-215-3p suppresses the growth, migration, and invasion of
colorectal cancer by targeting FOXM1. Technol Cancer Res Treat.
18(1533033819874776)2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Eymin B and Gazzeri S: Role of cell cycle
regulators in lung carcinogenesis. Cell Adh Migr. 4:114–123.
2010.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hong YG, Xin C, Zheng H, Huang ZP, Yang Y,
Zhou JD, Gao XH, Hao L, Liu QZ, Zhang W and Hao LQ: miR-365a-3p
regulates ADAM10-JAK-STAT signaling to suppress the growth and
metastasis of colorectal cancer cells. J Cancer. 11:3634–3644.
2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Al-Asadi S, Mansour H, Ataimish AJ,
Al-Kahachi R and Rampurawala J: MicroRNAs regulate tumorigenesis by
downregulating SOCS3 expression: An in silico approach. Bioinform
Biol Insights. 17(11779322231193535)2023.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hirota T, Date Y, Nishibatake Y, Takane H,
Fukuoka Y, Taniguchi Y, Burioka N, Shimizu E, Nakamura H, Otsubo K
and Ieiri I: Dihydropyrimidine dehydrogenase (DPD) expression is
negatively regulated by certain microRNAs in human lung tissues.
Lung Cancer. 77:16–23. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hou N, Han J, Li J, Liu Y, Qin Y, Ni L,
Song T and Huang C: MicroRNA profiling in human colon cancer cells
during 5-fluorouracil-induced autophagy. PLoS One.
9(e114779)2014.PubMed/NCBI View Article : Google Scholar
|
















