|
1
|
Fu M, Deng F, Chen J, Fu L, Lei J, Xu T,
Chen Y, Zhou J, Gao Q and Ding H: Current data and future
perspectives on DNA methylation in ovarian cancer (review). Int J
Oncol. 64(62)2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Geissler F, Nesic K, Kondrashova O,
Dobrovic A, Swisher EM, Scott CL and Wakefield MJ: The role of
aberrant DNA methylation in cancer initiation and clinical impacts.
Ther Adv Med Oncol. 16(17588359231220511)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dong Y, Zhao H, Li H, Li X and Yang S: DNA
methylation as an early diagnostic marker of cancer (review).
Biomed Rep. 2:326–330. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lyko F, Ramsahoye BH and Jaenisch R: DNA
methylation in Drosophila melanogaster. Nature. 408:538–540.
2000.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mattick JS and Gagen MJ: The evolution of
controlled multitasked gene networks: The role of introns and other
noncoding RNAs in the development of complex organisms. Mol Biol
Evol. 18:1611–1630. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rose AB: Introns as gene regulators. A
brick on the accelerator. Front Genet. 9(672)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gallegos JE and Rose AB: The enduring
mystery of intron-mediated enhancement. Plant Sci. 237:8–15.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Earp MA and Cunningham JM: DNA methylation
changes in epithelial ovarian cancer histotypes. Genomics.
106:311–321. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chmelarova M, Krepinska E, Spacek J, Laco
J, Beranek M and Palicka V: Methylation in the p53 promoter in
epithelial ovarian cancer. Clin Transl Oncol. 15:160–163.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cunningham JM, Winham SJ, Wang C, Weiglt
B, Fu Z, Armasu SM, McCauley BM, Brand AH, Chiew YE, Elishaev E, et
al: DNA methylation profiles of ovarian clear cell carcinoma.
Cancer Epidemiol Biomarkers Prev. 31:132–141. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Szewczuk W, Szewczuk O, Czajkowski K,
Gromadka R, Man YG, Waledziak M and Semczuk A: Methylation of the
selected TP53 introns in advanced-stage ovarian carcinomas. J
Cancer. 15:4040–4046. 2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Javadi S, Ganeshan DM, Qayyum A, Iyer RB
and Bhosale P: Ovarian cancer, the revised FIGO staging system, and
the role of imaging. AJR Am J Roentgenol. 206:1351–1360.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Comănescu M, Arsene D, Ardeleanu C and
Bussolati G: The mandate for a proper preservation in
histopathological tissues. Rom J Morphol Embryol. 53:233–242.
2012.PubMed/NCBI
|
|
15
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431.
2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sulewska A, Niklinska A, Kozlowski M,
Minarowski L, Naumnik W, Niklinski J, Dabrowska K and Chyczewski L:
DNA methylation in states of cell physiology and pathology. Folia
Histochem Cytobiol. 45:149–158. 2007.PubMed/NCBI
|
|
17
|
Tajlakhsh J, Mortazavi F and Gupta NK: DNA
methylation topology differentiates between normal and malignant in
cell models, resected human tissues, and exfoliated sputum cells of
lung epithelium. Front Oncol. 12(991120)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Didkowska J, Wojciechowska U, Michalek IM
and Dos Santos FLC: Cancer incidence and mortality in Poland in
2019. Sci Rep. 12(10875)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chi DS, Eisenhauer EL, Zivanovic O, Sonoda
Y, Abu-Rustum NR, Levine DA, Guile MW, Bristow RE, Aghajanian C and
Barakat RR: Improved progression-free and overall survival in
advanced ovarian cancer as a result of a change in surgical
paradigm. Gynecol Oncol. 114:26–31. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tavares V, Marques IS, Guerra de Melo I,
Assis J, Pereira D and Medeiros R: Paradigm shift: A comprehensive
review of ovarian cancer management in an era of advancements. Int
J Mol Sci. 25(1845)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kurman RJ and Shih IeM: Molecular
pathogenesis and extraovarian origin of epithelial ovarian
cancer-shifting the paradigm. Hum Pathol. 42:918–931.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vang R, Levine DA, Soslow RA, Zaloudek C,
Shih IM and Kurman RJ: Molecular alterations of TP53 are a defining
feature of ovarian high-grade serous carcinoma. A rereview of cases
lacking TP53 mutations in the cancer genome atlas ovarian study.
Int J Gynecol Pathol. 35:48–55. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Semczuk A, Gogacz M, Semczuk-Sikora A,
Jóźwik M and Rechberger T: The putative role of TP53 alterations
and p53 expression in borderline ovarian tumors-correlation with
clinicopathological features and prognosis: A mini-review. J
Cancer. 8:2684–2691. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tornaletti S and Pfeifer GP: Complete and
tissue-independent methylation of CpG sites in the p53 gene:
Implications for mutations in human cancers. Oncogene.
10:1493–1499. 1995.PubMed/NCBI
|
|
26
|
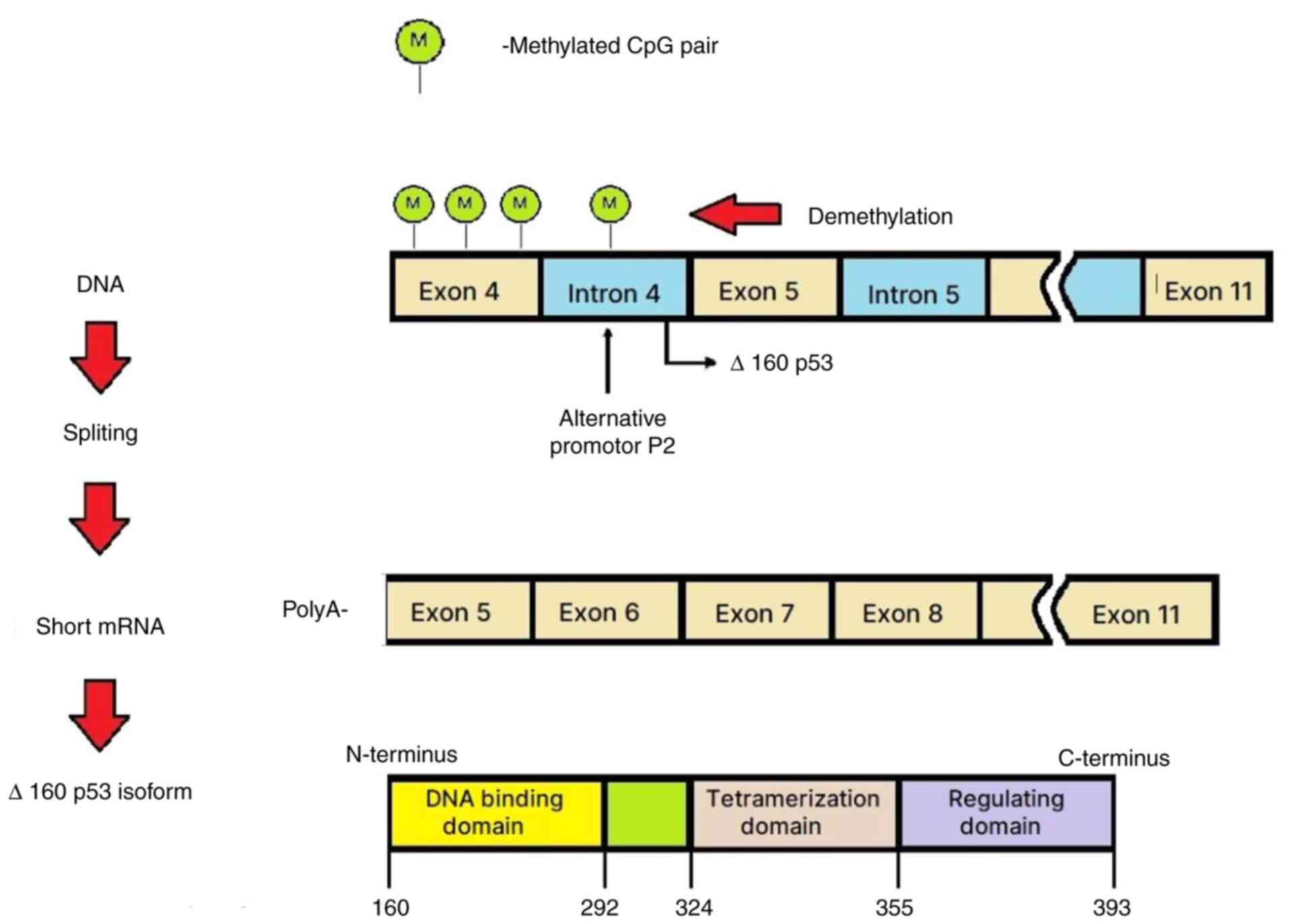
Blackburn J, Roden DL, Ng R, Wu J, Bosman
A and Epstein RJ: Damage-inducible intragenic demethylation of the
human TP53 tumor suppressor gene is associated with transcription
from an alternative intronic promoter. Mol Carcinog. 55:1940–1951.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Landry JR, Mager DL and Wilhelm BT:
Complex controls: The role of alternative promoters in mammalian
genomes. Trends Genet. 19:640–648. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bourdon JC, Fernandes K, Murray-Zmijewski
F, Liu G, Diot A, Xirodimas D, Saville MK and Lane DP: p53 isoforms
can regulate p53 transcriptional activity. Genes Dev. 19:2122–2137.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bourdon JC: p53 and its isoforms in
cancer. Br J Cancer. 97:277–282. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Khoury MP and Bourdon JC: p53 isoforms: An
intracellular microprocessor? Genes Cancer. 2:453–465.
2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Joruiz SM and Bourdon JC: p53 isoforms:
Key regulators of the cell fate decision. Cold Spring Harb Perspect
Med. 6(a026039)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lei J, Qi R, Tang Y, Wang W, Wei G,
Nussinov R and Ma B: Conformational stability and dynamics of the
cancer-associated isoform Δ133p53β are modulated by p53 peptides
and p53-specific DNA. FASEB J. 33:4225–4235. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Camus S, Ménendez S, Fernandes K, Kua N,
Liu G, Xirodimas DP, Lane DP and Bourdon JC: The p53 isoforms are
differentially modified by Mdm2. Cell Cycle. 11:1646–1655.
2012.PubMed/NCBI View Article : Google Scholar
|