Introduction
Epigenetic DNA modification mechanisms play a key
role in various cell cycle functions (1,2). High
methylation of certain fragments or whole chromosomes may be
associated with partial or complete transcriptional inactivation. A
total of ~5% of cytosine residues are continuously methylated in
mammals (3). The methylation
percentage differs between species; for example it is often 30% in
plants but does not appear in Drosophila melanogaster
(4).
In mammals, introns constitute up to 95% of the
primary gene transcripts (5).
Although the introns do not encode proteins, they participate in
important cellular functions (6).
Firstly, introns enhance the expression of corresponding genes and
must be present in the transcribed region to enhance gene
expression (7,8). Secondly, the introns increase
transcript initiation upstream and may represent a downstream
regulatory element for genes transcribed by RNA polymerase II
(8).
DNA methylation mechanisms have been reviewed in the
context of ovarian cancer development and progression (1,9).
However, a limited number of studies have analysed the CpG island
TP53 methylation status at the promoter region and within
introns (10,11). For example, the TP53 promoter
is methylated in 51.5% of ovarian carcinoma (OC) samples and 29.7%
of patients with healthy ovaries. However, no clinicopathological
parameters are associated with the TP53 methylation pattern.
These data revealed a significant difference in promoter
TP53 methylation between OC and control samples, implying
the influence of TP53 methylation on ovarian tumorigenesis
(10).
In our recent study, the TP53 methylation
status was investigated in a cohort of 80 patients with stage III
OC. Intron 1 was un-methylated in all samples, whereas introns 3
and 4 were methylated (12). In the
present study, 10 CpG exon 4 pairs were analysed in primary OC,
corresponding metastases, healthy samples and the A2780 ovarian
endometroid adenocarcinoma. The aim of the present study was to
compare the TP53 methylation status of exon and intron 4 in
advanced-stage OC samples.
Materials and methods
Samples
The tissue samples were collected from 80 patients
aged 55-65 who had undergone surgical treatment for advanced-stage
OC at the Department of Gynecology and Gynecology Oncology at the
Military Institute of Medicine, Warsaw, Poland, between January
2014 and December 2018. The present study included only patients
with FIGO (International Federation of Gynecology and Obstetrics)
stages IIIA-C (13).
Chemotherapy, hormonal therapy or radiotherapy were
not administered prior to the operation. In total, the study
included samples from 40 patients with serous G2/3 and 40 patients
with endometrioid G2/3 OC, corresponding metastatic samples and
healthy tissue (skin) samples from each patient. The detailed
clinicopathological features of the patients have been previously
presented (12). The human ovarian
cancer A2780 cell line (Merck KGaA), established from an ovarian
endometrioid-type adenocarcinoma in an untreated patient, was also
included. Control samples (skin) were sourced from 80 patients aged
35-45 years (50% male and 50% women), who had never had cancer who
and underwent bariatric surgical operations at the Department of
Surgery, Military Institute of Medicine, Warsaw, Poland, between
January 2014 and December 2018. The present study was approved by
the Bioethics Committee of the Military Institute of Medicine,
Warsaw, Poland with the informed consent of the patients (approval
no. N25/WIM/2013).
Following fixation in 10% buffered formalin (pH 7.4)
at room temperature for 24 h, hematoxylin and eosin-stained slides
(room temperature for 1 h) were prepared from paraffin-embedded
tissue (12). Thickness of sections
were 4 µm. The slides were analysed with light microscope by the
anatomopathologist to confirm the primary diagnosis (14).
DNA isolation and bisulfite
conversion
Total genomic DNA was isolated using the ExtractMe
DNA Tissue Isolation kit (cat. nr 51404; Qiagen GmbH, Germany). The
quantity and quality of DNA was evaluated spectrophotometrically
(DeNovix DS-11; DeNovix Inc.). Bisulfite DNA conversion was
performed using the Methyl Code Bisulfite Conversion kit (cat. nr.
1024702; Invitrogen; Thermo Fisher Scientific, Inc.) as previously
reported (12).
Primer sequences
Gene-specific primer sequences for exon 4/intron 4
were designed based on the TP53 sequence published in
National Center for Biotechnology Information (NC_000017.10) using
MethPrimer-Design software version no. 1 (urogene.org.) (15).
Primer sequences were as follows: Forward,
5'-TTGTGTAGTTGTGGGTTGATTTTATAT-3', and reverse,
5'-AAAAACCTAAAAACCCTAAACAACC-3'. The product size was 193 bp. In
total, 11 CpG pairs were investigated, of which one spanned the
TP53 intron 4 and 10 the TP53 exon 4 (Fig. 1).
DNA amplification and sequencing
The amplification of the TP53 exon 4/intron 4
sequences was performed using MyTaqHS Red Mix (cat. no. BIO-25048;
Blirt) as described previously (12). Thermocycling conditions were as
follows: Initial denaturation at 95˚C for 1 min, followed by
denaturation at 95˚C for 15 sec, annealing at 56˚C for 15 sec,
extension at 72˚C for 15 sec, number of cycles 45, and final
extension at 72˚C for 4 min. PCR products were verified by
electrophoresis on 1.5% agarose gels (1 ug/line) using Midori Green
nuclear staining dye (cat. no. MG04; Nippon Genetics Europe GmbH).
The product was extracted for cloning with the use of ExtractMe DNA
Gel-out kit (cat. no. 28706; Qiagen GmbH). The ligation between the
plasmid vector and the PCR product was performed using Qiagen PCR
Cloning kit (cat. no. 231124; Qiagen GmbH). Finally, the vectors
were transformed into E. coli high efficiency competent
cells according to the manufacturer's instructions (New England
BioLabs Ltd.). The cells were incubated at 37˚C overnight on LB
Agar Miller (Medium A&A Biotechnology) with ampicillin,
isopropyl-β-1-thiogalactopiranozyd) and X-gal (cat. no. MBO2501;
Blirt, Poland). A blue-white screening colony selection method was
used to select a recombinant white clone followed by PCR
amplification (as aforementioned) with MyTaqHS Red Mix (cat. no.
BIO-25047; Meridian Biosc., USA) of the colony to confirm the
cloning with the gene segments of interest. Only white colonies of
recombinant plasmid isolation from the bacteria growing in liquid
Luria Broth medium plate were selected and cultured in LB medium at
37˚C overnight. The plasmid was isolated using Plasmid Mini DNA
Isolation kit (cat. no. 020-50 A&A Biotechnology, Poland). The
results of the recombinant DNA were examined using 1.5% agarose gel
electrophoresis (1 µg/lane). The clones containing right inserts
were subjected to direct bidirectional sequencing using an AB3130
genetic analyser with T7/SP6 primers and an Big Dye®
Terminator v3.1 Cycle Sequencing kit (cat. no. 4337455; Thermo
Fisher Scientific, Inc.). All KITs were used according to the
manufacturer's instructions.
Results
Exon 4 TP53 CpG pairs were all methylated in
neoplastic ovarian samples collected from patients with
advanced-stage OC and in the corresponding metastatic samples. The
tissue samples from healthy people who had never had cancer were
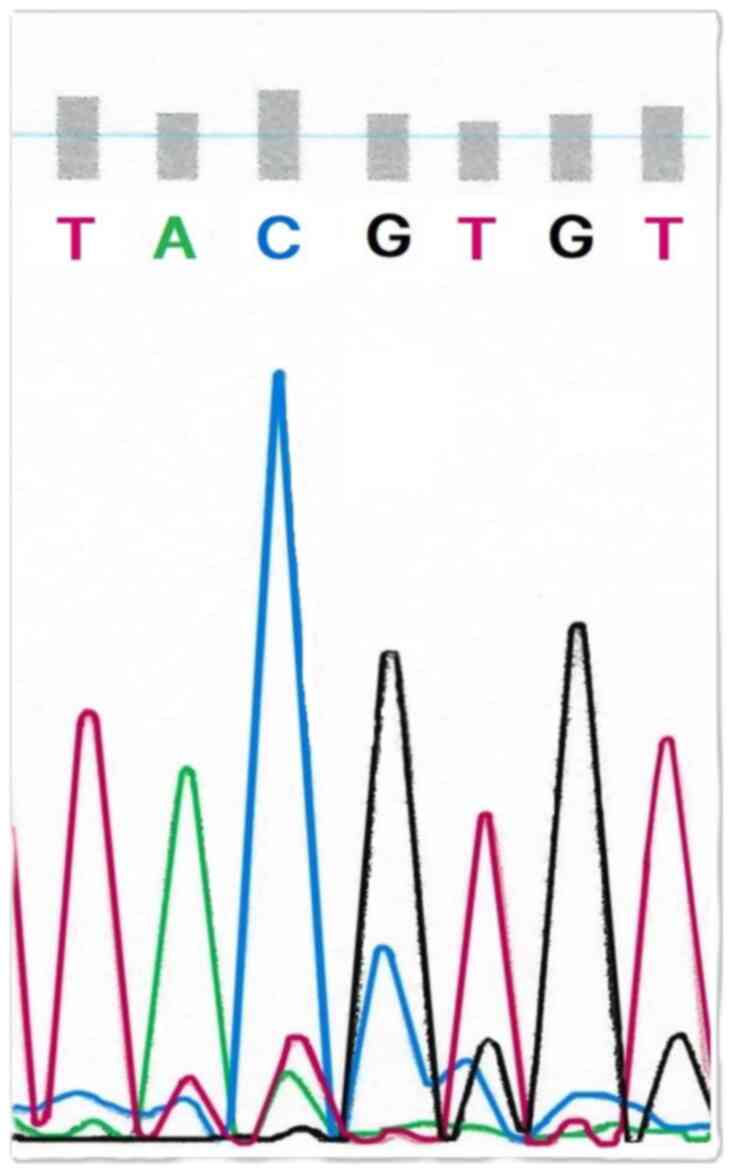
also all methylated. An example of the TP53 exon 4
methylation in the sequencing samples of primary stage IIIC
endometrioid-type OC is presented in Fig. 2.
Similarly, one intron 4 TP53 CpG pair
revealed methylation in all samples. Moreover, the A2780 human
ovarian cancer cell line revealed TP53 exon 4/intron 4
methylation (data not shown).
Finally, no differences were noted in the
exon/intron 4 methylation statuses of TP53 based on
clinicopathological OC features (Table
I).
 | Table IClinical and pathological variables
of patients with ovarian carcinoma. |
Table I
Clinical and pathological variables
of patients with ovarian carcinoma.
| Characteristic | Samples (%) |
|---|
| Age, (years) | |
|
<50 | 1 (1.5) |
|
50-60 | 18 (22.5) |
|
>60 | 61(76) |
| Menopause
status | |
|
Pre-menopause | 3(4) |
|
Post-menopause | 77(96) |
| Clinical stage | |
|
IIIA | 5(6) |
|
IIIB | 36(45) |
|
IIIC | 39(49) |
| Histological
type | |
|
Serous | 40(50) |
|
Endometrioid | 40(50) |
| Histological
grade | |
|
G1 | 0 (0) |
|
G2 | 37(46) |
|
G3 | 43(54) |
Discussion
DNA methylation serves a crucial role in normal
embryonic development, aging, gene regulation and specific cells
functions (16). In general, CpG
islands located within gene promoters are unmethylated in normal
human cells, except for the inactive genes spanning the X
chromosome and those subjected to genomic imprinting (the process
where one paternal copy of the certain gene is silenced) (16,17).
However, silencing of tumour suppressor gene activity in most cases
occurs through methylation of promoter regions at all stages of
human carcinogenesis, including the development of OC (1,2,9-12,17).
OC is one of the most insidious and dangerous female
genital tract malignancies due to its high aggressiveness (18,19).
The majority of OC is reported at advanced clinical stages of the
disease. The 5-year survival rate of stages III and IV is still
unsatisfactory, despite the use of extensive surgical procedures
and the development of anti-cancer therapy treatments (20,21).
The pathogenesis of OC is still debatable. Kurman and Shih
(22) proposed that fallopian tube
epithelium (benign or malignant) that implants on the ovary is the
source of low- and high-grade serous carcinoma rather than the
ovarian surface epithelium. Two OC histological subtypes (type I,
low-grade serous or endometrioid, clear cell, mucinous and
transitional OC, and type II: high-grade serous, undifferentiated,
and carcinosarcoma) differ significantly regardless of the
TP53 alterations and p53 immunoreactivity (23). Different TP53 alterations and
p53 expression patterns have been described in human borderline
ovarian tumours (24). However,
data regarding the role of TP53 exogenic/intragenic
methylation status during ovarian carcinogenesis are scarce
(9-12,25).
Although the role of extragenic methylation on
TP53 has not been fully resolved, CpG pairs are vulnerable
to methylation/demethylation mechanisms (7). Altered exonic CpG methylation modifies
promoter initiation sites, resulting in the expression of different
protein isoforms (26,27). Exogenic CpG sequences in TP53
are methylated in various cancer types (for example colon and lung
cancer) (25). Genetic TP53
alterations are associated with transitions (G:C->A:T), which
are frequently found in human tissue and normal cell lines, for
example in lung epithelial cells, mammary epithelial cells, or in
colonic mucosa cells (25)
Moreover, non-CpG methylation in CC and CCC sequences is associated
with methylation at repetitive TP53 genetic sequences
(28).
Hydrolytic deamination of 5-mC is typically
considered as the mechanism responsible for the high incidence of
TP53 C-T transition mutations within CpG dinucleotides
(3,25). C-T transitions at CpG may also
result from methyltransferase-catalysed cytosine deamination. The
frequency and types of TP53 mutations at CpG dinucleotides
vary between human tumours (1-3,23).
For example, in colon carcinoma, 47% of mutations are reported at
CpG islands, with 17% are reported in skin cancer and 9% in lung
cancer (1-3).
Although the methylation status of introns 1-4 in
TP53 in advanced-stage OC has been previously studied
(12), to the best of our
knowledge, exon 4 has not been previously investigated. The present
study examined exon/intron 4 TP53 and demonstrated that 11
CpG islands were all methylated. Most studies that have examined
comprehensive DNA methylation have focused on the promoter regions
of genes, and, in the majority of the cases, an inverse association
between gene expression and methylation has been found (1,3,25).
Methylation in downstream exon sequences generally is not
associated with expression or lack of expression of p53 in various
tissues. The TP53 sequences along exons 5-8 are completely
methylated at each CpG, including 46 different sites on both DNA
strands (25). This methylation
pattern is tissue-independent, suggesting that tissue-specific
methylation does not contribute to the differential mutation
pattern in various tumours. A total of nine types of normal human
tissues and cell lines, including skin fibroblasts, keratinocytes,
lung and mammary epithelial and colonic mucosa cells, have been
investigated (25). However, it is
unclear whether complete methylation of all CpG sites is unique to
TP53.
Although TP53 CpG dinucleotides are prone to
methylation-dependent mechanisms (25), the regulatory role of methylation
mechanism affecting CpG sites has yet to be clarified. In previous
studies, DNA damage and cell aging are both associated with
site-specific CpG demethylation in exon 5 accompanied by the
induction of expression of truncated protein isoforms regulated by
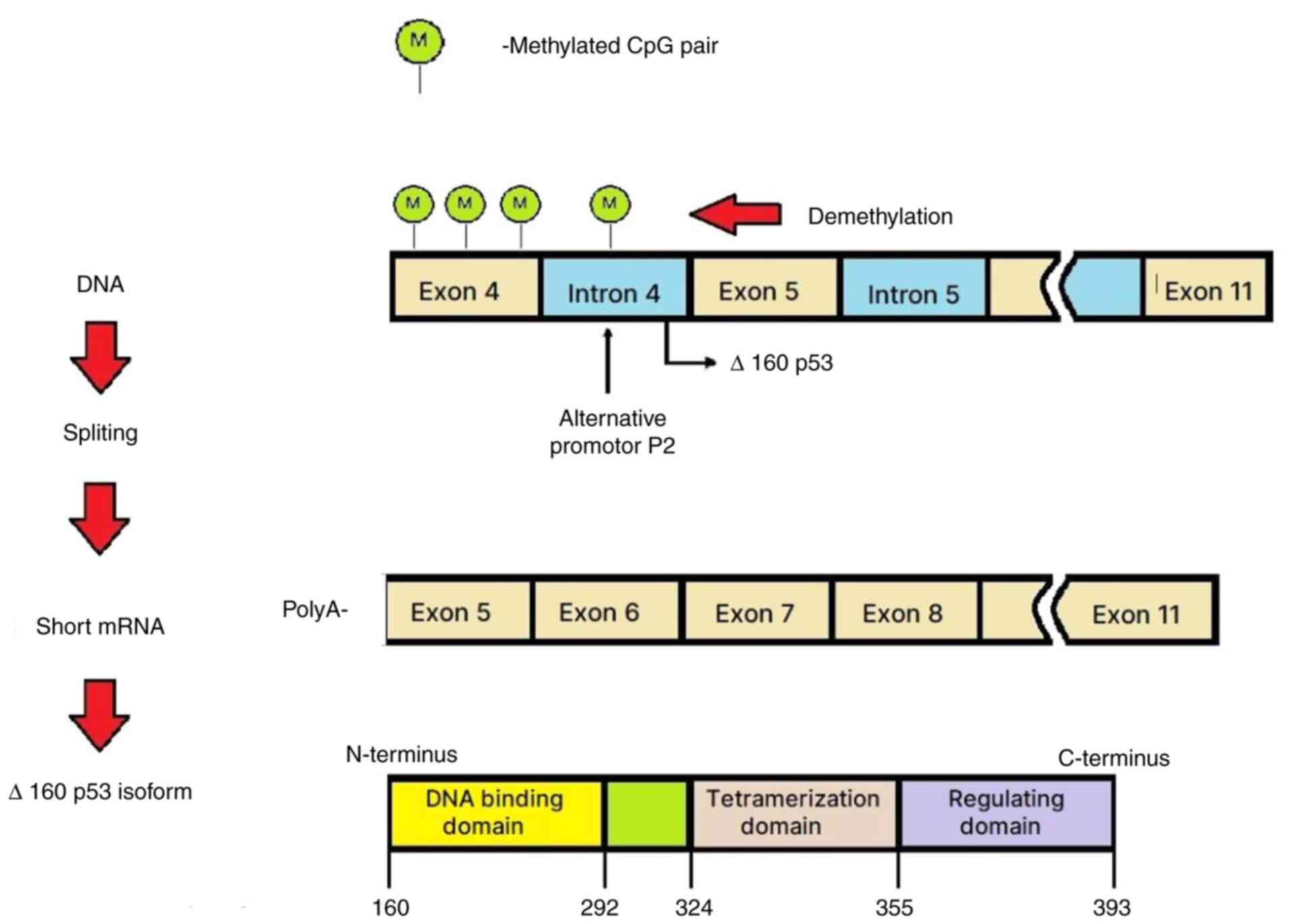
an adjacent intronic P2 promoter (spanning intron 4) (25,26).
The changes in the levels of intragenic TP53 CpG methylation
are extrinsically inducible, suggesting that cancer progression is
mediated, in part, by dysregulation of damage-inducible intragenic
CpG demethylation (26).
The role of alternative promoters in mammalian
genomes has been reviewed by Landry et al (27). Bourdon et al (28,29)
reported that TP53 has transcriptional start sites that span
exon 1 and contain a transcription initiation site at intron 4. The
aforementioned study revealed that TP53 had a complex
transcriptional regulatory pattern encoding different p53 mRNA
variants through the use of alternative splicing and an internal
(intron 4) promoter. The alternative promoter leads to the
expression of N-terminally truncated proteins. The two distinct
TP53 promoters (P1, upstream of exon 1, and P2, within
intron 4) and alternative splicing process and translation
initiation sites of the different mRNAs result in formation of
various p53 isoforms (30).
The transcription of TP53 mRNA can initiate
the formation of the Δ133p53 and Δ160p53 isoforms from the internal
P2 promoter. Considering alternative splicing at intron 9, these
transcripts lead to the production of various isoforms (Δ133p53α,
Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β and Δ160p53γ) (31). Despite truncations in the DNA
binding domain, the Δ133p53 and Δ160p53 isoforms have a stable 3D
conformation (31). The alternative
promoter located in intron 4 becomes active following DNA
demethylation in this region. Therefore, the generated p53 isoforms
are shorter, lack the mouse double minute homolog 2 binding site,
have a longer life-span and can potentially induce apoptosis
(28,29,32,33).
In conclusion, the present findings suggest the
existence of intragenic mechanisms responsible for the regulation
of the TP53 activity, based on demethylation/methylation status
(Fig. 3).
The intragenic demethylation-methylation mechanism
provides the ability to switch the cellular response from cell
cycle arrest to apoptosis by manipulating only the expression of
p53 isoforms and is damaged in solid cancer. Therefore, it has been
hypothesized that demethylation of the TP53 promoter in
intron 4 may be a target for the potential treatment of solid
tumours.
The present study had limitations. Firstly, other
histopathological OC subtypes, apart from serous and endometrioid
subtypes, should be investigated to explore the intron/exon 4
TP53 methylation pattern. Secondly, survival of patients
with OC in relation the TP53 intron 4/exon 4 methylation was
not analysed. Future studies should explore potential interactions
between TP53 methylation patterns and other epigenetic
modifiers (such as histone modification). Moreover, TP53
mutational analysis in advanced-stage OC should be performed.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Military
Institute of Medicine, Warsaw, Poland (grant number 7/8826, project
302).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WS conceived and designed the study, and performed
the experiments. OS made substantial contributions to the
conception of the study, design of the figures, and analysis and
interpretation of the data. KC analyzed and interpreted the data.
RG performed and analyzed the sequencing. MW participated in sample
collection, and analysis and interpretation of the data. MS
analyzed and interpreted the data. AS analyzed and interpreted the
data, revised the manuscript and prepared the final version of the
article. WS and AS confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Bioethics
Committee of the Military Institute of Medicine, Warsaw, Poland
(approval no. N25/WIM/2013). All participants read and signed an
informed consent form prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fu M, Deng F, Chen J, Fu L, Lei J, Xu T,
Chen Y, Zhou J, Gao Q and Ding H: Current data and future
perspectives on DNA methylation in ovarian cancer (review). Int J
Oncol. 64(62)2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Geissler F, Nesic K, Kondrashova O,
Dobrovic A, Swisher EM, Scott CL and Wakefield MJ: The role of
aberrant DNA methylation in cancer initiation and clinical impacts.
Ther Adv Med Oncol. 16(17588359231220511)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dong Y, Zhao H, Li H, Li X and Yang S: DNA
methylation as an early diagnostic marker of cancer (review).
Biomed Rep. 2:326–330. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lyko F, Ramsahoye BH and Jaenisch R: DNA
methylation in Drosophila melanogaster. Nature. 408:538–540.
2000.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mattick JS and Gagen MJ: The evolution of
controlled multitasked gene networks: The role of introns and other
noncoding RNAs in the development of complex organisms. Mol Biol
Evol. 18:1611–1630. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rose AB: Introns as gene regulators. A
brick on the accelerator. Front Genet. 9(672)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gallegos JE and Rose AB: The enduring
mystery of intron-mediated enhancement. Plant Sci. 237:8–15.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Earp MA and Cunningham JM: DNA methylation
changes in epithelial ovarian cancer histotypes. Genomics.
106:311–321. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chmelarova M, Krepinska E, Spacek J, Laco
J, Beranek M and Palicka V: Methylation in the p53 promoter in
epithelial ovarian cancer. Clin Transl Oncol. 15:160–163.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cunningham JM, Winham SJ, Wang C, Weiglt
B, Fu Z, Armasu SM, McCauley BM, Brand AH, Chiew YE, Elishaev E, et
al: DNA methylation profiles of ovarian clear cell carcinoma.
Cancer Epidemiol Biomarkers Prev. 31:132–141. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Szewczuk W, Szewczuk O, Czajkowski K,
Gromadka R, Man YG, Waledziak M and Semczuk A: Methylation of the
selected TP53 introns in advanced-stage ovarian carcinomas. J
Cancer. 15:4040–4046. 2024.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Javadi S, Ganeshan DM, Qayyum A, Iyer RB
and Bhosale P: Ovarian cancer, the revised FIGO staging system, and
the role of imaging. AJR Am J Roentgenol. 206:1351–1360.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Comănescu M, Arsene D, Ardeleanu C and
Bussolati G: The mandate for a proper preservation in
histopathological tissues. Rom J Morphol Embryol. 53:233–242.
2012.PubMed/NCBI
|
|
15
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431.
2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sulewska A, Niklinska A, Kozlowski M,
Minarowski L, Naumnik W, Niklinski J, Dabrowska K and Chyczewski L:
DNA methylation in states of cell physiology and pathology. Folia
Histochem Cytobiol. 45:149–158. 2007.PubMed/NCBI
|
|
17
|
Tajlakhsh J, Mortazavi F and Gupta NK: DNA
methylation topology differentiates between normal and malignant in
cell models, resected human tissues, and exfoliated sputum cells of
lung epithelium. Front Oncol. 12(991120)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Didkowska J, Wojciechowska U, Michalek IM
and Dos Santos FLC: Cancer incidence and mortality in Poland in
2019. Sci Rep. 12(10875)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chi DS, Eisenhauer EL, Zivanovic O, Sonoda
Y, Abu-Rustum NR, Levine DA, Guile MW, Bristow RE, Aghajanian C and
Barakat RR: Improved progression-free and overall survival in
advanced ovarian cancer as a result of a change in surgical
paradigm. Gynecol Oncol. 114:26–31. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tavares V, Marques IS, Guerra de Melo I,
Assis J, Pereira D and Medeiros R: Paradigm shift: A comprehensive
review of ovarian cancer management in an era of advancements. Int
J Mol Sci. 25(1845)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kurman RJ and Shih IeM: Molecular
pathogenesis and extraovarian origin of epithelial ovarian
cancer-shifting the paradigm. Hum Pathol. 42:918–931.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vang R, Levine DA, Soslow RA, Zaloudek C,
Shih IM and Kurman RJ: Molecular alterations of TP53 are a defining
feature of ovarian high-grade serous carcinoma. A rereview of cases
lacking TP53 mutations in the cancer genome atlas ovarian study.
Int J Gynecol Pathol. 35:48–55. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Semczuk A, Gogacz M, Semczuk-Sikora A,
Jóźwik M and Rechberger T: The putative role of TP53 alterations
and p53 expression in borderline ovarian tumors-correlation with
clinicopathological features and prognosis: A mini-review. J
Cancer. 8:2684–2691. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tornaletti S and Pfeifer GP: Complete and
tissue-independent methylation of CpG sites in the p53 gene:
Implications for mutations in human cancers. Oncogene.
10:1493–1499. 1995.PubMed/NCBI
|
|
26
|
Blackburn J, Roden DL, Ng R, Wu J, Bosman
A and Epstein RJ: Damage-inducible intragenic demethylation of the
human TP53 tumor suppressor gene is associated with transcription
from an alternative intronic promoter. Mol Carcinog. 55:1940–1951.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Landry JR, Mager DL and Wilhelm BT:
Complex controls: The role of alternative promoters in mammalian
genomes. Trends Genet. 19:640–648. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bourdon JC, Fernandes K, Murray-Zmijewski
F, Liu G, Diot A, Xirodimas D, Saville MK and Lane DP: p53 isoforms
can regulate p53 transcriptional activity. Genes Dev. 19:2122–2137.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bourdon JC: p53 and its isoforms in
cancer. Br J Cancer. 97:277–282. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Khoury MP and Bourdon JC: p53 isoforms: An
intracellular microprocessor? Genes Cancer. 2:453–465.
2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Joruiz SM and Bourdon JC: p53 isoforms:
Key regulators of the cell fate decision. Cold Spring Harb Perspect
Med. 6(a026039)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lei J, Qi R, Tang Y, Wang W, Wei G,
Nussinov R and Ma B: Conformational stability and dynamics of the
cancer-associated isoform Δ133p53β are modulated by p53 peptides
and p53-specific DNA. FASEB J. 33:4225–4235. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Camus S, Ménendez S, Fernandes K, Kua N,
Liu G, Xirodimas DP, Lane DP and Bourdon JC: The p53 isoforms are
differentially modified by Mdm2. Cell Cycle. 11:1646–1655.
2012.PubMed/NCBI View Article : Google Scholar
|