Introduction
The total number of patients with type 1 diabetes
mellitus (T1DM) is steadily rising. In 2025, an estimated 9.5
million people worldwide are living with T1DM, representing a 13%
increase compared with the 8.4 million reported in 2021. Since the
genetic transmission of this disease has remained relatively
constant, this growing prevalence suggests the involvement of
alternative risk factors, including environmental triggers,
lifestyle influences, and changes in early-life exposures. These
include pollutants, endocrine disruptors, vitamin D deficiency and
pathological colonization of the gastrointestinal tract (1,2).
The gastrointestinal microbiota (the colonization of
the gastrointestinal tract) comprises all bacteria, viruses and
fungi that inhabit this environment. An imbalance of this
microbiota (dysbiosis) with more proinflammatory bacteria that
affect the immune system is hypothesized to explain the global rise
in T1DM, especially the early onset of this disease (1-3).
The balance of the gut microbiota is disrupted by
various factors, leading to dysbiosis, which serves a vital role in
the development and progression of numerous types of disease,
including type 2 diabetes mellitus, autoimmune diseases such as
Hashimoto's thyroiditis and Chon's disease (4). The onset of T1DM, especially with
onset at younger ages, represents a global health challenge, as it
does not affect all individuals with genetic susceptibility
(5). The alterations in microbiota
and chronic inflammation, due to environmental influences, are
hypothesized as explanations for the shifts in the prevalence of
this disease (5,6).
Several mouse studies have provided evidence of a
potential link between microbiota and T1DM (7-9).
For example, exposure to antibiotics in the prenatal period or in
the first weeks after birth influences the gut microbiota and the
T1DM incidence by reducing the pancreatic inflammation and levels
of pro-inflammatory cytokines (10-14).
In the context of T1DM, changes in the gut
microbiota have been observed prior to the emergence of systemic
signs of islet autoimmunity (15,16).
This shift in microbiota could be attributed to the fact that
previous studies primarily identified these modifications through
gene analysis of 16S rRNA, which may not capture specific
structural and functional characteristics potentially involved in
disease progression (17,18). Later studies used specialized
designs to control all known factors influencing T1DM
susceptibility and analyzed microbiome characteristics using
longitudinal metagenomic sequencing of stool samples (19,20).
Patients with T1DM harbor a proinflammatory environment,
irrespective of geographical location, with a higher abundance of
Bacteroidetes and a lower abundance of Firmicutes
(21,22).
Decreased levels of Firmicutes (comprising strains
of Clostridium and Eubacterium) may pose a risk to
the host as this phylum encompasses numerous producers of the
short-chain fatty acid (SCFA) butyrate (23). Butyrate is key for intestinal
homeostasis, serving as an energy source for colonocytes and
promoting the secretion of mucins. This aids in reducing gut
permeability by facilitating the formation of tight junctions
(13,17,18,22,23)
and protecting against pathogens and harmful substances (21,24,25).
The Bacteroidetes phylum contains strains of Bacteroides and
Prevotella. Studies have shown that T1DM is characterized by
a dominance of Bacteroides, taxa associated with intestinal
inflammation, while levels of protective Prevotella are
decreased (26-28).
Species within the Bacteroides genus ferment
glucose and lactate to produce propionate, acetate and succinate,
but they lack the ability to generate butyrate (29). Additionally, lactate-producing
bacteria, including certain probiotic strains such as
Lactobacillus rhamnosus, L. reuteri, L. johnsonii N6.2,
L. plantarum and Bifidobacterium lactis, synthesize
butyrate, thereby reinforcing the intestinal barrier function
(26). These findings underscore
the role of the gut microbiota in T1DM and suggest that targeting
specific microbial components may have potential for interventions
aimed at supporting intestinal health in individuals with T1DM
(1,21,27-30).
Similar to autoimmune conditions, T1DM prevalence
exhibits a geographical pattern: The EURODIAB Autoimmune
Complications of Type 1 Diabetes study (1989-1990) reported
incidence rates of ~42.9 per 100,000/year in Finland, compared with
4.6/100,000/year in northern Greece (31). Climate, particularly sunlight
exposure, influences microbiota and impacts the immune system.
Vitamin D deficiencies and disturbances in the circadian rhythm
alter the gut microbiota, contributing to the development of
autoimmune disease (2,28,30).
Evidence suggests that vitamin D levels are lower in
newly diagnosed patients with T1DM compared with population
controls (32,33). Moreover, the supplementation of this
vitamin and the risk of T1DM show a dose-response effect (30).
The composition of early-life microbiota undergoes
frequent changes influenced by maternal and environmental microbes,
as well as exposure to food and animal-borne antigens. The method
of birth, whether cesarean or vaginal, has an impact on the
neonatal microbiota; findings from the TEDDY study indicate that
children delivered vaginally exhibit higher levels of
Bacteroides, which are associated with increased diversity
and accelerated maturation of gut intestinal flora (14,34).
An increasing number of studies underscore the role
of environmental factors in the rising incidence of T1DM,
particularly among younger children (35,36);
to the best of our knowledge, however, there are few studies from
Romania or Eastern Europe (37-39).
Given the geographical variation in T1DM, the data available may
not entirely align with the actual pathogenesis of T1DM.
Investigating the association between T1DM onset and
gut microbiota dysbiosis may foster collaborations between
researchers, clinicians and policymakers, leading to the
development of novel diagnostic tools, therapeutic interventions
and tailored public health policies.
Considering the cultural, dietary and environmental
changes that have occurred in Eastern Europe over the past three
decades, it has been hypothesized that alterations in the gut
microbiome may contribute to a decreased age at onset of T1DM, as
well as a more severe clinical presentation. Longitudinal
microbiome studies have shown that changes in microbial composition
and function, including a reduction in short-chain fatty acid
producers and enrichment of pro-inflammatory taxa, occur prior to
or around the development of islet autoimmunity and overt T1DM
(11,14). In parallel, epidemiological studies
from Eastern Europe, including Romania and Poland, have reported an
increase in T1DM incidence since the 1990s, with diagnosis
occurring at progressively younger ages (37,40).
At the time of diagnosis, a notable proportion of patients present
with diabetic ketoacidosis, reflecting both delayed recognition and
a more severe onset of disease, which may be associated with
environmental and microbial risk factors (41). Moreover, it has been well documented
that T1DM frequently coexists with other autoimmune diseases, such
as thyroid autoimmunity and celiac disease, further supporting a
role for shared environmental drivers, including gut microbiome
dysregulation, in shaping disease course and comorbidity (42,43).
The aim of the present study was to analyze the gut
microbiota composition in Romanian children with newly diagnosed
T1DM, and to investigate its potential associations with age of
onset, severity at presentation, glycemic control, and co-existing
autoimmune conditions.
Materials and methods
Study population
The present retrospective case-control pilot study
involved 31 pediatric patients (age, 1-18 years, 16 female and 15
male) diagnosed with T1DM within the past 6 months, based on the
criteria set by the American Diabetes Association (44).
The inclusion criteria for the patients were as
follows: i) Positive clinical T1DM diagnosis and ii) onset <6
months prior to admission. The exclusion criteria were as follows:
i) Antibiotic/probiotic administration in the 2 weeks prior to the
admission; ii) T1DM diagnosed >6 months previously and iii)
other types of diabetes mellitus.
These inclusion and exclusion criteria for patients
in the study were established to identify relevant changes in the
gut microbiota in patients with type 1 diabetes, in light of
observational evidence showing that individuals with type 1
diabetes exhibit altered fecal and oral microbiota composition,
reduced butyrate-producing species, and lower plasma levels of
acetate and propionate (45).
Patients were referred either by their primary care
physician or transferred from other pediatric hospitals in the
southern region of Romania to the Pediatric Endocrinology and
Diabetes Department at Elias Emergency and University Hospital,
Bucharest (Romania). Being one of three referral centers for
evaluating and initiating continuous glucose monitor devices and
insulin pumps reimbursed by the National Health System in the
southern region, most patients with recently diagnosed T1DM sought
treatment here. All participants were of Caucasian ethnicity, as
defined by Thomson et al (46). All eligible pediatric patients,
admitted between January 2019 and December 2021, were enrolled in
chronological order of admission. The study adhered to the Helsinki
Declaration of 2013, with parental approval obtained through
written informed consent for the use of medical records in
scientific research.
Patients with newly diagnosed T1DM were matched with
healthy children of similar age (4-18 years), sex, BMI and urban
environment. First-degree relatives with known T1DM were also
evaluated to assess the influence of genetic and environmental
factors. A comprehensive medical history, including demographics,
family history of autoimmune diseases and/or diabetes, personal
medical history, birth details, breastfeeding practices, infection
history and treatment details, was obtained from all participants.
Anthropometric measurements were conducted, including age, weight,
height, BMI, blood pressure, Tanner stage (47) and waist-to-hip ratio. Data were
collected from medical records and laboratory reports, and compiled
in an Excel Spreadsheet (version 16.0 (Microsoft Corporation)
Laboratory procedures
A 10 ml peripheral blood sample was obtained 8 h
after the last meal to assess hematological and biochemical
parameters. The complete blood count and blood chemistry analyses
included measurements of glucose, creatinine, total cholesterol,
high- and low-density lipoprotein-cholesterol, triglycerides,
glycated hemoglobin (HbA1c), hepatic function and reactive
C-protein (CRP). Additionally, levels of anti-thyroglobulin and
anti-thyroperoxidase (for autoimmune thyroid disorders) and
anti-tissue transglutaminase antibodies (for celiac disease) were
measured to assess autoimmune conditions. Thyroid function was
evaluated based on thyroid-stimulating hormone (TSH) and free
thyroxine (fT4) levels. Furthermore, assessments were made for
insulin-like growth factor 1 (IGF1), 25-hydroxy vitamin D [25(OH)
vitamin D], and phosphorus-calcium metabolism, including
measurements of total serum calcium, phosphorus and parathyroid
hormone (PTH).
ELISA
The levels of 25(OH) vitamin D, anti-thyroglobulin,
anti-thyroperoxidase and anti-tissue transglutaminase antibodies
were determined using ELISA on a Chemwell 2010 ELISA system
(Awareness Technology, Inc.) using commercially available kits as
follows: 25(OH) Vitamin D ELISA kit (Immunodiagnostic Systems, cat.
no. AC-57SF1), anti-thyroglobulin (catalog no. ABIN649023,
Antibodies-online), anti-thyroperoxidase ELISA kit (cat. no.
MBS3800854), and anti-tissue transglutaminase ELISA kit (both
MyBioSource, cat. no. MBS7208318). All assays were performed in
strict accordance with the manufacturer's instructions.
Chemiluminometric method
The determination of serum PTH levels were
determined using the COBAS E 411 analyzer (Roche Diagnostics), a
fully automated, random-access, multichannel instrument designed
for immunological analysis.
Spectrophotometric methods
Serum biochemical parameters were assessed using
standard enzymatic colorimetric methods. Lipids [total cholesterol
(cat. no. 72291UD00), HDL-C- kit number: 67136UQ10, triglycerides-
kit number:71068UD00), glucose (cat. no. 67921UQ02, creatinine- kit
number: 71560UD00, aspartate transaminase (AST; cat. no. 70068UD00,
alanine transaminase (ALT) - kit number: 72494UD00, C-reactive
protein (CRP)- kit no. 50519Y600, and calcium - kit number:
74372UD00; all Abbott Diagnostics) were measured on the Architect
c8000 Clinical Chemistry Analyzer (Abbott Diagnostics, Abbott Park,
IL, USA). Assays were performed according to the manufacturer's
instructions and quality control procedures.
Serum phosphorus was determined using the Vitros 5,1
FS Chemistry Analyzer and reagent kit (cat. no. 1203-0398-1506,
Ortho Clinical Diagnostics).
Flow cytometry
Complete blood count was determined using a Sysmex
XN 1000 hematology analyzer (Sysmex Corporation). Flow cytometry
was performed on whole blood samples. Cells were washed twice with
PBS and incubated with blocking buffer (2% FBS in PBS) for 15 min
at 4˚C. Surface markers were detected with directly conjugated
monoclonal antibodies, including CD3-FITC (clone UCHT1; cat. no.
555332; BD Biosciences), CD4-PE (clone RPA-T4; cat. no. 555347; BD
Biosciences), and CD8-APC (cat. no. 555369; BD Biosciences). For
apoptosis analysis, cells were stained with the Annexin V-FITC
Apoptosis Detection kit (cat. no. 556547; BD Biosciences) in
combination with propidium iodide (PI; cat. no. P4864;
Sigma-Aldrich; Merck KGaA). Where secondary detection was required,
biotinylated primary antibodies were visualized using
streptavidin-PE (cat. no. 554061; BD Biosciences). Data acquisition
was performed on a BD FACSCanto II flow cytometer (BD Biosciences),
and results were analyzed using FlowJo software, version 10.8 (Tree
Star Inc.).
ECLIA IGF-1, TSH and fT4 concentrations were
evaluated by ECLIA on a Roche COBAS® e 411 analyzer (Roche
Diagnostics). Commercial reagent kits were provided by
Immunodiagnostic Systems, including the IDS-iSYS IGF-1 assay (cat.
no. IS-3900), IDS-iSYS TSH assay (cat. no. IS-3100), and IDS-iSYS
free T4 assay (cat. no. IS-3300). All assays were performed
according to the manufacturer's instructions.
High-performance liquid chromatography
(HPLC)
HbA1c was measured using the Bio-Rad D-10 Hemoglobin
Testing System (Bio-Rad Laboratories, Inc.). The separation was
performed on the D-10 cation-exchange cartridge (Bio-Rad
Laboratories, Inc.; cat. no. 220-0101) at a controlled temperature
of 25˚C. Whole blood samples (5 µl) were injected. The mobile phase
consisted of two buffers (Buffer A: phosphate buffer, pH 6.5;
Buffer B: phosphate buffer with increased ionic strength). The
system was operated at a flow rate of 1.5 ml/min with a programmed
gradient increase of Buffer B according to the manufacturer's
protocol. An internal calibrator and quality control material
(Bio-Rad Laboratories, Inc.; cat. no. 220-0102 and 220-0103) were
included, and all procedures were carried out strictly according to
the manufacturer's instructions. β-cell function and insulin
resistance were evaluated using the homeostasis model assessment
indices (HOMA-B and HOMA-IR). HOMA-B was calculated as [20 x
fasting insulin [µU/ml])/(fasting glucose [mmol/L] - 3.5), and
HOMA-IR as (fasting insulin [µU/ml] x fasting glucose
[mmol/L])/22.5, according to Matthews et al (48)
Microbiota analysis
Stool samples were gathered during hospitalization
or at home using a standardized procedure that involved antiseptic
handling, collection in sterile tubes (without culture media) and
immediate freezing at -20˚C. Subsequently, fecal DNA was extracted
utilizing the PureLink Microbiome Purification kit (cat. Number
A29790, Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. The concentration of DNA was assessed
using a Qubit 4 fluorometer (Thermo Fisher Scientific, Inc.). DNA
samples were diluted in DNase-free water to a concentration of 3
ng/µl. Quantitative PCR was performed to determine the relative
abundance of intestinal microorganisms in stool DNA isolated from
both patients with T1DM and healthy controls, using a ViiA7© Fast
Real-Time instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Bacterial or fungal group-specific primers (16S and 18S
rRNA, respectively; Table I) were
used at their designated annealing temperatures. For all other
primers, each set was designed to target the 16S rRNA gene, which
is used as a molecular marker for bacterial identification and
quantification (49). The 16S rRNA
gene contains both highly conserved regions, enabling the design of
universal primers, and variable regions, allowing species-level
discrimination. The control was18S rRN) gene, which serves as a
conserved and widely used molecular marker for fungi. These primers
were designed to recognize a broad range of clinically and
environmentally relevant yeasts, with specificity toward
Candida and Saccharomyces spp. The primers were
previously described in Trandafir et al 2024, validated for
specificity toward the bacterial taxa of interest, ensuring
accurate amplification and minimizing off-target effects (49). Each PCR reaction comprised 2.5 nM
forward and reverse primers, 9 ng DNA and 2X SYBR Green Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Negative
controls were samples without DNA templates. Thermocycling
conditions were as follows: Initial denaturation at 95˚C for 5 min,
followed by 40 cycles of 95˚C for 10 sec, 60˚C for 30 sec and 72˚C
for 1 sec. Universal 16S rRNA and 18S rRNA primers were used for
normalization and relative abundance of bacterial abundance was
calculated using the 2-ΔΔCq method (50,51).
 | Table ISequences of the primers. |
Table I
Sequences of the primers.
| Target | Forward primer
(5'→3') | Reverse primer
(5'→3') | Target gene |
|---|
|
Eubacteria |
ACTCCTACGGGAGGCAGCAGT |
ATTACCGCGGCTGCTGGC | 16S rDNA |
| Bacteroides
spp. | CCTACGATGGATAGGGGT
T |
CACGCTACTTGGCTGGTTCAG | 16S rDNA |
|
Butyricicoccus spp. |
ACCTGAAGAATAAGCTCC |
GATAACGCTTGCTCCCTACGT | 16S rDNA |
| Gamma
proteobacteria |
GCTAACGCATTAAGTACCCCG |
GCCATGCAGCACCTGTCT | 16S rDNA |
| Akkermansia
muciniphila |
GCGTAGGCTGTTTCGTAAGTC GTGTGTGAAAG |
GAGTGTTCCCGATATCTACGC ATTTCA | 16S rDNA |
|
Lactobacillus spp. |
ACGAGTAGGGAAATCTTCCA |
CACCGCTACACATGGAG | 16S rDNA |
| Clostridium
leptum |
GCACAAGCAGTGGAGT |
CTTCCTCCGTTTTGTCAA | 16S rDNA |
| Clostridium
coccoides | GACGCCGCGTGAAGG
A |
AGCCCCAGCCTTTCACATC | 16S rDNA |
| Faecalibacterium
prausnitzii |
CCCTTCAGTGCCGCAGT |
GTCGCAGGATGTCAAGAC | 16S rDNA |
| rRNA 18S universal
primers |
ATTGGAGGGCAAGTCTGGTG |
CCGATCCCTAGTCGGCATAG | rRNA 18S |
|
Saccharomyces spp. |
AGGAGTGCGGTTCTTTG |
TACTTACCGAGGCAAGCTACA | rRNA 18S |
| Candida
spp. |
TTTATCAACTTGTCACACCAGA |
ATCCCGCCTTACCACTACCG | rRNA 18S |
Metabolite analysis
Fecal content pellets (0.2 g) were suspended in 1 ml
sterile saline solution and incubated at room temperature for 2
min. Subsequently, the sample was vigorously shaken for 4 min to
create a slurry. After centrifugation at 1,790 x g for 1 h at 4˚C,
the supernatant was collected and subjected to further
centrifugation at 28,600 x g for 30 min at 4˚C.
Fecal content pellets (0.2 g) were suspended in 1 ml
sterile saline solution and incubated at room temperature for 2
min. Subsequently, the sample was vigorously shaken for 4 min to
create a slurry. The resulting supernatant was transferred to a new
tube and filtered using a minisart-GF filter membrane (Sartorius
AG) with a 1 ml sterile plastic syringe. Finally, another
filtration step was performed using a Whatman-25 mmGD/X0 filter
(Merck Millipore) and a 1 ml sterile plastic syringe. Metabolite
levels (including butyrate, acetate, propionate, taurine, succinate
and lactate) were determined using commercial kits following the
manufacturer's instructions (Abbexa, cat. no. abx258338 for
butyrate quantification and Sigma-Aldrich for the other
metabolites- Cat. Number MAK355, MAK184, MAK065, and MAK086).
Optical densities were measured using a spectrophotometer at 455 nm
(Mulsiskan FC, Thermo Fisher Scientific, Inc.) and converted to
µg/g feces.
Statistical analysis
All data are presented as the mean ± SEM or SD and
were analyzed using the GraphPad Prism 9.0 software (Dotmatics). A
total of three independent repeats were performed. Power analysis
was initially performed with a set power (1) of 0.90 and 0.05 for two groups (Control
and T1DM) tested using difference in means and SD as parameters.
Standardized statistical test methods were used to analyze the
results of demography and laboratory tests. The analysis was
performed by a normality test (P<0.05 was considered to be
normal and homogeneous) followed by parametric testing (unpaired
t-test) and Welch's post hoc correction. Spearman correlation
analysis of the association between proinflammatory taxa and early
onset T1DM, high-level onset glycaemia and ketoacidosis, insulin
dose, inflammation, thyroid autoimmunity, IGF-1 and other clinical
parameters regarding growth was performed using SPSS Windows v.
17.0 (SPSS, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Among the 31 patients, the ratio male: female was
1.11 and the mean BMI was 17.48 kg/m2. The mean age of
the onset of the disease was 9.0±0.5 years and the mean glycaemia
was 350±98 mg/dl (Table II). The
vaginal: caesarian birth ratio was 0.36, (13/31) and 10/31 patients
with T1DM had a family history of autoimmune disease.
 | Table IIBiochemical and immunological
parameters. |
Table II
Biochemical and immunological
parameters.
| Parameter | Mean | SEM | Std. Deviation | Median | Normal values |
|---|
| Leukocytes,
x103/µl | 6.68 | 0.40 | 2.22 | 6.15 | 4.60-15.50 |
| Neutrophils,
x103/µl | 3.37 | 0.30 | 1.69 | 2.95 | 1.50-7.00 |
| Lymphocytes,
x103/µl | 2.62 | 0.19 | 1.09 | 2.31 | 1.50-10.50 |
| Hemoglobin,
g/dl | 13.42 | 0.21 | 1.15 | 13.00 | 11.00-15.30 |
| CRP, mg/dl | 3.58 | 2.11 | 8.71 | 0.55 | <0.50 |
| ALT, U/l | 19.29 | 1.77 | 9.84 | 16.00 | 13.00-26.00 |
| AST, U/l | 23.45 | 1.28 | 7.12 | 22.00 | 8.00-25.00 |
| Creatinine,
mg/dl | 0.66 | 0.03 | 0.15 | 0.63 | 0.57-0.86 |
| Total cholesterol
(mg/dl) | 172.23 | 8.16 | 44.70 | 164.50 | 140.00-200.00 |
| Triglycerides
(mg/dl) | 53.23 | 3.31 | 18.15 | 51.50 | >40.00 |
| HDL-cholesterol,
mg/dl | 63.48 | 2.45 | 13.64 | 61.00 | <130.00 |
| LDL-cholesterol,
mg/dl | 102.90 | 5.75 | 32.00 | 96.40 | 35.00-150.00 |
| Anti-thyroglobulin
antibodies, IU/ml | 60.87 | 29.48 | 161.44 | 10.00 | 1.00-16.00 |
| Anti-thyroid
peroxidase antibodies (IU/ml) | 51.66 | 31.90 | 177.60 | 9.00 | <20.00 |
|
Anti-transglutaminase antibodies,
IU/ml | 0.91 | 0.40 | 1.90 | 0.40 | <10.00 |
| TSH, µIU/ml | 2.507 | 0.22 | 1.18 | 2.36 | 0.30-3.60 |
| Free T4, ng/dl | 1.185 | 0.03 | 0.16 | 1.20 | 0.8-1.48 |
| 250H vitamin D,
ng/ml | 28.16 | 2.10 | 11.49 | 24.28 | >30.00 |
| Total calcium,
mg/dl | 9.57 | 0.07 | 0.37 | 9.60 | 8.40-10.20 |
| Phosphorus,
mg/dl | 4.83 | 0.19 | 0.65 | 4.85 | 2.50-4.50 |
| PTH, pg/ml | 31.36 | 2.52 | 13.31 | 27.89 | 15.00-65.00 |
| Insulin, IU/24
h/kg | 19.23 | 4.08 | 21.58 | 11.00 | 0.30-1.00 |
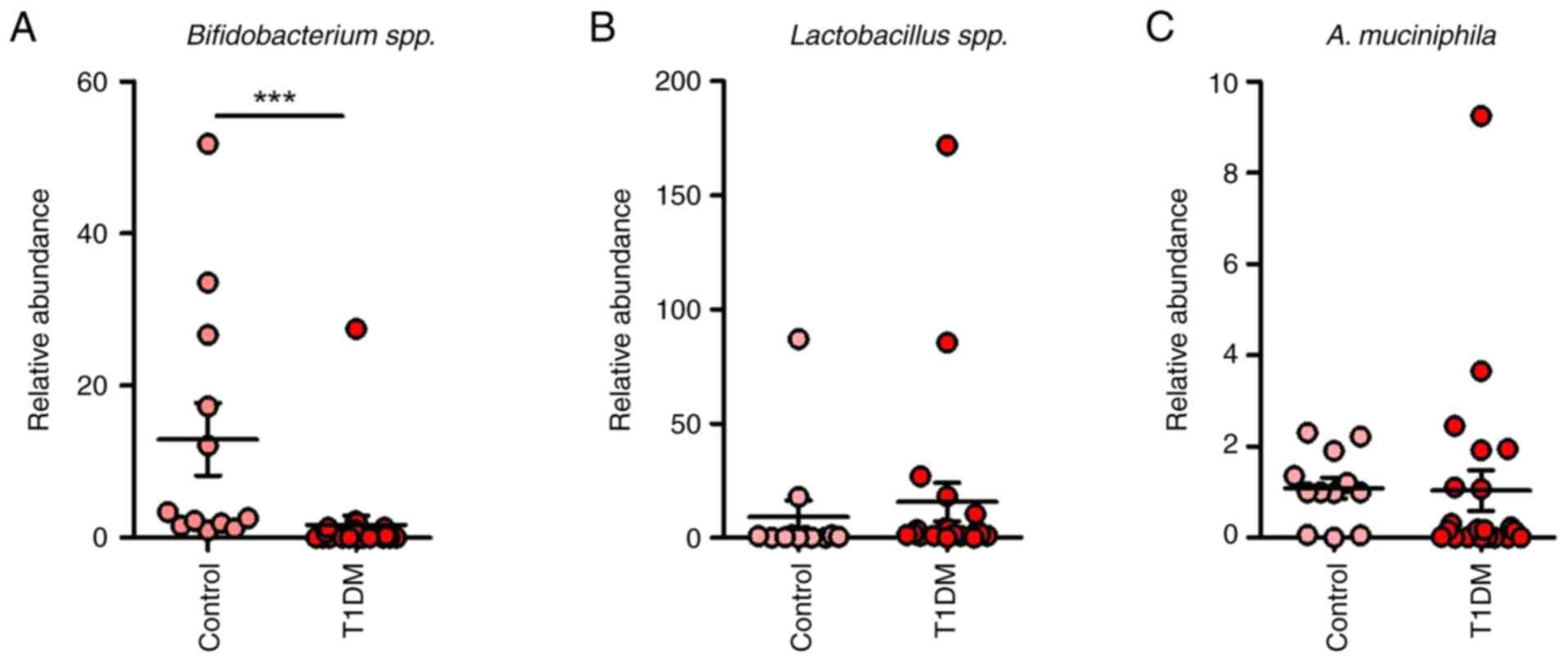
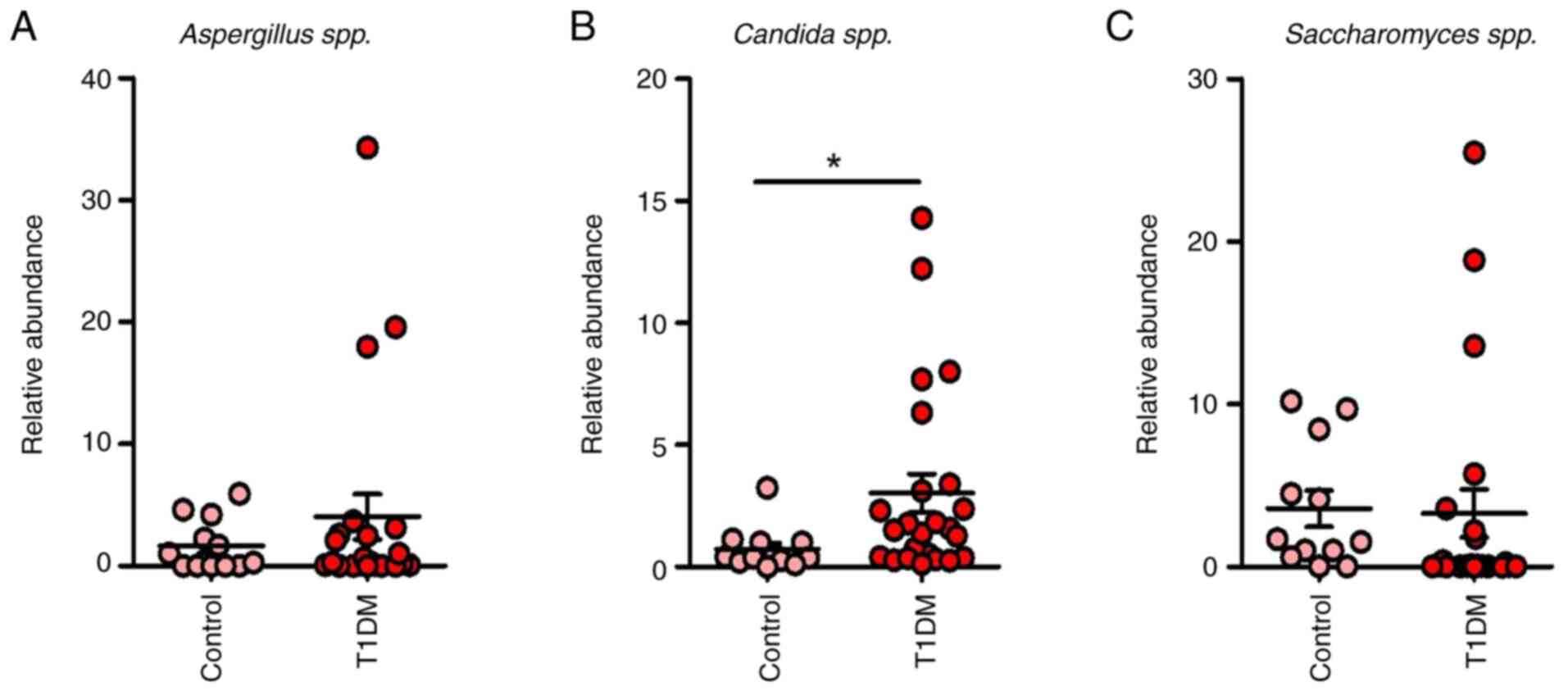
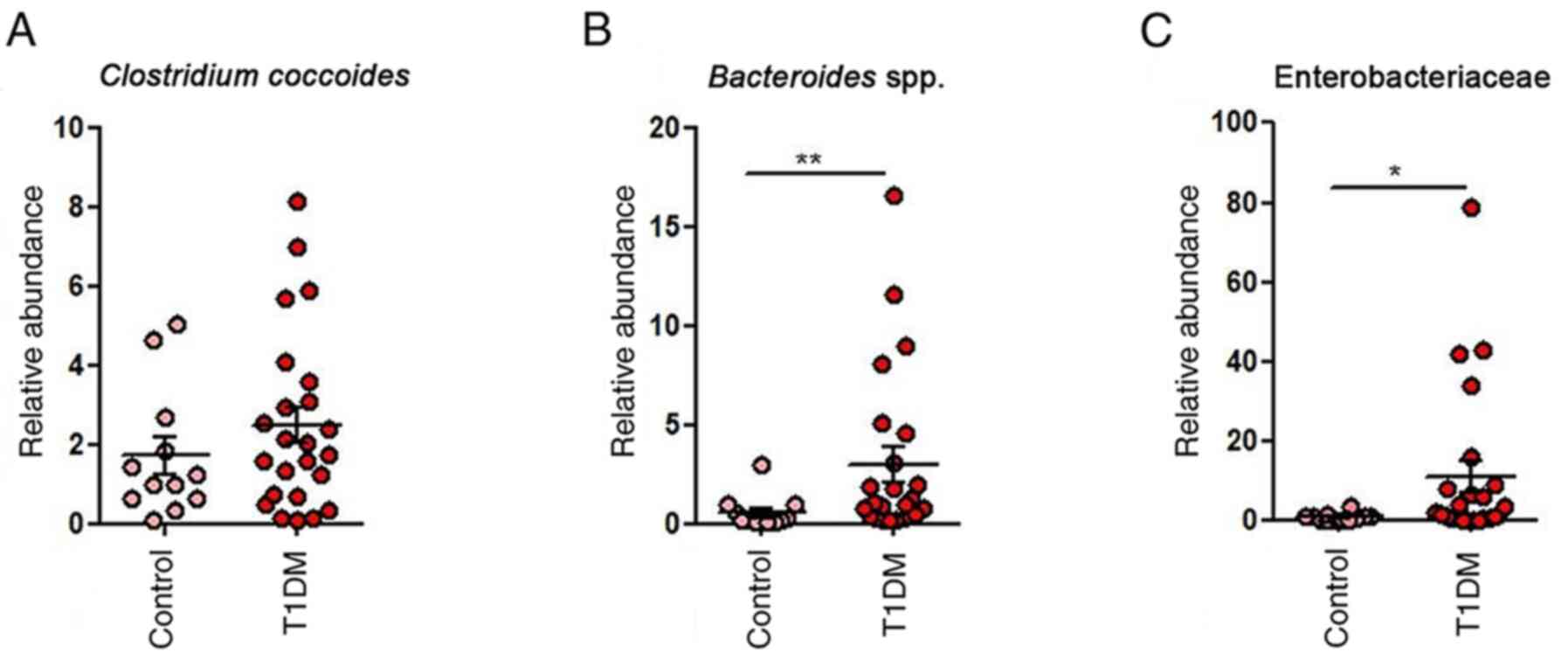
In the T1DM group, dysbiosis was evident,
characterized by an overabundance of detrimental bacteria such as
Clostridium coccoides, Faecalibacterium,
Bacteroides and Enterobacteriaceae, as well as fungi
including Candida, Saccharomyces and Aspergillus
spp., in contrast to the healthy control group (Fig. 1, Fig.
2 and Fig. 3). Furthermore, a
notable decrease in Bifidobacterium spp. and elevated
presence of A. muciniphila within the beneficial taxa were
observed among patients with T1DM compared with the healthy control
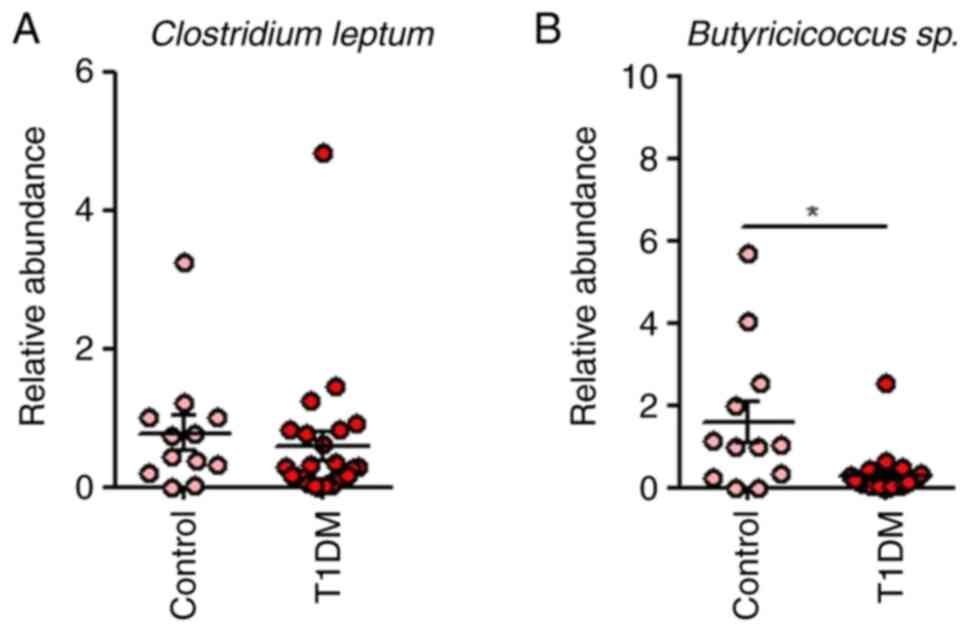
group (Figs. 1 and 4).
A decrease in butyrate-producing bacteria was
observed in the T1DM group with dysbiosis with increased abundance
of Enterobacteriaceae in T1DM (P=0.0193; Fig. 3). This pattern underscores the
alteration in the microbial composition associated with T1DM,
suggesting a potential role of gut microbiota dysregulation in the
pathogenesis of the disease.
Correlation analysis identified significant
correlations between specific gut microbiota taxa and clinical
parameters in pediatric T1DM (Fig.
5). The relative abundance of Butyricicoccus was
positively associated with younger age at onset (r=0.6276,
P=0.0018) but negatively linked to onset within 6 months (r=-0.517,
P=0.0164), pancreatic β cell reserve (HOMA-B, r=-0.5962, P=0.0034),
C-peptide (r=-0.5005, P=0.0344) and phosphorus levels (r=-0.5937,
P=0.0036). Bacteroides relative abundance showed negative
correlations with early-stage diabetes (r=-0.4431, P=0.0442) and
inflammation (indicated by CRP levels; r=-0.4331; P=0.0498). The
relative abundance of Clostridium leptum was inversely
associated with recent diagnosis (r=-0.6278, P=0.0023) but
positively correlated with hemoglobin (r=0.4765, P=0.025) and PTH
levels (r=0.6047, P=0.0029). Antibiotic intake was correlated
positively with the relative abundance of Candida albicans
(r=0.4431, P=0.0442), while the inflammatory index was linked
positively to Enterobacteriaceae (r=0.4331, P=0.0441) and
negatively with C. coccoides (r=-0.4241, P=0.0492).
Hemoglobin was also positively associated with
Butyricicoccus (r=0.5857, P =0.0042), highlighting potential
microbiota roles in T1DM progression and inflammation.
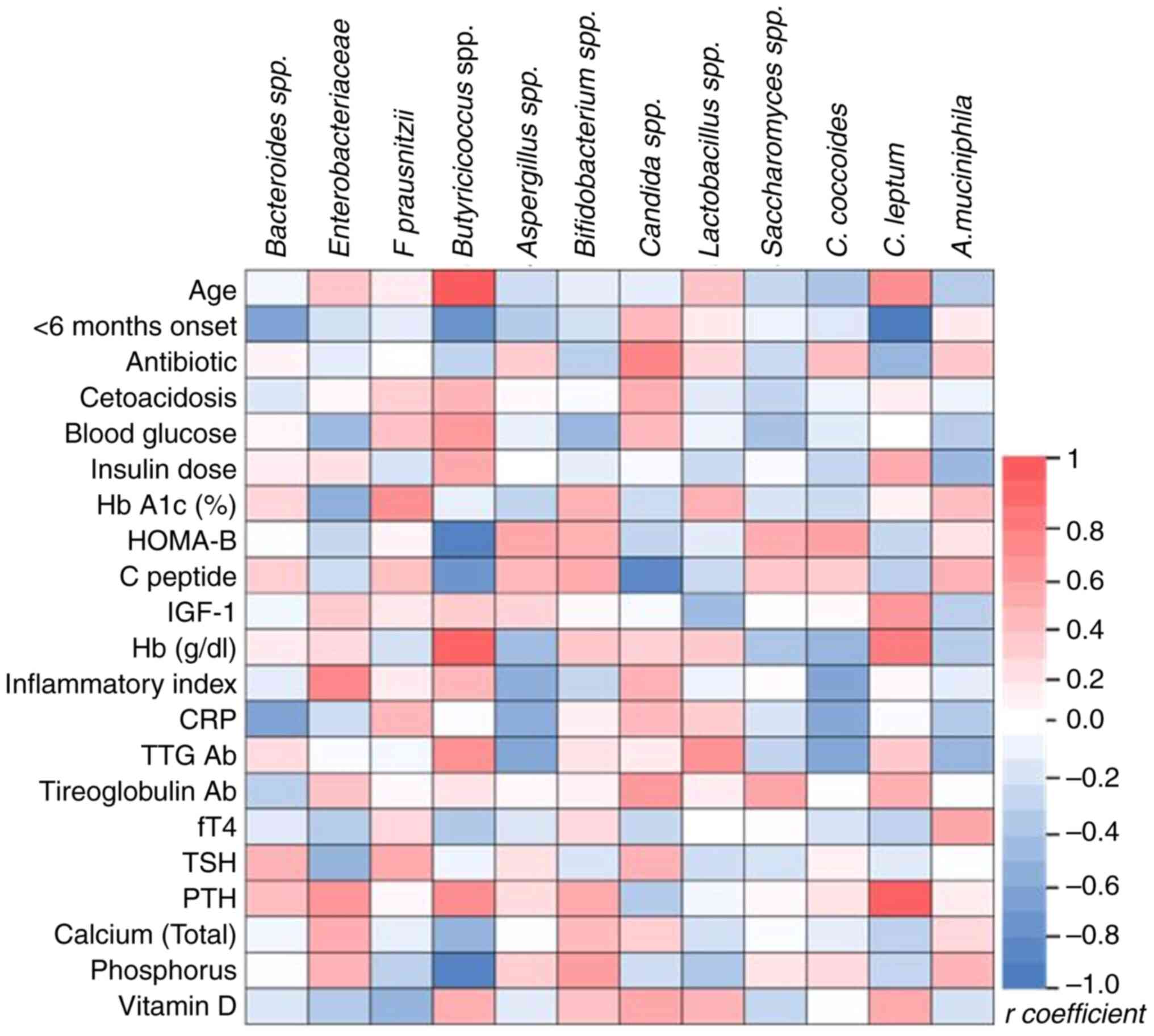 | Figure 5Spearman correlation coefficients
between clinical parameters (and relative abundance of bacterial
taxa in the microbiome. Red, positive correlation; blue, negative
correlations; intensity of color corresponds to the magnitude of
the correlation. HbA1C, Hemoglobin A1c (glycated hemoglobin);
HOMA-B, Homeostatic Model Assessment of β-cell function; IGF,
Insulin-like Growth Factor; CRP, C-reactive Protein; TTG Ab, Tissue
Transglutaminase Antibodies; fT4, Free Thyroxine; TSH,
Thyroid-Stimulating Hormone; PTH, Parathyroid Hormone. |
The microbiota findings were compared with the
metabolite levels in stool samples from both patients with T1DM and
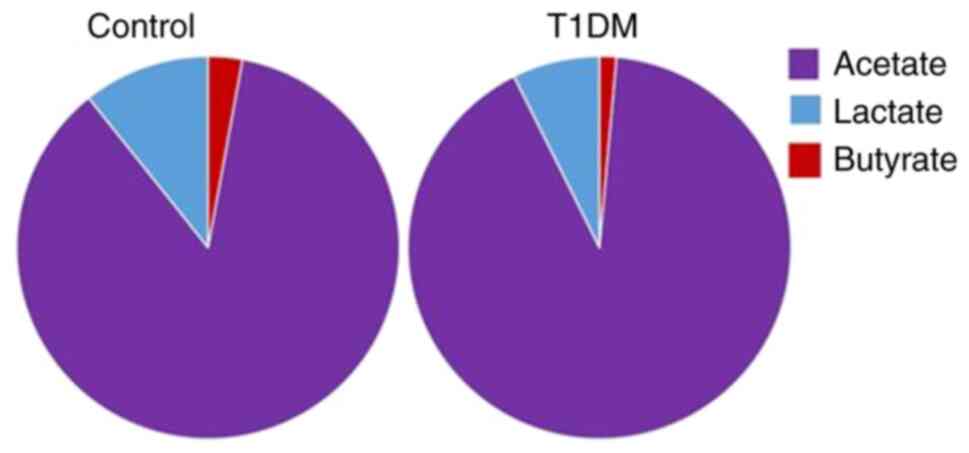
healthy controls (Fig. 6). Patients
with T1DM exhibited notably decreased levels of butyrate and
lactate compared with the healthy controls. Conversely, acetate
levels were higher in the T1DM cohort.
Discussion
The present retrospective case-control study was
conducted in Romania between January 2019 and December 2021,
involving 31 children. The timeline of analysis and manuscript
preparation was substantially affected by the coronavirus Disease
2019 (COVID-19) pandemic. Institutional priorities during this
period were redirected toward pandemic-related clinical and
research activities, and access to laboratories and clinical
facilities was intermittently restricted. These factors contributed
to an extended delay in data processing, analysis, and publication.
Nevertheless, all study data were securely stored, curated, and
analyzed in accordance with the approved protocol, ensuring the
integrity and validity of the findings.
The role of microbiota in T1DM has garnered
increasing attention and investigation: Previous studies have
linked specific bacterial taxa such as Bacteroides,
Bifidobacterium and Ruminococcus with heightened
cytokine levels, pancreatic inflammation, autoimmune reaction and
the onset of T1DM (1,2,4). The
present study demonstrated dysbiosis characterized by an
overabundance of proinflammatory bacteria (including C.
coccoides, Faecalibacterium, Bacteroides and
Enterobacteriaceae) and fungi in pediatric patients with
T1DM from Romania, in comparison with healthy controls.
Contrary to the healthy control group, patients with
T1DM exhibited a microbiota predominantly composed of
Bacteroides taxa, despite the presence of relatively
abundant protective bacteria such as Bifidobacterium and
Lactobacillus spp., and A. muciniphila. Notably,
earlier onset and more severe presentation of T1DM were positively
associated with specific proinflammatory bacterial species,
including Bacteroides and Butyricicoccus spp., in
alignment with the literature (16,52).
C. leptum is another taxon correlated with a younger age at
onset and although its role may vary depending on the context, host
health, and microbiota composition its altered abundance may
contribute to disease susceptibility and progression (53). In gut dysbiosis, its abundance may
shift, and interactions with other microbial taxa could influence
systemic inflammation indirectly. However, it is primarily reported
as a contributor to a balanced, healthy gut environment rather than
as a proinflammatory agent (21-23).
Studies conducted in a Romanian center on patients
with type 2 diabetes mellitus (T2DM) or metabolic syndrome have
revealed similar microbial compositions, particularly the presence
of Enterobacteriaceae, which is correlated with inflammation
and other chronic diseases (53,54).
The Spearman correlation analysis demonstrated
associations between specific microbial taxa and clinical markers,
including those of inflammation, metabolic status and endocrine
function, suggesting a potential role of the gut microbiome in
modulating health outcomes in the context of metabolic
disorder.
The presence of proinflammatory bacteria such as
Enterobacteriaceae showed a positive association with the
systemic inflammatory index; relative abundance of
anti-inflammatory bacteria as Bacteroides and C.
coccoides showed a negative association with CRP levels and
with the systemic inflammatory index (23-25).
Antibiotic use during acute infections in the 2
months preceding diagnosis in children with T1DM may have perturbed
the microbiota, resulting in an overabundance of Candida spp
(45,53,54).
Additionally, T1DM is associated with an increased susceptibility
to recurrent infections and alterations in the immune system within
the first year of the disease, as well as lower complement
component 4) levels and a higher helper/suppressor T lymphocyte
ratio (55,56). These findings are consistent with
those reported by other authors (55-57).
Assessing growth and bone density is key in children
with T1DM and T2DM, given the link between impaired bone health and
dysbiosis characterized by an elevated presence of
Lactobacilli (58). In the
present study, dysbiosis impacted calcium-phosphorous metabolism;
phosphorus levels negatively correlated with Butyricicoccus
and PTH levels were positively correlated with C. leptum
(56,59,60).
The role of Bacteroides in intestinal
homeostasis, particularly in T1DM, is complex and
context-dependent. Bacteroides spp. exhibit both
pro-inflammatory and anti-inflammatory properties, depending on
species, strain and host factors. On one hand, certain
Bacteroides strains (such as B. fragilis, producing
polysaccharide A) have been shown to exert anti-inflammatory
effects by promoting regulatory T cell response and maintaining
mucosal barrier integrity (61,62) On
the other hand, other species or imbalances in Bacteroides
abundance are associated with pro-inflammatory states, including
increased intestinal permeability and heightened immune activation,
which are relevant to the pathogenesis of T1DM (11).
Here, the enrichment of Bacteroides in
patients with T1DM may reflect a shift toward a dysbiotic profile
that contributes to inflammation and autoimmune activation.
However, not all Bacteroides are pathogenic, and their
impact may vary with microbial context, age and diet. Future
strain-level or functional metagenomic analyses may clarify the
specific contributions of Bacteroides in the T1DM gut
microbiome.
Patients with T1DM exhibited notably decreased
levels of butyrate and lactate compared with healthy controls, with
higher acetate levels in the T1DM cohort, diverging from other
studies that found decreased plasma levels of acetate and
propionate and similar levels of plasma butyrate in controls
compared with patients with T1DM (11,24).
Measurements of SCFA levels (butyrate, lactate and acetate) were
obtained from fecal samples, which primarily reflect microbial
fermentation activity in the colon and the unabsorbed SCFA
fraction. This contrasts with other studies reporting SCFA
concentrations in plasma, which are influenced not only by gut
microbial production but also by host absorption efficiency,
hepatic metabolism and systemic circulation dynamics (62,63).
Therefore, direct comparisons between fecal and plasma SCFA levels
must be interpreted with caution. The increase in fecal acetate
alongside decreased butyrate and lactate in the present T1DM cohort
may indicate altered microbial metabolism or impaired SCFA
absorption in these patients, rather than a contradiction of plasma
findings reported in previous studies (63,64).
The present study demonstrated the role of gut
microbiota in pediatric patients with T1DM. It highlighted
dysbiosis characterized by an overabundance of proinflammatory
bacteria such as Bacteroides, Enterobacteriaceae and
C. coccoides, alongside fungi such as Candida spp, in
patients with T1DM compared with healthy controls. This aligns with
existing research, underscoring the association of these taxa with
inflammation, autoimmune reaction and metabolic imbalances
(11,14). The study also demonstrated
correlations between microbial taxa and clinical markers, such as
the systemic inflammatory index, CRP levels and calcium-phosphorous
metabolism, emphasizing the microbiota influence on systemic
inflammation, bone health and disease progression.
Moreover, the perturbation of microbiota following
antibiotic use and the increased prevalence of recurrent infections
in patients with T1DM highlight the dynamic interactions between
external factors, immune function and the gut microbiome. By
revealing specific microbiota compositions associated with earlier
disease onset, severity and metabolic markers, the present study
demonstrated how gut dysbiosis impacts T1DM pathophysiology. These
findings may facilitate research into microbiota-targeted
interventions, such as dietary modification or probiotic therapy,
to improve disease management and patient outcomes in T1DM.
However, the present study had limitations,
including the heterogeneous composition of the study group, its
retrospective nature with a small patient cohort and the
commencement of enrollment a few months before the onset of the
COVID-19 pandemic and lockdown, primarily involving an urban
population. Additionally, the microbiota study excluded patients
who received antibiotics following infections, particularly those
with diabetic ketoacidosis.
Future research directions include proteomics and
metabolomics profiling to evaluate functional changes in the
microbiome of patients with T1DM. Longitudinal studies observing
disease onset and progression in patients with dysbiosis are also
warranted. Furthermore, the associations between dysbiosis and
other autoimmune diseases (such as Hashimoto's thyroiditis and
celiac disease) need to be investigated with regard to the
correlation between antibody positivity and clinical disease
onset.
The present data may be used to estimate the risk of
developing T1DM and other autoimmune diseases in the siblings of
these patients. Risk calculators that consider specific genetic
traits (human leukocyte antigen haplotypes), antibody presence and
family history of the disease may predict the likelihood of
developing diabetes before symptoms manifest. Early intervention,
when a reasonable number of insulin-producing β cells are present
in the pancreas, may positively influence immune responses by
modifying gut microbiota, potentially slowing disease progression
and improving outcomes.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Executive Agency for
Higher Education, Research, Development and Innovation Funding
(project ID no. PN-III-P1-1.1-PD-2019-0499, grant no.
224/2021).
Availability of data and materials
The data generated in the present study are not
publicly available due to participant privacy but may be requested
from the corresponding author.
Authors' contributions
AAI, SF and GGP conceived the study. AAI and GGP
confirm the authenticity of all the raw data. SI designed the
methodology. TP analyzed data. AAI and GGP performed experiments.
AAI wrote and edited the manuscript, constructed figures and
supervised the study. GGP edited the manuscript. SF and GGP
acquired funding. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
guidelines of The Declaration of Helsinki and was approved by the
Ethics Committee of University of Bucharest (protocol code CEC reg.
no. 235/9.10.2019) and the Institutional Review Board of the Elias
Hospital (approval no. 1695, 12.03.2019; both Bucharest, Romania).
Informed consent was obtained from the parents of patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ogle GD, Wang F, Haynes A, Gregory GA,
King TW, Deng K, Dabelea D, James S, Jenkins AJ, Li X, et al:
Global type 1 diabetes prevalence, incidence, and mortality
estimates 2025: Results from the International Diabetes Federation
Atlas, 11th Edition, and the T1D Index Version 3.0. Diabetes Res
Clin Pract. 225(112277)2025.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dedrick S, Sundaresh B, Huang Q, Brady C,
Yoo T, Cronin C, Rudnicki C, Flood M, Momeni B, Ludvigsson J and
Altindis E: The role of gut microbiota and environmental factors in
type 1 diabetes pathogenesis. Front Endocrinol (Lausanne).
11(78)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Penders J, Thijs C, Vink C, Stelma FF,
Snijders B, Kummeling I, Van den Brandt PA and Stobberingh EE:
Factors influencing the composition of the intestinal microbiota in
early infancy. Pediatrics. 118:511–521. 2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Gradisteanu Pircalabioru G, Corcionivoschi
N, Gundogdu O, Chifiriuc MC, Marutescu LG, Ispas B and Savu O:
Dysbiosis in the development of type I diabetes and associated
complications: From mechanisms to targeted gut microbes
manipulation therapies. Int J Mol Sci. 22(2763)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rampanelli E and Nieuwdorp M: Gut
microbiome in type 1 diabetes: The immunological perspective.
Expert Rev Clin Immunol. 19:93–109. 2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Abuqwider J, Corrado A, Scidà G, Lupoli R,
Costabile G, Mauriello G and Bozzetto L: Gut microbiome and blood
glucose control in type 1 diabetes: A systematic review. Front
Endocrinol (Lausanne). 14(1265696)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wen L, Ley RE, Volchkov PY, Stranges PB,
Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA,
et al: Innate immunity and intestinal microbiota in the development
of type 1 diabetes. Nature. 455:1109–1113. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Markle JG, Frank DN, Mortin-Toth S,
Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD,
Macpherson AJ and Danska JS: Sex differences in the gut microbiome
drive hormone-dependent regulation of autoimmunity. Science.
339:1084–1088. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hansen CH, Krych L, Nielsen DS, Vogensen
FK, Hansen LH, Sørensen SJ, Buschard K and Hansen AK: Early life
treatment with vancomycin propagates Akkermansia muciniphila and
reduces diabetes incidence in the NOD mouse. Diabetologia.
55:2285–2294. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Maffeis C, Martina A, Corradi M, Quarella
S, Nori N, Torriani S, Plebani M, Contreas G and Felis GE:
Association between intestinal permeability and faecal microbiota
composition in Italian children with beta cell autoimmunity at risk
for type 1 diabetes. Diabetes Metab. Res Rev. 32:700–709.
2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kostic AD, Gevers D, Siljander H, Vatanen
T, Hyötyläinen T, Hämäläinen AM, Peet A, Tillmann V, Pöhö P,
Mattila I, et al: The dynamics of the human infant gut microbiome
in development and in progression toward type 1 diabetes. Cell Host
Microbe. 17:260–273. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ziegler AG, Hoffmann GF, Hasford J,
Larsson HE, Danne T, Berner R, Penno M, Koralova A, Dunne J and
Bonifacio E: Screening for asymptomatic β-cell autoimmunity in
young children. Lancet Child Adolesc Health. 3:288–290.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vaarala O: Gut microbiota and type 1
diabetes. Rev Diabet Stud. 9:251–259. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Vatanen T, Franzosa EA, Schwager R,
Tripathi S, Arthur TD, Vehik K, Lernmark Å, Hagopian WA, Rewers MJ,
She JX, et al: The human gut microbiome in early-onset type 1
diabetes from the TEDDY study. Nature. 562:589–594. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bäckhed F, Ley RE, Sonnenburg JL, Peterson
DA and Gordon JI: Host-bacterial mutualism in the human intestine.
Science. 307:1915–1920. 2025.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Murri M, Leiva I, Gomez-Zumaquero JM,
Tinahones FJ, Cardona F, Soriguer F and Queipo-Ortuño MI: Gut
microbiota in children with type 1 diabetes differs from that in
healthy children: A case-control study. BMC Med.
11(46)2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Arts RJW, Joosten LAB and Netea MG: The
potential role of trained immunity in autoimmune and
autoinflammatory disorders. Front Immunol. 9(298)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Marchesi JR and Ravel J: The vocabulary of
microbiome research: A proposal. Microbiome. 3(31)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jung TH, Han KS, Park JH and Hwang HJ:
Butyrate modulates mucin secretion and bacterial adherence in LoVo
cells via MAPK signaling. PLoS One. 17(e0269872)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Vatanen T, de Beaufort C, Marcovecchio ML,
Overbergh L, Brunak S, Peakman M, Mathieu C and Knip M: INNODIA
consortium. Gut microbiome shifts in people with type 1 diabetes
are associated with glycaemic control: An INNODIA study.
Diabetologia. 67:1930–1942. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Flint HJ, Scott KP, Duncan SH, Louis P and
Forano E: Microbial degradation of complex carbohydrates in the
gut. Gut Microbes. 3:289–306. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hague A, Butt AJ and Paraskeva C: The role
of butyrate in human colonic epithelial cells: An energy source or
inducer of differentiation and apoptosis? Proc Nutr Soc.
55:937–943. 1996.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lopetuso LR, Scaldaferri F, Petito V and
Gasbarrini A: Commensal Clostridia: Leading players in the
maintenance of gut homeostasis. Gut Pathog. 5(23)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Leiva-Gea I, Sánchez-Alcoholado L,
Martín-Tejedor B, Castellano-Castillo D, Moreno-Indias I,
Urda-Cardona A, Tinahones FJ, Fernández-García JC and Queipo-Ortuño
MI: Gut microbiota differs in composition and functionality between
children with type 1 Diabetes and MODY2 and healthy control
subjects: A case-control study. Diabetes Care. 41:2385–2395.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Brown CT, Davis-Richardson A, Giongo A,
Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip
M, et al: Gut microbiome metagenomics analysis suggests a
functional model for the development of autoimmunity for type 1
diabetes. PLoS One. 6(e25792)2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Valladares R, Sankar D, Li N, Williams E,
Lai KK, Abdelgeliel AS, Gonzalez CF, Wasserfall CH, Larkin J,
Schatz D, et al: Lactobacillus johnsonii N6.2 mitigates the
development of type 1 diabetes in BB-DP rats. PLoS One.
5(e10507)2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Human Microbiome Project Consortium.
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hypponen E: Vitamin D and increasing
incidence of type 1 diabetes-evidence for an association? Diabetes
Obes Metab. 12:737–743. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chia LW, Mank M, Blijenberg B, Aalvink S,
Bongers RS, Stahl B, Knol J and Belzer C: Bacteroides
thetaiotaomicron fosters the growth of butyrate-producing
Anaerostipes caccae in the presence of lactose and total human milk
carbohydrates. Microorganisms. 8(1513)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zipitis CS and Akobeng AK: Vitamin D
supplementation in early childhood and risk of type 1 diabetes: A
systematic review and meta-analysis. Arch Dis Child. 93:512–517.
2008.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Karvonen M, Viik-Kajander M, Moltchanova
E, Libman I, LaPorte R and Tuomilehto J: Incidence of childhood
type 1 diabetes worldwide. Diabetes Mondiale (DiaMond) Project
Group. Diabetes Care. 23:1516–1526. 2000.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pozzilli P, Manfrini S, Crinò A, Picardi
A, Leomanni C, Cherubini V, Valente L, Khazrai M and Visalli N:
IMDIAB group. Low levels of 25-hydroxyvitamin D3 and
1,25-dihydroxyvitamin D3 in patients with newly diagnosed type 1
diabetes. Horm Metab Res. 37:680–683. 2005.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Borkar VV, Devidayal Verma S and Bhalla
AK: Low levels of vitamin D in North Indian children with newly
diagnosed type 1 diabetes. Pediatr Diabetes. 11:345–50.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hagopian WA, Erlich H, Lernmark Å, Rewers
M, Ziegler AG, Simell O, Akolkar B, Vogt R Jr, Blair A, Ilonen J,
et al: The environmental determinants of diabetes in the young
(TEDDY): Genetic criteria and international diabetes risk screening
of 421,000 infants. Pediatr Diabetes. 12:733–743. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lönnrot M, Lynch KF, Elding Larsson H,
Lernmark Å, Rewers MJ, Törn C, Burkhardt BR, Briese T, Hagopian WA,
She JX, et al: TEDDY study group: Respiratory infections are
temporally associated with initiation of type 1 diabetes
autoimmunity: The TEDDY study. Diabetologia. 60:1931–1940.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rewers M and Ludvigsson J: Environmental
risk factors for type 1 diabetes. Lancet. 387:2340–2348.
2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vlad A, Serban V, Green A, Möller S, Vlad
M, Timar B and Sima A: ONROCAD Study Group. Time trends, regional
variability and seasonality regarding the incidence of type 1
diabetes mellitus in Romanian children aged 0-14 years, between
1996 and 2015. J Clin Res Pediatr Endocrinol. 10:92–99.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ionescu-Tîrgoviște C, Guja C, Călin A and
Moţa M: An increasing trend in the incidence of type 1 diabetes
mellitus in children aged 0-14 years in Romania-ten years
(1988-1997) EURODIAB study experience. J Pediatr Endocrinol Metab.
17:983–991. 2004.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gyürüs EK, Patterson C and Soltész G:
Hungarian Childhood Diabetes Epidemiology Group. Twenty-one years
of prospective incidence of childhood type 1 diabetes in
Hungary-the rising trend continues (or peaks and highlands?).
Pediatr Diabetes. 13:21–25. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jarosz-Chobot P, Polańska J, Szadkowska A,
Kretowski A, Bandurska-Stankiewicz E, Ciechanowska M, Deja G,
Mysliwiec M, Peczynska J, Rutkowska J, et al: Rapid increase in the
incidence of type 1 diabetes in Polish children from 1989 to. 2004,
and predictions for 2010 to 2025. Diabetologia. 54:508–515.
2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Usher-Smith JA, Thompson M, Ercole A and
Walter FM: Variation between countries in the frequency of diabetic
ketoacidosis at first presentation of type 1 diabetes in children:
A systematic review. Diabetologia. 55:2878–2894. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Triolo TM, Armstrong TK, McFann K, Yu L,
Rewers MJ, Klingensmith GJ, Eisenbarth GS and Barker JM: Additional
autoimmune disease found in 33% of patients at type 1 diabetes
onset. Diabetes Care. 34:1211–1213. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Akhtadze E, Cilio CM, Hansen I, Gerdes AM,
Mortensen HB, Pociot F and Nerup J: Distribution of autoimmune
diseases in Danish children and adolescents with type 1 diabetes
mellitus. Horm Res. 65:193–198. 2006.
|
|
44
|
ElSayed NA, Aleppo G, Aroda VR, Bannuru
RR, Brown FM, Bruemmer D, Collins BS, Hilliard ME, Isaacs D,
Johnson EL, et al: 2. Classification and diagnosis of diabetes:
Standards of Medical Care in Diabetes-2023. Diabetes Care. 46
(Suppl 1):S19–S40. 2023.PubMed/NCBI View Article : Google Scholar
|
|
45
|
de Groot PF, Belzer C, Levels JH, Aalvink
S, Boot F, Holleman F, Van Raalte DH, Scheithauer TP, Simsek S,
Schaap FG, et al: Distinct fecal and oral microbiota composition in
human type 1 diabetes, an observational study. PLoS One.
12(e0188475)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Thomson G, Valdes AM, Noble JA, Kockum I,
Grote MN, Najman J, Erlich HA, Cucca F, Pugliese A, Steenkiste A,
et al: Relative predispositional effects of HLA class II DRB1-DQB1
haplotypes genotypes on type 1 diabetes: a meta-analysis. Tissue
Antigens. 70:110–127. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Emmanuel M and Bokor BR: Tanner Stages.
2022 Dec 11. In StatPearls. StatPearls Publishing, Treasure Island,
FL, 2025. https://www.ncbi.nlm.nih.gov/books/NBK470280/.
|
|
48
|
Matthews DR, Hosker JP, Rudenski AS,
Naylor BA, Treacher DF and Turner RC: Homeostasis model assessment:
Insulin resistance and β-cell function from fasting plasma glucose
and insulin concentrations in man. Diabetologia. 28:412–419.
1985.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Trandafir M, Pircalabioru GG and Savu O:
Microbiota analysis in individuals with type two diabetes mellitus
and end-stage renal disease: A pilot study. Exp Ther Med.
27(211)2024.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Srinivasan R, Karaoz U, Volegova M,
MacKichan J, Kato-Maeda M, Miller S, Nadarajan R, Brodie EL and
Lynch SV: Use of 16S rRNA gene for identification of a broad range
of clinically relevant bacterial pathogens. PLoS One.
10(e0117617)2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Giongo A, Gano KA, Crabb DB, Mukherjee N,
Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyöty H, et al:
Toward defining the autoimmune microbiome for type 1 diabetes. ISME
J. 5:82–91. 2011.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Gradisteanu Pircalabioru G, Chifiriuc MC,
Picu A, Petcu LM, Trandafir M and Savu O: Snapshot into the
type-2-diabetes-associated microbiome of a romanian cohort. Int J
Mol Sci. 23(15023)2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Gradisteanu Pircalabioru G, Ilie I, Oprea
L, Picu A, Petcu LM, Burlibasa L, Chifiriuc MC and Musat M:
Microbiome, mycobiome and related metabolites alterations in
patients with metabolic syndrome-a pilot study. Metabolites.
12(218)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hull CM, Peakman M and Tree TIM:
Regulatory T cell dysfunction in type 1 diabetes: what's broken and
how can we fix it? Diabetologia. 60:1839–1850. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Alnek K, Tagoma A, Metsküla K, Talja I,
Janson H, Mandel M, Vorobjova T, Oras A, Sepp H, Pruul K, et al:
Comparison of immunological and immunogenetic markers in
recent-onset type 1 diabetes among children and adults. Sci Rep.
15(15491)2025.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Mazzoni MB, Roversi M, Maltoni G, Biasucci
G, Berardi A, Tonielli E, Cicognani A and Salardi S: Gualandi.
Recurrent infections in children and adolescents with type 1
diabetes mellitus: A case-control study. BMC Infect Dis.
18(599)2018.
|
|
58
|
Hygum K, Starup-Linde J, Harsløf T,
Vestergaard P and Langdahl BL: Mechanisms in endocrinology:
Diabetes mellitus, a state of low bone turnover-a systematic review
and meta-analysis. Eur J Endocrinol. 176:R137–R157. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Knudsen JK, Leutscher P and Sørensen S:
Gut microbiota in bone health and diabetes. Curr Osteoporos Rep.
19:462–479. 2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Janner M and Saner C: Impact of type 1
diabetes mellitus on bone health in children. Horm Res Paediatr.
95:205–214. 2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Round JL and Mazmanian SK: Inducible
Foxp3+ regulatory T-cell development by a commensal bacterium of
the intestinal microbiota. Proc Natl Acad Sci USA. 107:12204–12209.
2010.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Mazmanian SK, Round JL and Kasper DL: A
microbial symbiosis factor prevents intestnal inflammatory disease.
Nature. 453:620–625. 2008.PubMed/NCBI View Article : Google Scholar
|
|
63
|
den Besten G, van Eunen K, Groen AK,
Venema K, Reijngoud DJ and Bakker BM: The role of short-chain fatty
acids in the interplay between diet, gut microbiota and host energy
metabolism. J Lipid Res. 54:2325–2340. 2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
de Groot PF, Belzer C, Aydin Ö, Levin E,
Levels JH, Aalvink S, Boot F, Holman R, Reitsma PH, Gerdes VE and
Nieuwdorp M: Distinct fecal and oral microbiota composition in
human type 1 diabetes, an observational study. PLoS ONE.
12(e0188475)2017.PubMed/NCBI View Article : Google Scholar
|