Introduction
Cardiovascular diseases (CVDs) are responsible for
the majority of illnesses and deaths worldwide, accounting for 49%
of female and 40% of male deaths (1). Acute coronary syndrome (ACS), a type
of CVD, encompasses acute ST-segment elevation myocardial
infarction (STEMI), acute non-STEMI ST-segment elevation myocardial
infarction (NSTEMI), and unstable angina pectoris (UA). These
conditions arise from the rupture or invasion of plaques in
coronary arteries affected by atherosclerosis (2,3). ACS
often serves as a significant risk factor for acute heart failure
(4). Despite advancements in
treatment options, such as coronary revascularization,
antithrombotic agents, and anticoagulants, approximately one-fifth
of patients with ACS experience recurrent adverse cardiovascular
events within 3 years following the initial diagnosis (5,6).
Therefore, a deeper understanding of the molecular mechanisms
underlying ACS is crucial for improving patient outcomes.
Long non-coding RNAs (lncRNAs) are a type of ncRNA
consisting of over 200 nucleotides that cannot encode proteins.
They have been scientifically proven to influence various
biological processes, including epigenetics, transcription, and
post-transcriptional regulation in living organisms (7). Several studies have highlighted the
involvement of numerous lncRNAs in ACS diagnosis and development of
ACS. Chen et al (8)
conducted a microarray analysis of serum samples obtained from
individuals with ACS and identified 353 dysregulated lncRNAs.
Subsequent investigations revealed that the modulation of
arachidonate 15-lipoxygenase expression by a specific lncRNA
ENST00000538705.1 could expedite the progression of ACS (8). Additionally, Barbalata et al
(9) found an association between
two distinct lncRNAs (LIPCAR and MALAT1) and hyperglycemia in
patients with UA, identifying them as reliable prognostic
indicators of adverse outcomes in patients with STEMI (9). Another study revealed a negative
association between increased levels of lncRNA PELATON and ACS
prognosis (10). Notably,
transcription factor AP-2α (TFAP2A)-AS1 is a relatively
understudied lncRNA located on chromosome 6q24.3. Current research
on lncRNA TFAP2A-AS1 has primarily focused on its impact on the
pathogenesis and progression of various human cancers. For
instance, elevated expression of TFAP2A-AS1 has been associated
with improved prognosis in gastric cancer (GC) and breast cancer
(BC), whereas overexpression of TFAP2A-AS1 in GC and BC cells
suppressed cell proliferation, migration, and invasion (11,12).
Conversely, TFAP2A-AS1 was shown to be highly expressed in
non-small cell lung cancer (NSCLC) and oral squamous cell carcinoma
(OSCC); however, its absence inhibited tumor development (13,14).
Nevertheless, the precise biological role and mechanism of action
of TFAP2A-AS1 in ACS remain poorly understood.
The aim of the present study was to explore the
mechanism of action of TFAP2A-AS1 in the pathogenesis of ACS and to
identify potential molecular targets for therapeutic intervention
in patients with ACS.
Materials and methods
Patients and diagnostic criteria
The present research adhered to the principles
outlined in the Declaration of Helsinki and received approval
(approval ID: KY-2021053) from the Ethics Committee of Wenzhou
People's Hospital (Wenzhou, China). From September 2021 to October
2022, a total of 5 patients diagnosed with ACS were admitted to
Wenzhou People's Hospital. Concurrently, 5 healthy individuals of
comparable age were recruited to serve as controls. The diagnostic
criteria for ACS included confirmed stenosis (at least one major
coronary artery ≥50%) through coronary angiography, and/or meeting
the acute myocardial infarction criteria (typical clinical
symptoms, elevated cardiac enzyme levels, and representative
electrocardiography findings). The inclusion criteria were as
follows: i) Meeting the diagnostic criteria; ii) age between 35-75
years; and iii) providing informed consent. The exclusion criteria
were as follows: i) The presence of comorbid conditions such as
cardiomyopathy, valvular heart disease, severe arrhythmia, heart
failure or other concurrent ailments; ii) challenges in data
collection due to religious or language barriers; and iii) being
pregnant or breastfeeding. Venous blood samples (5 ml) were
collected from both patients with ACS and healthy controls. The
samples were then centrifuged at 3,000 x g at a temperature of 4˚C
for a duration of 5 min. Following centrifugation, the samples were
stored at -80˚C for subsequent experiments. The clinicopathological
factors of healthy controls and patients with ACS are summarized in
Table I.
 | Table IClinicopathological factors of
healthy controls and patients with ACS. |
Table I
Clinicopathological factors of
healthy controls and patients with ACS.
|
Characteristics | Healthy controls
(n=5) | Patients with ACS
(n=5) | P-value |
|---|
| Age, years | 64.00±3.03 | 62.40±10.07 | 0.7425 |
| BMI,
kg/m2 | 24.66±4.21 | 25.12±3.28 | 0.8520 |
| Sex
(male/female) | 2/3 | 2/3 | N/A |
| Smoking,
(N/total) | 2/5 | 2/5 | N/A |
| Drinking,
(N/total) | 2/5 | 2/5 | N/A |
| Hypertension,
(N/total) | 3/5 | 3/5 | N/A |
| Cardiac troponin I,
µg/l | 0.014±0.0001 | 4.57±0.21 | <0.0001 |
Reagents
Human primary coronary artery endothelial cells
(HCAECs), mouse coronary artery endothelial cells (MCAECs) and 293
cells were sourced from Pricella Biotechnology Co., Ltd. The
utilization of primary cells was approved (approval ID:
KZ-20240521) by the Ethics Committee of Wenzhou People's Hospital.
Endothelial basal medium 2 (EBM-2) and fetal bovine serum (FBS)
were obtained from Lonza Group, Ltd. CCK-8 solution was provided by
Abbkine Scientific Co., Ltd. BD Biosciences provided the Matrigel.
Genomeditech (Shanghai) Co., Ltd. Synthesized TFAP2A-AS1 small
interfering (si)RNA1/2/3, TFAP2A siRNA1/2/3, lentiviral vectors
integrated with TFAP2A-AS1 shorth hairpin (sh)RNA/TFAP2A shRNA, and
the corresponding negative controls (NC siRNA or NC shRNA). Vazyme
Biotech Co., Ltd. supplied an Annexin V-FITC/PI Apoptosis Detection
kit. Beyotime Institute of Biotechnology provided the hematoxylin
and eosin (H&E) staining kit, the RIPA lysis buffer, the
enhanced chemiluminescence (ECL) kit and the bicinchonic acid (BCA)
kit. Thermo Fisher Scientific, Inc. provided the Lipofectamine
RNAiMAX transfection agent and Pierce Magnetic RNA-Protein
Pull-Down kit (cat. no. 20164). Beijing Solarbio Science &
Technology Co., Ltd. supplied the Total cholesterol (TC) Content
Assay Kit (cat. no. BC1980). Nanjing Jiancheng Bioengineering
Institute provided the high-density lipoprotein-cholesterol (HDL-C)
assay kit (cat. no. A112-1-1) and low-density
lipoprotein-cholesterol (LDL-C) assay kit (cat. no. A113-1-1).
Takara Biotechnology Co., Ltd. supplied the MiniBEST Universal RNA
Extraction kit (cat. no. 9767). Invitrogen; Thermo Fisher
Scientific, Inc. supplied the PrimeScript RT reagent and SYBR Green
Premix for the experiment. Primary antibodies TFAP2A (cat. no.
13019-3-AP), IgG control (cat. no. 30000-0-AP) and GAPDH (cat. no.
60004-1-Ig) were purchased from Proteintech Group, Inc., along with
horseradish peroxidase (HRP)-conjugated secondary antibodies (cat.
no. SA00001-2). The Magna RIP RNA-Binding Protein
Immunoprecipitation kit (cat. no. 17-700) was obtained from
MilliporeSigma.
Cell culture
The HCAECs were cultured in EBM-2 supplemented with
15% FBS, under conditions of 5% CO2 and a temperature of
37˚C. For functional experiments, cells in the phase of logarithmic
growth were collected.
Cell transfection
Cell transfection was performed in strict accordance
with the protocols of Lipofectamine RNAiMAX transfection reagent.
In brief, HCAECs were individually transfected with 50 nM of
TFAP2A-AS1 siRNA1/2/3 (TFAP2A-AS1 siRNA1 forward,
5'-UGCUAAUGAGGCGAUUAGGCU-3' and reverse,
5'-CCUAAUCGCCUCAUUAGCAUA-3'; TFAP2A-AS1 siRNA2 forward,
5'-AUUUCUAAUAAAAUUGCACGG-3' and reverse,
5'-GUGCAAUUUUAUUAGAAAUCA-3'; TFAP2A-AS1 siRNA3 forward,
5'-UGUUUUUAGGCUCAACUUCAG-3' and reverse,
5'-GAAGUUGAGCCUAAAAACAGA-3'); TFAP2A siRNA1/2/3 (TFAP2A siRNA1
forward, 5'-AUUUAAUCCUAUUUUGUCCAG-3' and reverse,
5'-GGACAAAAUAGGAUUAAAUCU-3'; TFAP2A siRNA2 forward,
5'-UUGCAUAUCUGUUUUGUAGCC-3' and reverse,
5'-CUACAAAACAGAUAUGCAAAG-3'; TFAP2A siRNA3 forward,
5'-UGCUUUUGGCGUUGUUGUCCG-3' and reverse,
5'-GACAACAACGCCAAAAGCAGU-3'); and NC siRNA forward,
5'-GAAAUGUUUAGGGCCAGUGCU-3' and reverse,
5'-AGCACUGGCCCUAAACAUUUC-3' for 48 h at 37˚C using Lipofectamine
RNAiMAX transfection reagent. Following transfection, cells were
collected for subsequent experiments.
Flow cytometric analysis
The apoptosis of HCAECs was assessed by employing a
commercially available Annexin V-FITC/PI Apoptosis Detection kit.
Briefly, HCAECs (2x105) were suspended in 500 µl of
binding buffer, and subsequently stained with PI and Annexin V-FITC
(both 5 µl) at 4˚C for 15 min in the absence of light. Following
this step, cell apoptosis was evaluated using a FACScan flow
cytometer (version 3.0; Becton, Dickinson and Company) and analyzed
using BD CellQuest Pro software (version 5.2.1; Becton, Dickinson
and Company). Cells were classified as viable cells (lower left
quadrant), necrotic cells (upper left quadrant), early apoptotic
cells (lower right quadrant) and late apoptotic cells (upper right
quadrant). The apoptosis rate was estimated by calculating the
total percentages of both early and late apoptotic cells.
CCK-8 assay
HCAECs were cultured in 96-well plates at a density
of 3,500 cells per well and incubated for various time intervals
(24, 36, 48, and 72 h). Subsequently, each well was supplemented
with 10 µl of CCK-8 solution and further incubated for 2 h at 37˚C.
Following the addition of 10 µl of stop solution to every well,
cell viability was assessed using a microplate reader (DR-3518G;
Wuxi Hiwell Diatek Instruments Co., Ltd.), which measured the
absorbance at a wavelength of 450 nm.
Transwell assay
The upper chamber of Transwell insert was pre-coated
with 50 µl of Matrigel at 37˚C for 30 min, which had been diluted
5-fold in serum-free EBM-2. HCAECs were incubated for 24 h
following digestion, after which the culture medium was removed by
centrifugation (1,500 x g; at 4˚C for 5 min). Subsequent to washing
with PBS, 1x105 resuspended cells were seeded into the
Transwell chamber and incubated for an additional 24 h. The cells
were then fixed with 4% paraformaldehyde at 37˚C for 15 min,
washed, and stained with crystal violet at 37˚C for 15 min. The
invasive cells were observed under a light microscope (Leica
Microsystems GmbH), and images were captured from three randomly
selected fields of view.
Wound healing assay
HCAECs were cultured in 6-well plates at a density
of 3x105 cells per well. Once the cells attained 100%
confluence, a scratch was created on the monolayer using a 10 µl
pipette tip. Subsequently, HCAECs were incubated in serum-free
EBM-2 at 37˚C for 48 h. The widths of the scratches at both 0 h
(initial width) and 48 h (final width) were observed using a light
microscope (Leica Microsystems GmbH). To assess the migratory
abilities of HCAECs, ImageTool software version 3.0 (UTHSCSA, San
Antonio, USA) was employed with the following formula: (Initial
width-final width)/initial width x100%.
RNA immunoprecipitation (RIP)
assay
The Magna RIP RNA-Binding Protein
Immunoprecipitation kit was used for RIP assays following the
manufacturer's protocol. Briefly, HCAECs were collected and lysed
using RIP lysis buffer. For each IP reaction, HCAECs
(2x107 cells) were resuspended in 1 ml of lysis buffer
and incubated on ice for 30 min. The samples were then centrifuged
at 14,000 x g for 20 min at 4˚C. Whole-cell extracts were incubated
with RIP buffer (at 4˚C for 30 min) containing magnetic beads
conjugated to human anti-TFAP2A antibody or the control IgG. The
beads were then washed five times with 1 ml wash buffer and
centrifuged at 1,000 x g for 1 min at 4˚C between washes.
Subsequently, 100 µl of elution buffer was added in the samples to
incubate for 10 min at 65˚C. Proteinase K was added to the samples
to digest the protein, and the immunoprecipitated RNA was isolated.
Purified RNA was used for reverse transcription-quantitative PCR
(RT-qPCR) analysis. The expressional level of GAPDH was used as the
control.
RNA pull-down assay
The interaction between TFAP2A-AS1 and TFAP2A
protein was examined using Pierce Magnetic RNA-Protein Pull-Down
kit according to the manufacturer's protocols. Proteins from HCAECs
were extracted using lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1%
NP-40, 10% glycerin, 2 mM EDTA, 1 mM PMSF, 1X Protease inhibitor
cocktail, and 40 U/ml RNase). For each reaction, HCAECs
(5x106 cells) were resuspended in 1 ml of lysis buffer
and incubated on ice for 60 min. Biotin-labeled TFAP2A-AS1 or
antisense RNA was co-incubated with protein extract of HCAECs and
Protein G-magnetic beads (40 µl). The generated bead-RNA-Protein
compound was collected by low-speed centrifugation (800 x g for 2
min at 4˚C). After being washed with 2X SDS loading buffer, the
bead compound was boiled for 10 min in SDS buffer, and the
retrieved protein was detected by western blotting with the
expressional level of GAPDH as the control.
Animal grouping and treatment
Animal experiments were conducted in accordance with
the Guidelines for the Use of Laboratory Animals and approved by
the Ethics Committee of the First Affiliated Hospital of Wenzhou
Medical University (approval no. xmsq2022-1230). A total of 24
C57BL/6J mice, weighting 18-22 g and aged 8 to 10 weeks, were
procured from Pengsheng Biological Technology Co., Ltd. The ACS
mouse model was established in accordance with a previously
described protocol (15). C57BL/6J
mice were randomly assigned to four groups: Sham group, ACS model +
NC shRNA group, ACS model + TFAP2A-AS1 shRNA group, and ACS model +
TFAP2A shRNA group. Each group consisted of six mice. Anesthesia
was induced through an intraperitoneal injection of pentobarbital
sodium at a dose of 50 mg/kg. The limbs of the mice were then
secured on a surgical table while maintaining them in a supine
position. A thoracotomy was performed to expose the heart, followed
by ligation of the anterior descending coronary artery using 6/0
surgical sutures. To prevent infection, each mouse was administered
a subcutaneous injection of penicillin at a dose of 50 mg/kg. By
contrast, the mice in the sham group underwent only threading
without ligation. Successful establishment of the ACS mouse model
was confirmed based on significantly elevated ST segment and/or
high T wave readings (data not shown). A three-plasmid system,
along with 9 µg of lentiviral vectors, was co-transfected into 293
cells using Lipofectamine RNAiMAX transfection agent at room
temperature. The ratio of lentiviral plasmid to packaging vector to
envelope was maintained at 1:1:1. The lentivirus-containing culture
medium was harvested 48 h post-transfection and subsequently used
to infect MCAECs (second passage; 5x106 cells per well)
with a multiplicity of infection set at 20. Subsequently, 48 h
after infection, the cells were subjected to selection using 1
µg/ml puromycin. Prior to surgery, mice in the ACS model + NC shRNA
group received an intramyocardial injection containing lentiviral
vectors integrated with the NC shRNA sequence (50 µg;
5'-GGCAGGGTGATGGGCAACATA-3'). By contrast, those in both the ACS
model + TFAP2A-AS1 shRNA and ACS model + TFAP2A shRNA groups were
administered with lentiviruses carrying either the TFAP2A-AS1 shRNA
sequence (5'-GGCTGGTGAAGAACCTGAAGG-3') or the TFAP2A shRNA sequence
(5'-GAGTTGCTTGACCCACTTCAA-3'), respectively, via intramyocardial
injection (both at a dose of 50 µg). Anesthesia was maintained for
30 min. The humane endpoints were rapid or labored breathing. After
a two-week period, euthanasia was conducted on all groups using an
overdose of pentobarbital sodium administered via intraperitoneal
injection at a dose of 200 mg/kg. Death was confirmed by cardiac
and respiratory arrest and a lack of response to tail clamping.
Following the confirmation of death, whole blood samples (300 µl;
collected from the eyeballs) and heart tissues were collected for
further analysis.
RT-qPCR
The extraction of RNA from heart tissues of mice,
HCAECs, and serum samples of patients with ACS, was conducted using
the MiniBEST Universal RNA Extraction Kit. Subsequently, reverse
transcription into cDNA was performed at 45˚C for 45 min using the
PrimeScript RT reagent kit. PCR analysis was then conducted in
accordance with the SYBR Green Premix protocols on a QuantStudio7
system (Thermo Fisher Scientific, Inc.). The reaction conditions
consisted of an initial denaturation step at 95˚C for 2 min,
followed by 40 cycles of denaturation at 95˚C for 10 sec, annealing
at 60˚C for 30 sec, and extension at 72˚C for 30 sec. The primers
used in the present study are listed in Table II. The relative expression levels
of TFAP2A-AS1 and TFAP2A were normalized against GAPDH and
calculated using the 2-ΔΔCq method (16).
 | Table IIReal-time PCR primer synthesis
list. |
Table II
Real-time PCR primer synthesis
list.
| Gene | Sequences |
|---|
| LncRNA
TFAP2A-AS1 | Forward
5'-GCATCCACGTCCTCTCTCTG-3' |
| | Reverse
5'-GCAGATTGTGGTACTGGCGA-3' |
| TFAP2A | Forward
5'-TCCGCTTCACGCTCGATTT-3' |
| | Reverse
5'-AATCCGTGTCTCCCCCTCTT-3' |
| GAPDH | Forward
5'-TCAAGAAGGTGGTGAAGCAGG-3' |
| | Reverse
5'-TCAAAGGTGGAGGAGTGGGT-3' |
Western blot analysis
The concentration of total proteins was determined
in HCAECs or heart tissues by extracting them with RIPA lysis
buffer, followed by measurement using a BCA kit. Subsequently, the
proteins were separated using 10% SDS polyacrylamide gel
electrophoresis and then transferred onto a PVDF membrane. The
membrane was blocked using 5% nonfat milk for 2 h at 25˚C.
Subsequently, primary antibodies (TFAP2A at a dilution of 1:5,000;
GAPDH at a dilution of 1:50,000) overnight at 4˚C, along with their
corresponding secondary antibodies (at a dilution of 1:1,000) at
37˚C for 1 h were incubated separately. For the purpose of protein
normalization, GAPDH was employed as a reference. The protein
signals were detected using an ECL kit and quantified with Alpha
Innotech software version 6.0 (Alpha Innotech Corporation).
Biochemical tests
The whole blood samples from mouse eyeballs were
maintained at ambient temperature for 2 h, followed by
centrifugation at a force of 1,000 x g for a 20 min at 4˚C. To
determine the serum levels of TC, HDL-C, and LDL-C, commercially
available kits were employed in conjunction with an automated
bioanalysis system (Gallery Plus Discrete Analyzer; Thermo Fisher
Scientific, Inc.).
H&E staining
The myocardial tissues of mice were subjected to
fixation (4% paraformaldehyde at 25˚C for 12 h), deparaffinization,
and hydration prior to embedding in paraffin. Subsequently, they
were sectioned into slices with a thickness of 4 µm. The sections
underwent a 3-min H&E staining process at 25˚C. Following the
application of neutral gum for mounting, the images were captured
using a Leica light microscope.
Statistical analysis
In vitro experiments were performed in
triplicate, and each experiment was repeated 3 times. In
vivo experiments were performed using six mice per group. The
differences between two groups were evaluated using unpaired
Student's t-test. To assess variations among multiple groups, a
one-way ANOVA followed by Tukey's post hoc multiple comparisons
test was utilized. Data analysis was performed using SPSS software
version 22.0 (Dotmatics). The data were presented as the mean ±
standard deviation, and P<0.05 was considered to indicate a
statistically significant difference.
Results
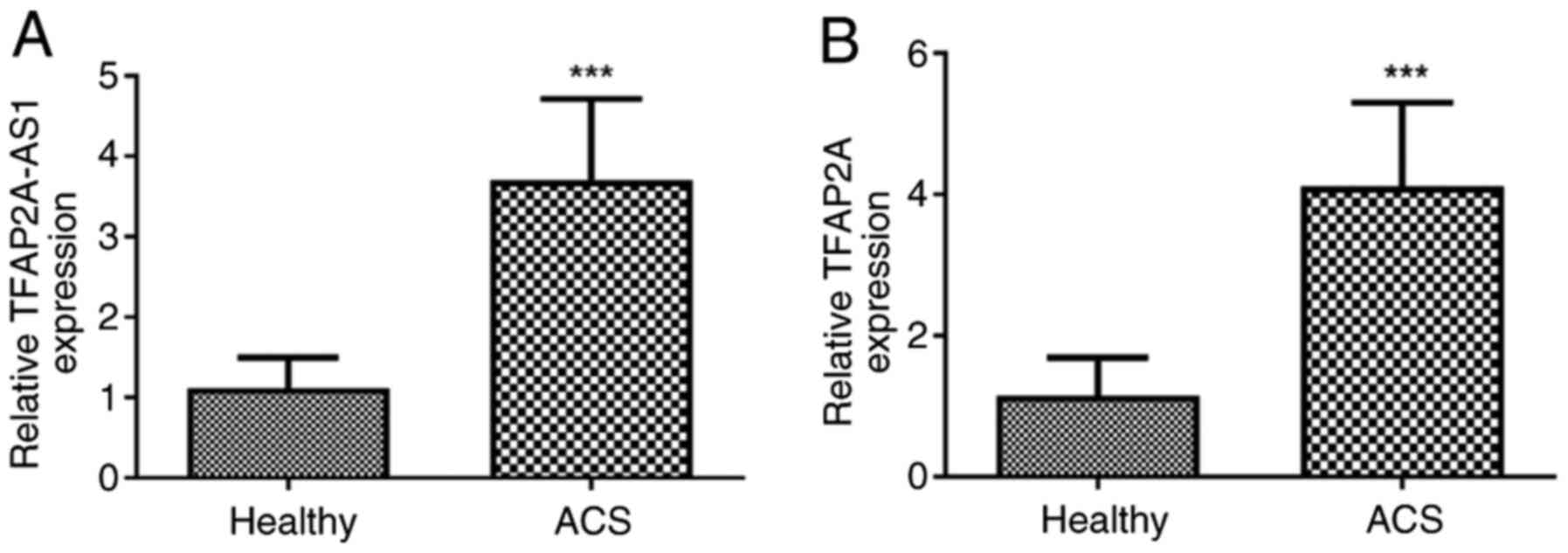
Serum from patients with ACS exhibits
elevated expression of TFAP2A-AS1 and TFAP2A
After obtaining the necessary consent from the
participants, venous blood samples were collected from individuals
diagnosed with ACS. Subsequently, centrifugation was conducted to
obtain serum samples, and then the expression levels of TFAP2A-AS1
and TFAP2A were determined using RT-qPCR. The findings, as depicted
in Fig. 1A, revealed a significant
upregulation of TFAP2A-AS1 expression in patients with ACS compared
with that observed in healthy individuals (P<0.001; Fig. 1B). Similarly, elevated levels of
TFAP2A were also detected in patients with ACS when compared with
controls without any health issues (P<0.001; Fig. 1B).
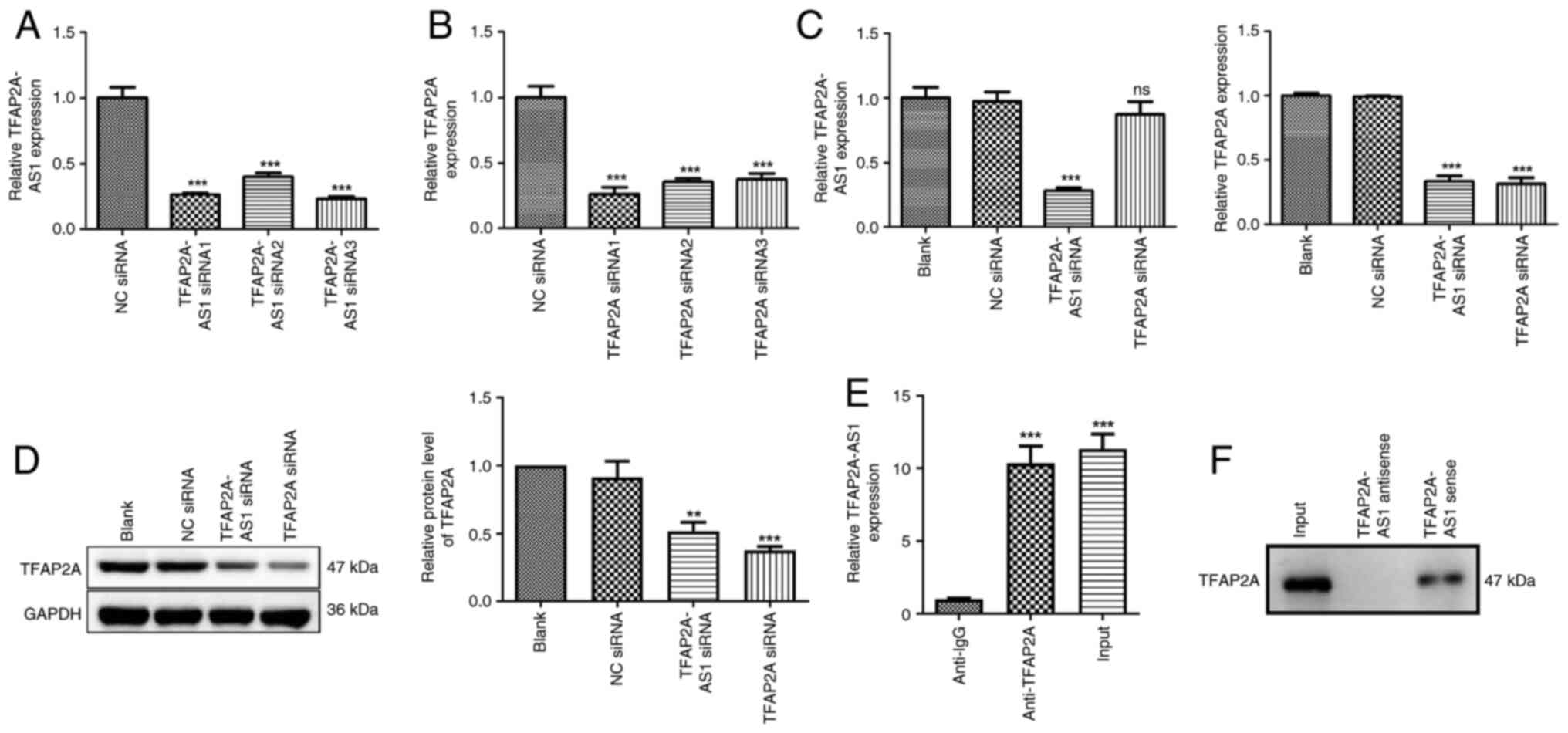
Silencing of TFAP2A-AS1 suppresses
TFAP2A expression
To investigate the roles of TFAP2A-AS1 and TFAP2A in
the progression of ACS in vitro, gene silencing techniques
were initially employed to downregulate the expression levels of
either TFAP2A-AS1 or TFAP2A in HCAECs. As illustrated in Fig. 2A, the HCAECs were transfected with
TFAP2A-AS1 siRNA1, TFAP2A-AS1 siRNA2, and TFAP2A-AS1 siRNA3
individually. It was revealed that the expression level of
TFAP2A-AS1 was significantly decreased following transfection
(P<0.001). The selection of TFAP2A-AS1 siRNA3 for subsequent
experiments was based on its relatively high silencing efficiency.
Similarly, TFAP2A siRNA transfection also significantly inhibited
TFAP2A expression, and TFAP2A siRNA1 was selected for the
subsequent experiments (P<0.001; Fig. 2B). The regulatory associations
between TFAP2A-AS1 and TFAP2A were then determined. The results of
RT-qPCR and western blotting demonstrated that transfection of
TFAP2A siRNA did not affect TFAP2A-AS1 expression, while
transfection of TFAP2A-AS1 siRNA markedly decreased the mRNA
expression and protein level of TFAP2A (P<0.01; Fig. 2C and D). The findings indicated that TFAP2A-AS1
possesses the capability to regulate the expression of TFAP2A;
however, the converse does not hold true. A RIP assay was performed
to confirm the functional interaction between TFAP2A-AS1 and
TFAP2A. RNA from RIP assays using an antibody against TFAP2A was
subjected to qPCR analysis, which demonstrated an enrichment of
TFAP2A-AS1 (P<0.001; Fig. 2E).
Next, whether TFAP2A-AS1 interacted with TFAP2A protein was
examined using an RNA pull-down assay, and it was revealed that the
expression level of TFAP2A interacting with biotin-labeled
TFAP2A-AS1 was higher than that with the antisense TFAP2A-AS1 group
(Fig. 2F). These data suggested the
interaction between TFAP2A-AS1 and TFAP2A protein.
Silencing of TFAP2A-AS1 and TFAP2A
inhibits the proliferative, migratory, and invasive capacities
while enhancing the apoptotic rate of HCAECs
The flow cytometric analysis showed that
transfection of both TFAP2A-AS1 siRNA and TFAP2A siRNA
significantly increased the apoptosis rate of HCAECs (P<0.001;
Fig. 3A). Starting from 24 h, the
viability of HCAECs was significantly reduced following the
silencing TFAP2A-AS1 or TFAP2A (P<0.001; Fig. 3B). Additionally, the effects of
TFAP2A-AS1 and TFAP2A on the invasive and migratory capacities of
HCAECs were also assessed. The relative number of invasive cells
was significantly decreased in the TFAP2A-AS1 siRNA and TFAP2A
siRNA groups compared with those in the NC siRNA group, as
demonstrated in Fig. 3C
(P<0.001). Similar patterns were observed in the effects of
TFAP2A-AS1 siRNA and TFAP2A siRNA on cell migration (P<0.001;
Fig. 3D).
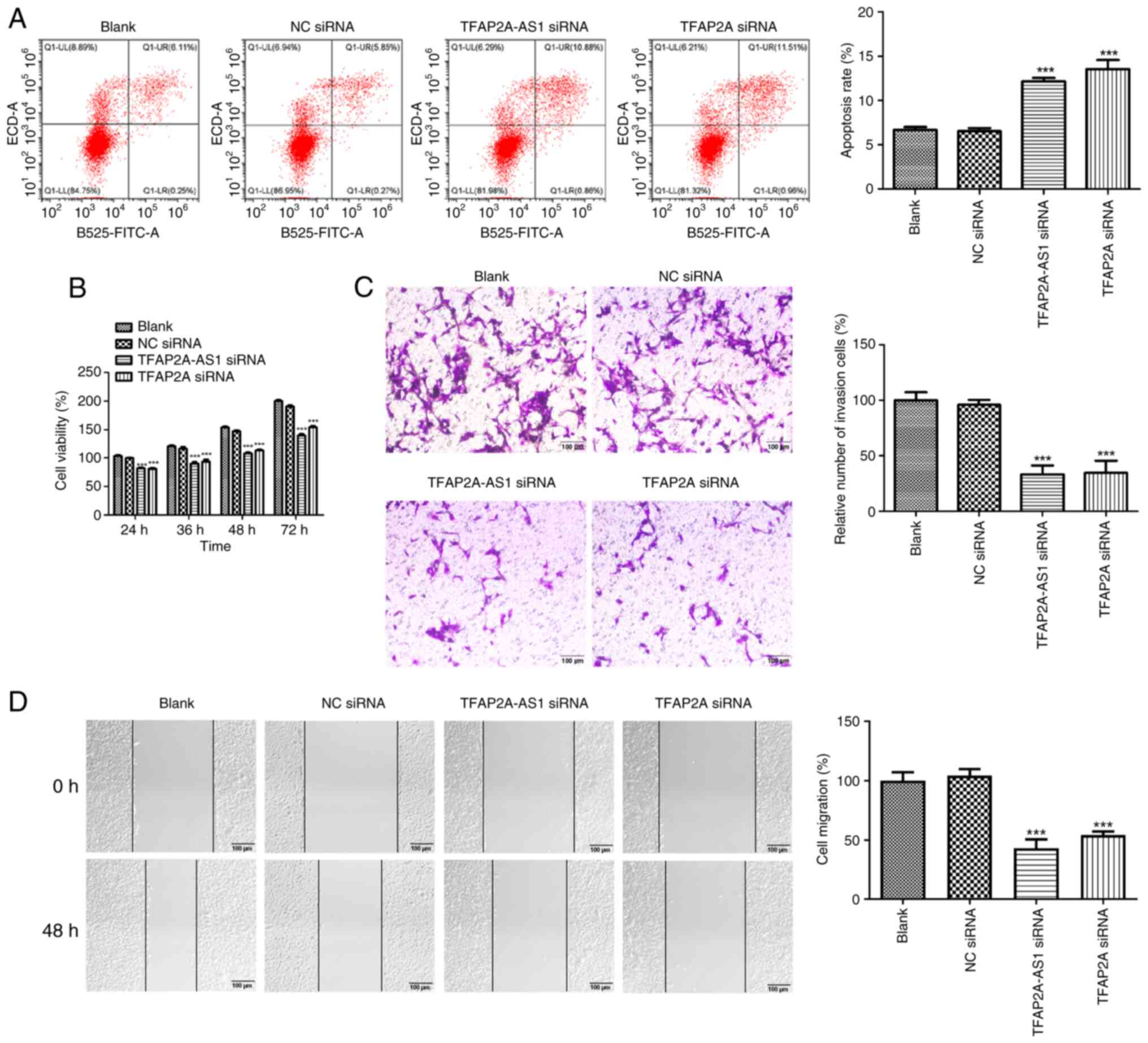 | Figure 3Silencing of TFAP2A-AS1 and TFAP2A
suppresses the proliferative, migratory, and invasive capacities
while enhancing the apoptotic rate of HCAECs. (A) The apoptosis
rate, (B) viability, (C) invasion and (D) migration of HCAECs
transfected TFAP2A-AS1 siRNA, TFAP2A siRNA or NC siRNA was assessed
by flow cytometric analysis, Counting Kit-8 assay, Transwell
invasion assay and wound healing assay, respectively. Scale bar,
100 µm. ***P<0.001 vs. NC siRNA. TFAP2A,
transcription factor AP-2α; HCAECs, human coronary artery
endothelial cells; siRNA, small interfering RNA; NC, negative
control. |
Knockdown of TFAP2A-AS1 and TFAP2A
leads to a decrease in the serum lipid levels and an improvement in
myocardial injury in a mouse model of ACS
A mouse model of ACS was established in the present
study. As demonstrated in Fig. 4A,
the expression of TFAP2A-AS1 and TFAP2A was significantly increased
in mice with ACS (P<0.001). However, their expression levels
were reduced after injection of TFAP2A-AS1 shRNA and TFAP2A shRNA,
respectively (P<0.001). Notably, injection of TFAP2A shRNA did
not affect the expression of TFAP2A-AS1, while injection of
TFAP2A-AS1 shRNA significantly decreased the mRNA expression of
TFAP2A in mice with ACS (P<0.01). Moreover, similar patterns
were observed for the protein level of TFAP2A (P<0.01; Fig. 4B). Furthermore, it was revealed that
compared with the sham group, the serum levels of TC and LDL-C were
significantly elevated in a mouse model of ACS (P<0.001;
Fig. 4C), while the levels of HDL-C
were significantly decreased (P<0.001). Notably, the
intramyocardial injection of both TFAP2A-AS1 shRNA and TFAP2A shRNA
not only led to a reduction in the levels of TC and LDL-C
(P<0.05), but also resulted in increased levels of HDL-C
(P<0.001) in mice with ACS. Subsequently, heart tissues from
mice across various groups were collected for the purpose of
pathological examination. As depicted in Fig. 4D, cardiomyocytes from the sham group
exhibited a normal morphology characterized by a well-organized
arrangement and an absence of breaks. By contrast, cardiomyocytes
derived from the ACS mice displayed signs of swelling and
thickening, accompanied by irregular morphology and disordered
arrangement. Following intramyocardial injection of both TFPAF-A21
shRNA and TFAF-A21 shRNA, a significant improvement in the
morphology and arrangement of cardiomyocytes was observed.
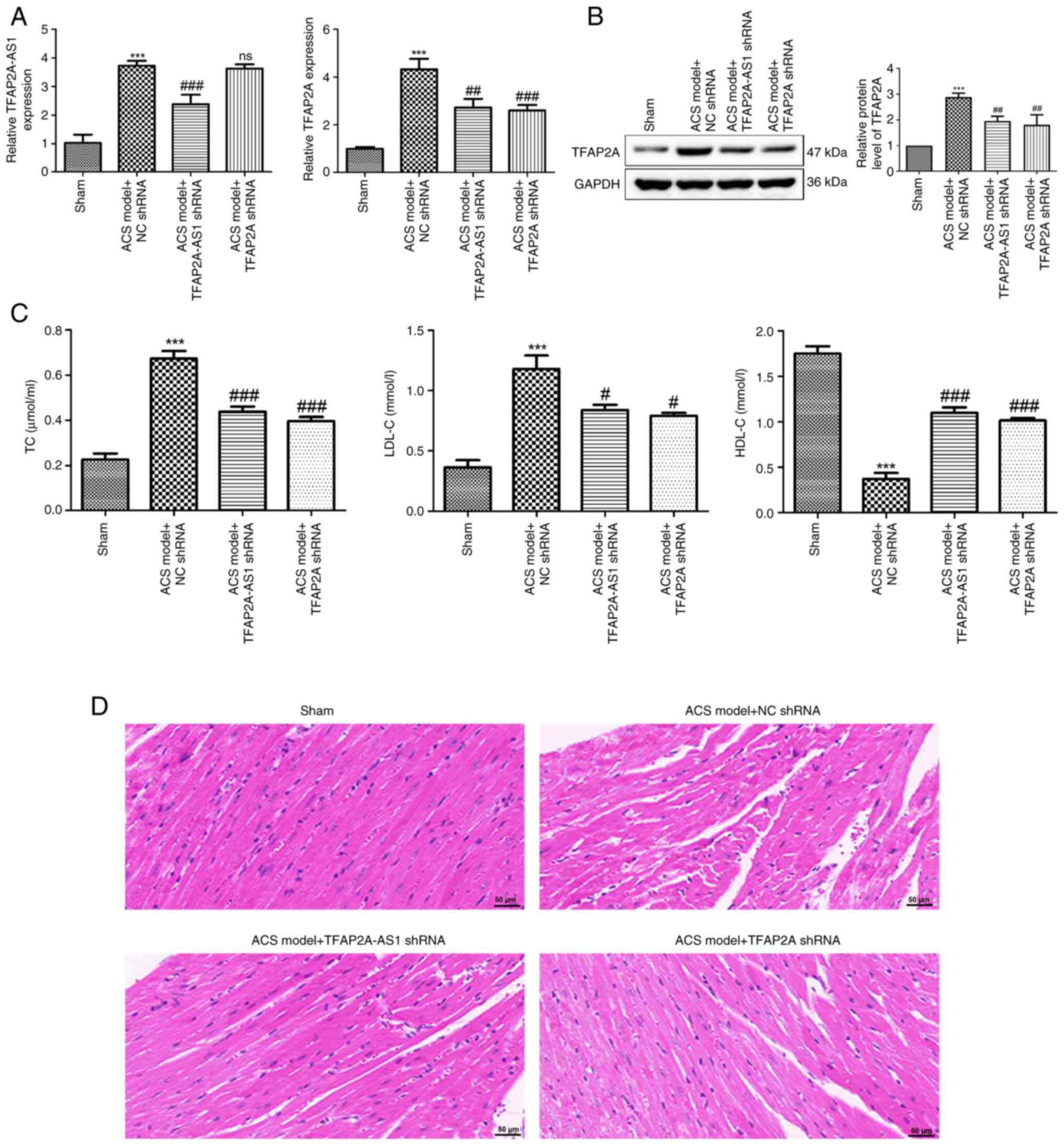 | Figure 4Knockdown of TFAP2A-AS1 and TFAP2A
leads to a reduction in serum lipid levels and an improvement in
myocardial injury in an ACS mouse model. (A) The expression of
TFAP2A-AS1 and TFAP2A in ACS mice after injection of TFAP2A-AS1
shRNA, TFAP2A shRNA or NC shRNA was detected by reverse
transcription-quantitative PCR. (B) The protein levels of TFAP2A in
ACS mice after injection of TFAP2A-AS1 shRNA, TFAP2A shRNA or NC
shRNA were determined using western blotting. (C) The levels of TC,
LDL-C and HDL-C in ACS mice after injection of TFAP2A-AS1 shRNA,
TFAP2A shRNA or NC shRNA were determined using biochemical tests.
(D) Hematoxylin and eosin staining was performed to observe the
pathological condition of myocardial tissues in different groups.
Scale bar, 50 µm. ***P<0.001 vs. sham;
#P<0.05, ##P<0.01, and
###P<0.001 vs. the ACS model + NC shRNA. TFAP2A,
transcription factor AP-2α; ACS, acute coronary syndrome; shRNA,
short hairpin RNA; ns, no significance. |
Discussion
Despite significant advancements in the diagnosis
and treatment of ACS, CVDs remain the leading cause of death
globally. Low- and middle-income countries bear a considerable
burden, with ~7 million deaths and 129 million cases of disability
reported annually (17-19).
Furthermore, despite a significant decline in mortality rates
associated with ACS, it is still estimated that ~40% of patients
will succumb to death within five years following a coronary event.
The risk of fatality increases 5 to 6-fold in individuals who
experience recurrent events (20,21).
The economic impact associated with ACS is also substantial; with
each patient in the U.S. incurring an estimated annual cost ranging
from $22,528 to $32,345 primarily due to hospitalizations (22). Consequently, ACS not only imposes a
significant burden on patients but also affects society at large.
Therefore, there is an urgent need to explore more effective
treatment options for this condition.
Most research on lncRNA TFAP2A-AS1 primarily focuses
on its role in cancer. Aberrant expression of TFAP2A-AS1 has been
shown to influence the occurrence and development of multiple human
cancers, including GC, BC, NSCLC and OSCC (11-14).
In the present study, the dysregulation of TFAP2A-AS1 was also
observed in both patients with ACS and corresponding mouse models,
suggesting that TFAP2A-AS1 may be significantly associated with the
progression of ACS. The significance of endothelial cells in the
pathogenesis of ACS should not be underestimated. The pathology of
ACS represents a complex and multifaceted process, with plaque
rupture identified as one of its primary triggers (3,23).
Following a myocardial infarction, significant alterations occur in
the behavior of endothelial cells, which are crucial for
angiogenesis. Consequently, the abnormal proliferation, migration
and invasion of these endothelial cells post-myocardial infarction
may potentially contribute to plaque instability and erosion,
ultimately leading to the onset and development of ACS (24,25).
In the present study, it was demonstated that knockdown of
TFAP2A-AS1 not only significantly suppressed the proliferative,
invasive and migrated potentials of HCAECs, but also markedly
increased the apoptosis rate. Furthermore, previous studies have
reported that elevated levels of TC and LDL-C are associated with
endothelial dysfunction (26,27).
Based on the findings of the present study which demonstrated that
silencing of TFAP2A-AS1 effectively inhibited the aberrant
proliferation, invasion, and migration of HCAECs, it is reasonable
to propose that the downregulation of TFAP2A-AS1 may play a pivotal
role in regulating blood lipid levels. In the animal experiments,
as anticipated, a significant reduction in the levels of TC and
LDL-C, accompanied by an elevation in the levels of HDL-C were
observed upon the injection of TFAP2A-AS1 shRNA in mice with ACS.
The aforementioned results indicated that the downregulation of
TFAP2A-AS1 may play a crucial role in the regulation of endothelial
dysfunction. Moreover, the myocardial injury symptoms observed in
ACS mice were significantly ameliorated following the injection of
TFAP2A-AS1 shRNA. Therefore, the findings indicated that the
absence of TFAP2A-AS1 may confer a beneficial effect on mitigating
the progression of ACS.
TFAP2A, a member of the AP-2 family, has been
identified as a transcription factor involved in various aspects of
development (28). In
branchio-oculo-facial syndrome, a rare autosomal-dominant disorder
characterized by cleft palate and craniofacial abnormalities, a
deletion in the TFAP2A gene has been observed (29). Additionally, studies have shown that
the expression of TFAP2A is upregulated and functions as an
oncogene in the progression of various cancers, including cervical
cancer, gallbladder carcinoma, and ovarian cancer (30-32).
Moreover, TFAP2A is essential for cardiac morphogenesis
specifically during outflow tract formation and cardiac septation
by regulating cell proliferation and terminal differentiation
(33,34). It was therefore hypothesized that
TFAP2A may also play a crucial role in ACS. In the present study,
an increased expression of TFAP2A in both patients with ACS and a
mouse model was observed Moreover, it was also demonstrated that
the knockdown of TFAP2A could modulate endothelial dysfunction by
inhibiting blood lipid levels in ACS mice, as well as suppressing
the aberrant proliferation, migration and invasion of HCAECs. All
these results supported our hypothesis that TFAP2A is associated
with the progression of ACS. Additionally, a recent study by Yang
et al (35) demonstrated
that the expression levels of both TFAP2A-AS1 and TFAP2A were
upregulated in OSCC, exhibiting a positive correlation between
their expression levels. This suggested that TFAP2A-AS1 can
interact with TFAP2A to affect OSCC progression. Based on the
observed effects of TFAP2A-AS1 and TFAP2A on HCAECs and mouse
models, a potential association between TFAP2A-AS1 and TFAP2A in
the progression of ACS was postulated. The findings demonstrated
that knockdown of TFAP2A-AS1 led to decreased expression of TFAP2A,
while no reciprocal effect was observed for TFAP2A on the
expression of TFAP2A-AS1. In addition, the RIP and RNA pull-down
assays further verified the interaction of TFAP2A-AS1 with TFAP2A.
These results suggest a positive regulatory role for TFAP2A-AS1 in
modulating the expression of TFAP2A. Consequently, the present
study concludes that the silencing of TFAP2A-AS1 may impede the
progression of ACS by inhibiting the expression of TFAP2.
Some limitations that must be acknowledged in this
study are as follows: Firstly, confirming whether TFAP2A-AS1
regulates TFAP2A expression in a post-transcriptional manner may
require a more rigorous approach. Secondly, the sample size of
patients was relatively limited, and increasing the sample size
would provide more robust data and enhance the statistical power of
the findings. Thirdly, the study was conducted on a specific
population in Wenzhou, China. Expanding the research to include
diverse populations in China, will be considered in future
studies.
In summary, the present research demonstrated that
TFAP2A-AS1 acts as a pathogenic lncRNA in ACS. The suppression of
TFAP2A-AS1 led to a significant decrease in TFAP2A expression,
thereby hindering the progression of ACS through the regulation of
endothelial dysfunction. This newly identified regulatory axis may
represent a potential therapeutic target for the treatment of
ACS.
Acknowledgements
Not available.
Funding
Funding: The present study was supported by Wenzhou Municipal
Science and Technology Plan Project (grant no. Y20210160).
Availability of data and materials
The data are available from the corresponding author
on reasonable request.
Authors' contributions
HC and SHu made substantial contributions to the
conception and design of the study. SHu, FG, FY, BY, and SHa made
substantial contributions to the acquisition, analysis and
interpretation of data for the study. SHu drafted the manuscript.
SHu, FG, FY, BY, SHa, and HC revised the manuscript critically for
important intellectual content. SHu, FG, FY, BY, SHa, and HC
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript and agree to be to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The patients/participants provided their written
informed consent to participate in this study. The human study was
conducted according to the guidelines of the Declaration of
Helsinki and approved (approval no. KY-2021053) by the Ethics
Committee of Wenzhou People's Hospital (Wenzhou, China). Animal
experiments were conducted in compliance with the Guidelines for
the Use of Laboratory Animals and approved (approval no.
xmsq2022-1230) by the Ethics Committee of the First Affiliated
Hospital of Wenzhou Medical University (Wenzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
WHO CVD Risk Chart Working Group. World
Health Organization cardiovascular disease risk charts: Revised
models to estimate risk in 21 global regions. Lancet Glob Health.
7:e1332–e1345. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Knuuti J, Wijns W, Saraste A, Capodanno D,
Barbato E, Funck-Brentano C, Prescott E, Storey RF, Deaton C,
Cuisset T, et al: 2019 ESC guidelines for the diagnosis and
management of chronic coronary syndromes. Eur Heart J. 41:407–477.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Collet JP, Thiele H, Barbato E, Barthélémy
O, Bauersachs J, Bhatt DL, Dendale P, Dorobantu M, Edvardsen T,
Folliguet T, et al: 2020 ESC guidelines for the management of acute
coronary syndromes in patients presenting without persistent
ST-segment elevation. Eur Heart J. 42:1289–1367. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kaul P, Ezekowitz JA, Armstrong PW, Leung
BK, Savu A, Welsh RC, Quan H, Knudtson ML and McAlister FA:
Incidence of heart failure and mortality after acute coronary
syndromes. Am Heart J. 165:379–385.e2. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
McKnight AH, Katzenberger DR and Britnell
SR: Colchicine in acute coronary syndrome: A systematic review. Ann
Pharmacother. 55:187–197. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Smith JN, Negrelli JM, Manek MB, Hawes EM
and Viera AJ: Diagnosis and management of acute coronary syndrome:
An evidence-based update. J Am Board Fam Med. 28:283–293.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang L and Jin Y: Noncoding RNAs as
biomarkers for acute coronary syndrome. Biomed Res Int.
2020(3298696)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen H, Huang S, Guan F, Han S, Ye F, Li X
and You L: Targeting circulating lncRNA ENST00000538705.1 relieves
acute coronary syndrome via modulating ALOX15. Dis Markers.
2022(8208471)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Barbalata T, Niculescu LS, Stancu CS,
Pinet F and Sima AV: Elevated levels of circulating lncRNAs LIPCAR
and MALAT1 predict an unfavorable outcome in acute coronary
syndrome patients. Int J Mol Sci. 24(12076)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen L and Huang Y: High expression of
lncRNA PELATON serves as a risk factor for the incidence and
prognosis of acute coronary syndrome. Sci Rep.
12(8030)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao X, Chen L, Wu J, You J, Hong Q and Ye
F: Transcription factor KLF15 inhibits the proliferation and
migration of gastric cancer cells via regulating the
TFAP2A-AS1/NISCH axis. Biol Direct. 16(21)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhou B, Guo H and Tang J: Long non-coding
RNA TFAP2A-AS1 inhibits cell proliferation and invasion in breast
cancer via miR-933/SMAD2. Med Sci Monit. 25:1242–1253.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang Y, Ma L, Zhang T, Li P, Xu J and
Wang Z: Long noncoding RNA TFAP2A-AS1 exerts promotive effects in
non-small cell lung cancer progression via controlling the
microRNA-548a-3p/CDK4 axis as a competitive endogenous RNA. Oncol
Res. 29:129–139. 2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jie G, Peng S, Cui Z, He C, Feng X and
Yang K: Long non-coding RNA TFAP2A-AS1 plays an important role in
oral squamous cell carcinoma: Research includes bioinformatics
analysis and experiments. BMC Oral Health. 22(160)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Luo XY, Zhu XQ, Li Y, Wang XB, Yin W, Ge
YS and Ji WM: MicroRNA-150 restores endothelial cell function and
attenuates vascular remodeling by targeting PTX3 through the NF-κB
signaling pathway in mice with acute coronary syndrome. Cell Biol
Int. 42:1170–1181. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vedanthan R, Seligman B and Fuster V:
Global perspective on acute coronary syndrome: A burden on the
young and poor. Circ Res. 114:1959–1975. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moran AE, Oliver JT, Mirzaie M,
Forouzanfar MH, Chilov M, Anderson L, Morrison JL, Khan A, Zhang N,
Haynes N, et al: Assessing the global burden of ischemic heart
disease: Part 1: Methods for a systematic review of the global
epidemiology of ischemic heart disease in 1990 and 2010. Glob
Heart. 7:315–329. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Forouzanfar MH, Moran AE, Flaxman AD, Roth
G, Mensah GA, Ezzati M, Naghavi M and Murray CJ: Assessing the
global burden of ischemic heart disease, part 2: Analytic methods
and estimates of the global epidemiology of ischemic heart disease
in 2010. Glob Heart. 7:331–342. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Rogers WJ, Canto JG, Lambrew CT,
Tiefenbrunn AJ, Kinkaid B, Shoultz DA, Frederick PD and Every N:
Temporal trends in the treatment of over 1.5 million patients with
myocardial infarction in the US from 1990 through 1999: The
national registry of myocardial infarction 1, 2 and 3. J Am Coll
Cardiol. 36:2056–2063. 2000.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Thom T, Haase N, Rosamond W, Howard VJ,
Rumsfeld J, Manolio T, Zheng ZJ, Flegal K, O'Donnell C, Kittner S,
et al: Heart disease and stroke statistics-2006 update: A report
from the American heart association statistics committee and stroke
statistics subcommittee. Circulation. 113:e85–e151. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Menzin J, Wygant G, Hauch O, Jackel J and
Friedman M: One-year costs of ischemic heart disease among patients
with acute coronary syndromes: Findings from a multi-employer
claims database. Curr Med Res Opin. 24:461–468. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Libby P and Pasterkamp G: Requiem for the
‘vulnerable plaque’. Eur Heart J. 36:2984–2987. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bonetti PO, Lerman LO and Lerman A:
Endothelial dysfunction: A marker of atherosclerotic risk.
Arterioscler Thromb Vasc Biol. 23:168–175. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhu F, Wang Q, Guo C, Wang X, Cao X, Shi
Y, Gao F, Ma C and Zhang L: IL-17 induces apoptosis of vascular
endothelial cells: A potential mechanism for human acute coronary
syndrome. Clin Immunol. 141:152–160. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Stein RA: Endothelial dysfunction,
erectile dysfunction, and coronary heart disease: The
pathophysiologic and clinical linkage. Rev Urol. 5 (Suppl
7):S21–S27. 2003.PubMed/NCBI
|
|
27
|
Ling L, Zhao SP, Gao M, Zhou QC, Li YL and
Xia B: Vitamin C preserves endothelial function in patients with
coronary heart disease after a high-fat meal. Clin Cardiol.
25:219–224. 2002.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yang YL and Zhao LY: AP-2 family of
transcription factors: Critical regulators of human development and
cancer. J Cancer Treat Diagn. 5:1–4. 2021.
|
|
29
|
Gestri G, Osborne RJ, Wyatt AW, Gerrelli
D, Gribble S, Stewart H, Fryer A, Bunyan DJ, Prescott K, Collin JR,
et al: Reduced TFAP2A function causes variable optic fissure
closure and retinal defects and sensitizes eye development to
mutations in other morphogenetic regulators. Hum Genet.
126:791–803. 2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang P, Hou Q and Yue Q:
MiR-204-5p/TFAP2A feedback loop positively regulates the
proliferation, migration, invasion and EMT process in cervical
cancer. Cancer Biomark. 28:381–390. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang HX, Yang G, Yang Y, Yan J, Tang XY
and Pan Q: TFAP2A is a novel regulator that modulates ferroptosis
in gallbladder carcinoma cells via the Nrf2 signalling axis. Eur
Rev Med Pharmacol Sci. 24:4745–4755. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu H, Wang L and Jiang X: Silencing of
lncRNA DLEU1 inhibits tumorigenesis of ovarian cancer via
regulating miR-429/TFAP2A axis. Mol Cell Biochem. 476:1051–1061.
2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hutson MR and Kirby ML: Model systems for
the study of heart development and disease. Cardiac neural crest
and conotruncal malformations. Semin Cell Dev Biol. 18:101–110.
2007.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hammer S, Toenjes M, Lange M, Fischer JJ,
Dunkel I, Mebus S, Grimm CH, Hetzer R, Berger F and Sperling S:
Characterization of TBX20 in human hearts and its regulation by
TFAP2. J Cell Biochem. 104:1022–1033. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yang K, Niu Y, Cui Z, Jin L, Peng S and
Dong Z: Long noncoding RNA TFAP2A-AS1 promotes oral squamous cell
carcinoma cell growth and movement via competitively binding
miR-1297 and regulating TFAP2A expression. Mol Carcinog.
61:865–875. 2022.PubMed/NCBI View
Article : Google Scholar
|