Introduction
Colorectal cancer (CRC) is one of the most common
malignancies and the second leading cause of cancer-related death
in Spain. An individual’s life-time risk of developing CRC is 6%,
with over 90% of cases occurring after the age of 50 years.
Together, these facts indicate that CRC is an important health
concern (1,2).
Population-based genetic association studies (case
control studies) are the most widely used study designs with which
to determine the impact of genetic variants on the risk of
developing a particular complex disease (3,4).
In this way, hundreds of association studies have
been performed in order to elucidate the genetic contribution in
complex diseases, such as cancer. Through these studies several low
penetrance genes have been found to behave as cancer risk
modifiers, contributing to the understanding of tumor formation in
many types of cancers and leading to advances in diagnosis and
therapy.
Due to the high frequency of genetic polymorphisms
in the human genome and the number of possible haplotypes in a
particular chromosomal region, it is difficult to establish the
level of similarity between different haplotypes (5). However, this problem was solved a few
years ago through the identification of an exceptionally abundant
Yin-Yang haplotype pattern extending 75–80% of the entire human
genome (6). This peculiar pattern
is defined as two mismatched haplotypes (Yin and Yang) representing
the majority of the existing haplotypes in a particular genomic
region. An increase of the recombination rate has been observed in
human genome regions with Yin-Yang haplotype structure (6). The human adenomatous polyposis coli
(APC) gene is localized on chromosome 5q21-227 and encodes a
multifunctional protein that participates in several cellular
processes: cell adhesion and migration, signal transduction,
microtubule assembly and chromosome segregation (7). A somatic mutation in APC is
the most common acquired genetic alteration in colorectal adenomas
(CRA) and carcinomas (CRC) and one of the earliest mutation events
in sporadic colon cancer (8–10).
Somatic inactivation occurs in more than 80% of sporadic CRC and it
follows the Knudson hypothesis, showing similar incidences of LOH
and gene mutation as second events (11,12).
These sporadic tumors, which do not present somatic inactivation of
APC (less than 20%), show tumor microsatellite instability
(MSI) (13) while its carcinogenic
pathway is different, being affected by the mismatch repair pathway
suppressor tumor genes (14).
The APC gene is our candidate gene, since it
functions as a gatekeeper of colorectal neoplasia. APC shows
a Yin-Yang haplotype pattern described in a European (CEU)
population (5) and also in a
Spanish population (15).
Several association studies have been published on
APC gene variants and CRC risk. Many of them analyzed
separate SNPs (16–20), while a few others analyzed the
combination of various SNPs (haplotypes) (21,22).
However, no studies have been published involving diplotype
structure and colon cancer risk.
In this study, we searched for other genetic factors
that act as risk modifiers apart from those affecting the DNA
sequence. For instance, the similarity between homologous
chromosomes at particular loci may affect the recombination rate in
these loci and, therefore, may favor the inactivation of particular
recessive genes. To test this hypothesis, we present an association
study using a specific approach in which this particular structural
characteristic (diplotype) acted as a genetic variant. The aim of
the present study was to investigate whether the APC
Yin-Yang homozygote diplotype is over-represented in patients with
stable microsatellite (MSS) sporadic CRC when compared to its
distribution in controls, and to establish whether this diplotype
is a risk factor for CRC in a Spanish population.
Patients and methods
Study populations
Cases and controls were recruited from two public
hospitals in Spain; Hospital Clínico San Carlos in Madrid (HCSC
samples) and Instituto Catalán de Oncología in Barcelona (ICO
samples).
HCSC samples included 157 CRC cases with a mean age
at diagnosis of 67 years (range 25–90) recruited between 1998 and
2005 at the Molecular Oncology Unit, and 405 controls with a mean
age of 65.8 years (range 25–90) recruited between 2001 and 2008 at
the Clinical Pathology Laboratory. Specifically, eligible
individuals had no personal or family history of cancer.
ICO samples included 221 CRC cases with a mean age
at diagnosis of 70.6 years (range 31–91) recruited between 1990 and
2005 at the Catalán Institute of Oncology, and 237 controls with a
mean age of 67.5 years (range 35–91) recruited between 1990 and
2001 at the National Blood Transfusion Centre.
Informed consent was obtained from all participants,
and the study was approved by the Institutional Review Board of
HCSC.
Cases and controls from HCSC and ICO were matched
for age and gender.
DNA isolation
Genomic DNA was isolated from peripheral blood
lymphocytes by a salting out procedure (23) or by automatic DNA extraction
(Magnapure®; Roche) according to the manufacturer’s
protocol. Tumor DNA was isolated from the dissection of tumor cells
from paraffin-embedded tissue (selected on the basis of an
H&E-stained slide), using the QIAamp DNA FFPE Tissue kit
extraction method. DNA was quantified using Nanodrop® R
(ND 1000) and diluted to a final concentration of 50 ng/μl for
genotyping.
Microsatellite instability (MSI)
MSI analysis was performed in all paired
tumor-normal tissue DNA samples testing the Bethesda (24) panel of five markers (D2S123,
D5S346, D17S250, BAT 25 and BAT 26). PCR amplifications were
performed with the HNPCC Microsatellite Instability kit (Roche
Diagnostic, Basel, Switzerland) according to the supplier’s
instructions. Products were analyzed in an ABI310 genetic analyzer.
Tumors were classified as MSS when none or just one of the five
Bethesda markers showed instability.
Genotyping
The APC Yin-Yang haplotype was described by
Ribas et al (15) in a
Spanish population. We chose three tagged SNPs (tgSNPs) (rs2019720,
rs2431512 and rs2546108) for the two frequent haplotypes. Although
we needed only one tgSNP to test our hypothesis, we used three to
confirm the Yin-Yang haplotype structure of our study population.
The three tgSNPs were selected through the HapMap Phased Haplotype
plot and distributed along the APC Yin-Yang region (Fig. 1). Their heterozygosity index was
close to 0.5 (Table I). Genotyping
was carried out by nuclease assay (TaqMan®). TaqMan
genotyping reagents were designed by Applied Biosystems as
Assay-by-Design (Table I). PCR
reaction was carried out according to the manufacturer’s
recommended protocols in an ABI PRISM® 7900HT System
(Applied Biosystems). The analysis was carried out with the SDS
2.2.1 ABI software.
 | Table I.Characteristics of the selected tgSNPs
(NCBI). |
Table I.
Characteristics of the selected tgSNPs
(NCBI).
| NCBI SNP
reference | rs2019720 | rs2431512 | rs2546108 |
|---|
| Location on gene
(chromosone 5) | Intron 1
(112.102.168) | Intron 6
(112.146.855) | Intron 11
(112.189.368) |
| Reference of SNP
alleles | A/G | C/T | A/C |
| Ancestral allele
(frequency in CEU) | A (0.508) | C (0.5) | C (0.492) |
| Dyes | VIC - allele
A
FAM - allele G | VIC - allele
C
FAM - allele T | VIC - allele
A
FAM - allele C |
| Heterozygosity ±
standard error | 0.494±0.055 | 0.495±0.047 | 0.495±0.048 |
| TaqMan assay | C_1120624_10 | C_3162979_10 | C_9389410_10 |
| Yin haplotype | G | T | A |
| Yang haplotype | A | C | C |
Statistical analysis
Deviations from Hardy-Weinberg equilibrium were
tested using the Chi-square test. Two-sided Chi-square tests were
used to assess the differences in APC diplotype frequency
distributions between cases and controls. Power calculations were
performed by Epi-Info 6 program.
Results
We performed a case-control study to investigate the
possible association of the three tgSNPs selected across the
APC gene (Fig. 1) with CRC
risk. Genotyping of the rs2019720, rs2431512 and rs2546108 was
carried out in 157 cases and 405 controls from the Hospital Clínico
San Carlos in Madrid, Spain (HCSC samples). All tgSNPs tested were
in Hardy-Weinberg equilibrium (P=0.17, 0.19 and 0.17,
respectively). The genotyping data are available upon request.
Yin-Yang haplotypes were established in the HCSC samples. The
frequencies in the CRC cases and controls were 52.25% for the GTA
haplotype (Yin) and 46.65% for the ACC haplotype (Yang). The
remaining 1.1% belonged to other haplotypes. The APC
Yin-Yang alleles (GTA and ACC, respectively) comprised 98.8% of all
of the haplotypes (Table II).
 | Table II.Haplotype frequencies in the case and
control groups. |
Table II.
Haplotype frequencies in the case and
control groups.
| Population | No. | GTA (Yin) no.
(%) | ACC (Yang) no.
(%) | Others no. (%) |
|---|
| HCSC | | | | |
| CRC | 314 | 166 (52.9) | 146 (46.5) | 2 (0.6) |
| Control | 810 | 418 (51.6) | 379 (46.8) | 13 (1.6) |
| ICO | | | | |
| CRC | 442 | 233 (52.7) | 203 (45.9) | 6 (1.4) |
| Control | 474 | 242 (51.1) | 226 (47.7) | 6 (1.3) |
| All samples | | | | |
| CRC | 756 | 399 (52.8) | 349 (46.2) | 8 (1.1) |
| Control | 1,284 | 660 (51.4) | 605 (47.1) | 19 (1.5) |
After genotyping, samples were divided into two
subgroups; the homozygote diplotype (GTA/GTA or ACC/ ACC) and the
heterozygote diplotype (GTA/ACC). Rare haplotypes were not detected
in homozygosis, so they were included in the heterozygote diplotype
group (Table III). We observed an
increase in the homozygote frequency in the HCSC cases when
compared to the HCSC-controls (OR=1.93; 95% CI 1.32–2.81; P=0.001).
Statistical power was >80%.
 | Table III.Diplotype frequencies in the case and
control groups. |
Table III.
Diplotype frequencies in the case and
control groups.
| Population | No. | Homo (%) | Hete (%) | OR | 95% CI | P-value |
|---|
| HCSC | | | | | | |
| CRC | 157 | 96 (61.1) | 61 (38.8) | 1.93 | 1.32–2.81 | 0.001 |
| Control | 405 | 182 (44.9) | 223 (55.1) | | | |
| ICO | | | | | | |
| CRC | 221 | 109 (49.3) | 112 (50.7) | 0.89 | 0.61–1.28 | 0.521 |
| Control | 237 | 124 (52.3) | 113 (47.7) | | | |
| All samples | | | | | | |
| CRC | 378 | 205 (54.2) | 173 (45.8) | 1.30 | 1.01–1.68 | 0.043 |
| Control | 642 | 306 (47.7) | 336 (52.3) | | | |
We subsequently decided to validate the positive
results in another Spanish group of cases and controls. We
genotyped 221 cases and 237 controls from the Instituto Catalan de
Oncologia in Barcelona (cases and controls are described in
Patients and methods). The SNPs were also in Hardy-Weinberg
equilibrium (P=0.33, 0.23 and 0.37, respectively). The frequencies
for the Yin-Yang haplotypes were 51.9 for GTA (Yin), 46.8 for ACC
(Yang) and 1.3 for other haplotypes (Table II). We found very similar haplotype
frequencies in the HCSC and ICO samples. In relation to the
diplotype results we did not find any significant difference in the
frequency of the homozygote diplotype between the cases and
controls (OR=0.89; 95% CI 0.61–1.28; P=0.521) (Table III).
When we combined the samples from HCSC and ICO to
analyze both populations (378 cases and 642 controls) the
homozygote diplotype frequency in the cases was 54.2% (Table III). A slightly significant result
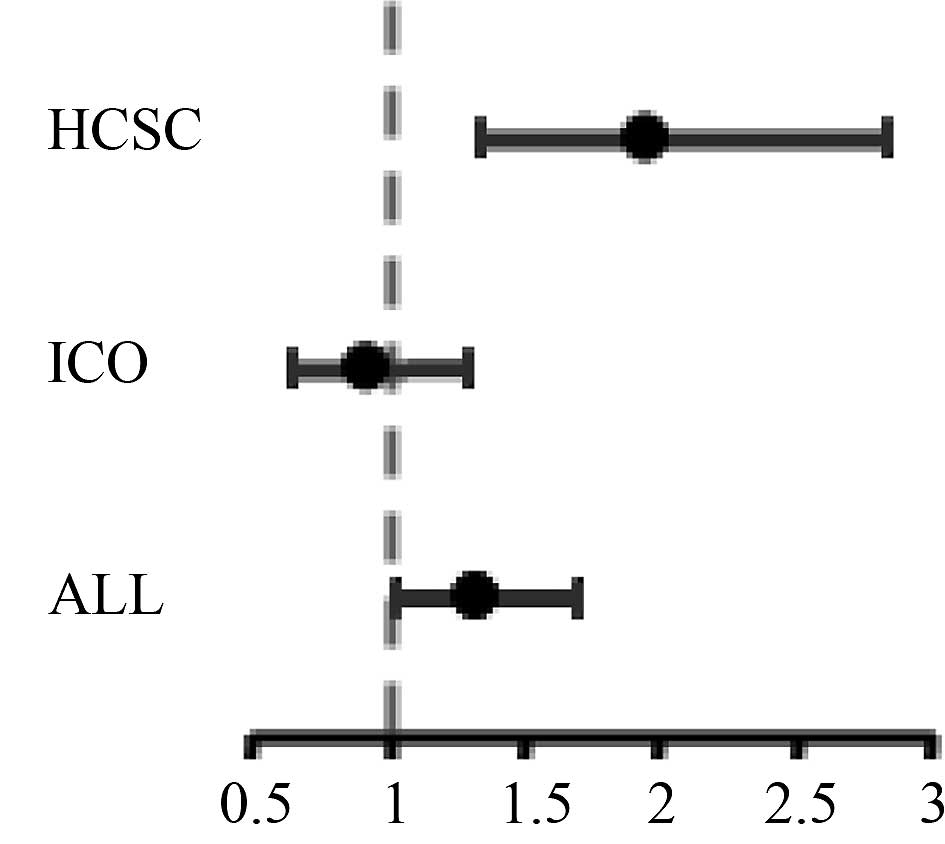
was noted (OR=1.3; 95% CI 1.01–1.68; P=0.043) (Table III, Fig. 2).
Discussion
With the aim of verifying our hypothesis that
over-representation of the APC Yin-Yang homozygote diplotype
occurs in MSS sporadic CRC populations, we genotyped three tgSNPs
spanning the gene (Fig. 1,
Table I).
We assessed the haplotype distribution in the cases
and controls of the HCSC and ICO populations. All groups showed
similar frequencies of Yin-Yang haplotypes (Table II), indicating no
over-representation of a single haplotype in our study group.
Moreover, we found similar frequencies of APC
Yin-Yang haplotypes (98.8%) as Ribas et al (25) in a Spanish population (84.1%).
We carried out a case-control analysis of the
Yin-Yang diplotype in the HCSC population and observed a
significantly increased frequency of the homozygote diplotype in
the case group; 61.1 vs. 44.9% in the control group (OR=1.93; 95%
CI 1.32–2.81; P=0.001).
In order to replicate our positive results, we
repeated the analysis in a second Spanish MSS sporadic CRC
population confirmed by ICO and compared this group to a control
group of the same origin. In this case, the ICO sample did not show
a difference between the cases and controls; 49.3 vs. 52.3%
(OR=0.89; 95% CI 0.61–1.28; P=0.521).
In the HCSC samples, we found differences in the
diplotype distribution between the cases and controls. In contrast,
we did not find any difference in the ICO samples. Since the
selection criteria for the cases were the same in both populations,
these discrepancies may be attibuted to the eligibility criteria
for the controls. The ICO controls were enrolled at the National
Blood Transfusion Centre, while the HCSC controls were enrolled at
the Clinical Pathology Laboratory after filling out a
questionnaire, and only those with no history of cancer were
chosen. Selection criteria for the ICO control group did not
exclude the enrollment of subjects affected with cancer. This fact
may also explain the similar homozygote frequencies found in our
study between the ICO cases and controls.
Environmental factors also affected the disease
prevalence in a different manner in both the populations, so these
may have contributed to the observed differences in the diplotype
distribution in both samples.
Finally, notably, enough statistical power (>80%)
was present to detect an OR as high as 1.9 in the HCSC sample,
while enough power was not present to detect a low OR in the ICO
samples. Therefore, we cannot discard a weaker effect of the
Yin-Yang homozygote diplotype in the ICO population, which could
not be detected with our sample size.
To resolve this problem, additional studies using
different sets of patients and control subjects are warranted,
since supporting evidence exits for an association between the
APC Yin-Yang homozygote diplotype and CRC risk in the HCSC
population.
Although the association between the APC
Yin-Yang homozygote diplotype and CRC risk remains to be confirmed,
a singular strategy to test the hypothesis has been presented. In
this study, a structural genetic characteristic (such as Yin-Yang
diplotype) was compared in different groups instead of genetic
variants and, therefore, other different mechanisms (such as
allelic recombination or gene conversion) were tested instead of
allelic functionality. Studies on SNPs included in the Yin-Yang
structure and CRC risk may provide more information than isolated
SNPs. The selection of cases and controls from different sources
must be based on identical criteria in order to compare both
populations. A more extensive study is required to support the
evidence for the association between the APC Yin-Yang
homozygote diplotype and CRC risk.
Acknowledgements
The present study was supported by the
Instituto de Salud Carlos III (ISCIII) research grant FIS07/0359,
Spanish Ministry of Science and Innovation. P.G. and A.R. are
ISCIII pre-doctoral researchers and M.H. is ISCIII senior
researcher. P.I. is on the staff of UCM. S.G. and G.C. are on the
staff of ICO. E.D.R. and T.C. are on the staff of HCSC.
References
|
1.
|
Fearon ER and Volgestein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
De la Chapelle A: Genetic predisposition
to colorectal cancer. Nat Rev Cancer. 4:769–779. 2004.
|
|
3.
|
Carlson CS, Eberle MA, Rieder MJ, Yi Q,
Kruglyak L and Nickerson DA: Selecting a maximally informative set
of single nucleotide polymorphisms for association analyses using
linkage disequilibrium. Am J Hum Genet. 74:106–120. 2004.
View Article : Google Scholar
|
|
4.
|
Gabriel SB, Scaffner SF, Nguyen H, et al:
The structure of haplotypes blocks in the human genome. Science.
296:2225–2229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
The International HapMap Consortium: A
haplotype map of the human genome. Nature. 437:1299–1320. 2005.
View Article : Google Scholar
|
|
6.
|
Zhang J, Rowe WL, Clark AG and Buetow KH:
Genomewide distribution of high-frequency, completely mismatching
SNP haplotype pairs observed to be common across human populations.
Am J Hum Genet. 73:1073–1081. 2003. View
Article : Google Scholar
|
|
7.
|
Bodmer WF, Bailey CJ, Bodmer J, et al:
Localization of the gene for familial adenomatous polyposis on
chromosome 5. Nature. 328:614–616. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Fodde R: The APC gene in colorectal
cancer. Eur J Cancer. 38:867–871. 2002. View Article : Google Scholar
|
|
9.
|
Senda T, Iizuka-Kogo A, Onouchi T and
Shimomura A: Adenomatous polyposis coli (APC) plays multiple roles
in the intestinal and colorectal epithelia. Med Mol Morphol.
40:68–81. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Powell SM, Zilz N, Beazer-Barclay Y, et
al: APC mutations occur early during colorectal tumorigenesis.
Nature. 359:235–237. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Rowan AJ, Lamlum H, Ilyas M, et al: APC
mutations in sporadic colorectal tumors: a mutational ‘hotspot’ and
interdependence of the ‘two hits’. Proc Natl Acad Sci USA.
97:3352–3357. 2000.
|
|
12.
|
Segditsas S, Sieber OM, Rowan A, et al:
Promoter hypermethylation leads to decreased APC mRNA expression in
familial polyposis and sporadic colorectal tumours, but does not
substitute for truncating mutations. Exp Mol Pathol. 85:201–206.
2008. View Article : Google Scholar
|
|
13.
|
Ionov Y, Peinado MA, Malkhosyan S, Shibata
D and Percho M: Ubiquitous somatic mutations in simple repeated
sequences reveal a new mechanism for colonic carcinogenesis.
Nature. 363:558–561. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Markowitz SD and Bertagnolli MM: Molecular
origins of cancer: molecular basis of colorectal cancer. N Engl J
Med. 361:2449–2460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ribas G, Gonzalez-Neira A, Salas A, et al:
Evaluating HapMap SNP data transferability in a large-scale
genotyping project involving 175 cancer-associated genes. Hum
Genet. 118:669–679. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Laken SJ, Petersen GM, Gruber SB, et al:
Familial colorectal cancer in Ashkenazim due to a hypermutable
tract in APC. Nat Genet. 17:79–83. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Gryfe R, Di Nicola N, Gallinger S and
Redston M: Somatic instability of the APC I1307K allele in
colorectal neoplasia. Cancer Res. 58:4040–4043. 1998.PubMed/NCBI
|
|
18.
|
Frayling IM, Beck NE, Ilyas M, et al: The
APC variants I1307K and E1317Q are associated with colorectal
tumors, but not always with a family history. Pro Natl Acad Sci
USA. 95:10722–10727. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Lamlum H, Al Tassan N, Jaeger E, et al:
Germline APC variants in patients with multiple colorectal
adenomas, with evidence for the particular inportance of E1317Q.
Hum Mol Genet. 9:2215–2221. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Azzopardi D, Dallosso AR, Eliason K, et
al: Multiple rare nonsynonymous variants in the adenomatous
polyposis coli gene predispose to colorectal adenomas. Cancer Res.
68:358–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Cui DH, Jiang KD, Jiang SD, Xu YF and Yao
H: The tumour suppressor adenomatous polyposis coli gene is
associated with susceptibility to schizophrenia. Mol Psychiatry.
10:669–677. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Egan JB, Jacobs ET, Martínez ME, Gerner
EW, Jurutkan PW and Thompson PA: Presence of a TA haplotype in the
APC gene containing the common 1822 polymorphism and colorectal
adenoma. Cancer Res. 68:6006–6013. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151998. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Umar A, Boland CR, Terdiman JP, et al:
Revised Bethesda Guidelines for hereditary nonpolyposis colorectal
cancer (Lynch Syndrome) and microsatellite instability. J Natl
Cancer Inst. 96:261–268. 2004. View Article : Google Scholar
|
|
25.
|
Ribas G, Milne RL, Gonzalez-Neira A and
Benitez J: Haplotype patterns in cancer-related genes with
long-range linkage disequilibrium: no evidence of association with
breast cancer or positive selection. Eur J Hum Genet. 16:252–260.
2008. View Article : Google Scholar
|