Introduction
Recently, concerns about an association with
assisted reproductive technologies (ART) and genomic imprinting
defects have been raised (1,2).
Genomic imprinting is a process of chemical modification of
nucleotides in which only one allele of a specific gene is
functioning and the other allele is silenced based on the
parent-of-origin. The most thoroughly studied epigenetic
modification is methylation of CpG dinucleo-tides in
cis-regulatory sequences (3). Allelic expression of imprinted genes
is controlled by differentially methylated regions (DMRs) which are
thought to function as imprinting control centers (4). DMRs are thought to be particularly
sensitive to disruption by environmental factors such as the large
amounts of gonadotropin and different culture mediums (5,6).
The effect of in vitro fertilization (IVF) on
DNA methylation in mouse embryos has been demonstrated in many
studies (5,7–9).
Small epidemiology studies in humans have suggested a 4- to 9-fold
increased incidence of Beckwith-Wiedemann syndrome (BWS) among
children conceived by IVF or intracytoplasmic sperm injection
(ICSI) (10–13). The loss of maternal DNA methylation
at the DMR of KCNQ1 represents the most frequent alternation
in BWS patients (11–13). Moreover, epimutations at other DMRs
in ART children resulting in BWS, such as the mesoderm-specific
transcript (MEST), or small nuclear
ribonucleoprotein-associated polypeptide N (SNRPN) have also
been reported (14).
However, in a large cohort study from Denmark
including 6,052 IVF singleton children and 442,349 normally
conceived singletons, there were no cases of imprinting-related
disease identified in the IVF group and 54 children with
imprinting-related diseases in the non-IVF cohort (15). An increased risk of the overall
imprinting diseases after IVF was not observed in the study
(15). A questionnaire-based
survey from the Republic of Ireland and Central England aimed to
detect children after ART with possible diagnosis of BWS or
Angelman syndrome (AS) (16). In
the study, data from 1,524 children were analyzed and only one case
of BWS was identified having hypomethylation at KvDMR1,
suggesting that although prevalence of BWS in children born after
ART may be higher, the absolute risk of having a child with BWS
conceived by ART is notably remote (16). Moreover, ten DMRs were analyzed in
185 phenotypically normal children by Tierling et al, and
they found no association with ART and imprinting (17). Notably, in a recent study conducted
by Gomes et al, aberrant imprinting was observed in 3 out of
18 clinically normal children conceived by ART (18), suggesting that the impact of ART on
the epigenetics is not completely understood.
In order to investigate the possible genetic risk of
DNA methylation defects associated with ART, we analyzed the
methylation patterns of six DMRs in 161 phenotypically normal
children, comprising 40 children conceived by ICSI, 61 by IVF and
60 conceived spontaneously. The DMRs analyzed included
KvDMR1, SNRPN, MEST, maternally expressed gene
3 (MEG3), transient neonatal diabetes mellitus (TNDM)
and X (inactive)-specific transcript (XIST).
Materials and methods
Study population and DNA samples
Informed consent and medical information release
documents were obtained as approved by the institutional review
board. Data regarding maternal age, gestational age, birth weight,
and birth length of the neonates without pathological findings were
documented. Umbilical cord blood samples from 101 children born
after ART and 60 children conceived spontaneously were taken
directly after birth and stored at −80°C. DNA purification was
performed using 3–7 ml EDTA-blood with the Genomic DNA purification
kit (Promega, USA).
Sodium bisulfite treatment and
methylation-specific PCR
The methylation assay was performed at the DMRs of
six imprinted genes (KvDMR1, SNRPN, MEST,
MEG3, TNDM and XIST). Bisulfite treatment of
genomic DNA was performed with the EpiTect Bisulfite kit (Qiagen,
Germany). Methylation-specific PCR (MS-PCR) utilizes this sodium
bisulfite treatment to distinguish methylated from unmethylated DNA
(19,20).
Purified, non-methylated and methylated human DNA
standard was used as the negative and positive control in the
methylation detection applications, which was performed with the
Human Methylated & Non-methylated DNA set (Zymo Research, USA).
Each sample was analyzed in two independent MS-PCR reactions. PCR
reactions were carried out with a GeneAmp PCR System 9700. The
25-μl PCR reaction mix contained 2X PCR HotStart Premix buffer
(Takara, Tokyo, Japan), 0.5 μM primer-M forward and 0.5 μM primer-M
reverse in the PCR reaction amplifying the methylated imprint
specifically or 0.5 μM primer-U forward and 0.5 μM primer-U reverse
in the unmethylated PCR, and 2 μl of bisulfite-modified DNA. Primer
sequences for KvDMR1 (21),
MEST (22), SNRPN
(23), MEG3 (24), XIST (25), and TNDM (26) were used as described, and PCR
programs were summarized in Table
I. PCR products were separated on a 2% agarose gel, stained
with ethidium bromide and visualized under UV illumination.
 | Table I.Analyzed differentially methylated
regions (DMRs) and methylation-specific PCR procedure. |
Table I.
Analyzed differentially methylated
regions (DMRs) and methylation-specific PCR procedure.
| DMRs | Chromosomal
location | Allelic
methylation | Ta
| Size (bp)
|
|---|
| M | U | M | U |
|---|
| KvDMR1 | 11p15.5 | Maternal | 60 | 58 | 170 | 170 |
| MEST | 7q32.2 | Maternal | 60 | 60 | 300 | 300 |
| SNRPN | 15q11-q13 | Maternal | 60 | 60 | 216 | 313 |
| MEG3 | 14q32 | Paternal | 60 | 60 | 160 | 120 |
| XIST | X | | 60 | 60 | 264 | 280 |
| TNDM | 6q24 | Maternal | 60 | 60 | 175 | 187 |
Statistical analysis
SPSS 16.0 was used to analyze the characteristics of
the study population. Continuous variables were presented as the
mean ± SD and were tested by two-sample t-test or analysis of
variance (ANOVA). The Chi-square test was used to compare the
categorical neonatal and maternal data. P<0.05 was considered
statistically significant.
Results
Clinical data
Blood samples were collected from 101 children born
after ART (61 conceived by IVF and 40 by ICSI) and 60 children
conceived spontaneously. IVF and ICSI procedures were carried out
following standard protocols (27). The ICSI was performed with
ejaculated spermatozoa in the majority of the cases (n=37, 92.5%),
with fresh testicular spermatozoa after testicular sperm extraction
(TESE) in 1 case (2.5%) and with epididymal spermatozoa after
microsurgical epididymal sperm aspiration (MESA) in 2 cases (5%).
The ICSI procedure was performed due to male factor infertility in
all couples.
A major difference between the three sample groups
concerns the frequency of twin pregnancies. Due to multiple embryo
transfer following ART twin pregnancies were very frequent in the
IVF and ICSI groups. Of the 101 babies from ART, 46 were delivered
from 23 twin pregnancies, and the remaining 55 children were born
from singleton pregnancies, while only 2 twin pregnancies were
among the 60 children in the spontaneously conceived group
(P<0.05). As compared with the infants in the spontaneous
pregnancy group, infants conceived by IVF and ICSI were more likely
to have low birth weight and to be born before term (P<0.05)
(Table II). In addition, maternal
age was significantly higher in the IVF group than that in the
spontaneous pregnancy and ICSI groups.
 | Table II.Neonatal and maternal characteristics
of the IVF, ICSI and spontaneously conceived pregnancies. |
Table II.
Neonatal and maternal characteristics
of the IVF, ICSI and spontaneously conceived pregnancies.
| Characteristic | Spontaneous
(n=60) | IVF (n=61) | ICSI (n=40) | P-value |
|---|
| Male gender, no.
(%) | 35 (58.3) | 39 (63.9) | 19 (47.5) | NS |
| Twins, no. (%) | 4 (6.7) | 26 (42.6) | 20 (50.0) | <0.001 |
| Gestational age at
delivery (week) | 38.8±1.3 | 36.3±2.8a | 37.0±1.6b | <0.001 |
| Preterm delivery
<37 weeks, no. (%) | 3 (5.0) | 24 (39.3) | 13 (32.5) | <0.001 |
| Birth weight
(g) | 3,124±386 | 2,554±631a | 2,785±451b | <0.001 |
| Maternal age
(year) | 28.2±4.3 |
31.7±4.5a,c | 28.9±3.3 | <0.001 |
All 161 children participated in a clinical
follow-up. None of the children (aged between 7 months and 3 years
in the follow-up) had clinical symptoms of any imprinting diseases,
e.g, feeding problems, reduced weight gain, mental retardation,
short stature, absence of speech and paroxysms of laughter.
DNA methylation patterns in children born
after ART and after spontaneous pregnancies
MS-PCR of five DMRs (KvDMR1, SNRPN,
MEST, MEG3, and TNDM) from normal individuals
generated two products from the methylated and unmethylated alleles
of genomic DNA. Regarding XIST, where in females, the DMR is
DNA methylated on one X chromosome whereas the other chromosome is
unmethylated; but in males, who have only one X chromosome, the
single locus is normally methylated, alternatively, only methylated
allele can be detected. All of the 161 children born after ART and
spontaneous pregnancies showed normal methylation patterns in the
six studied DMRs (Fig. 1).
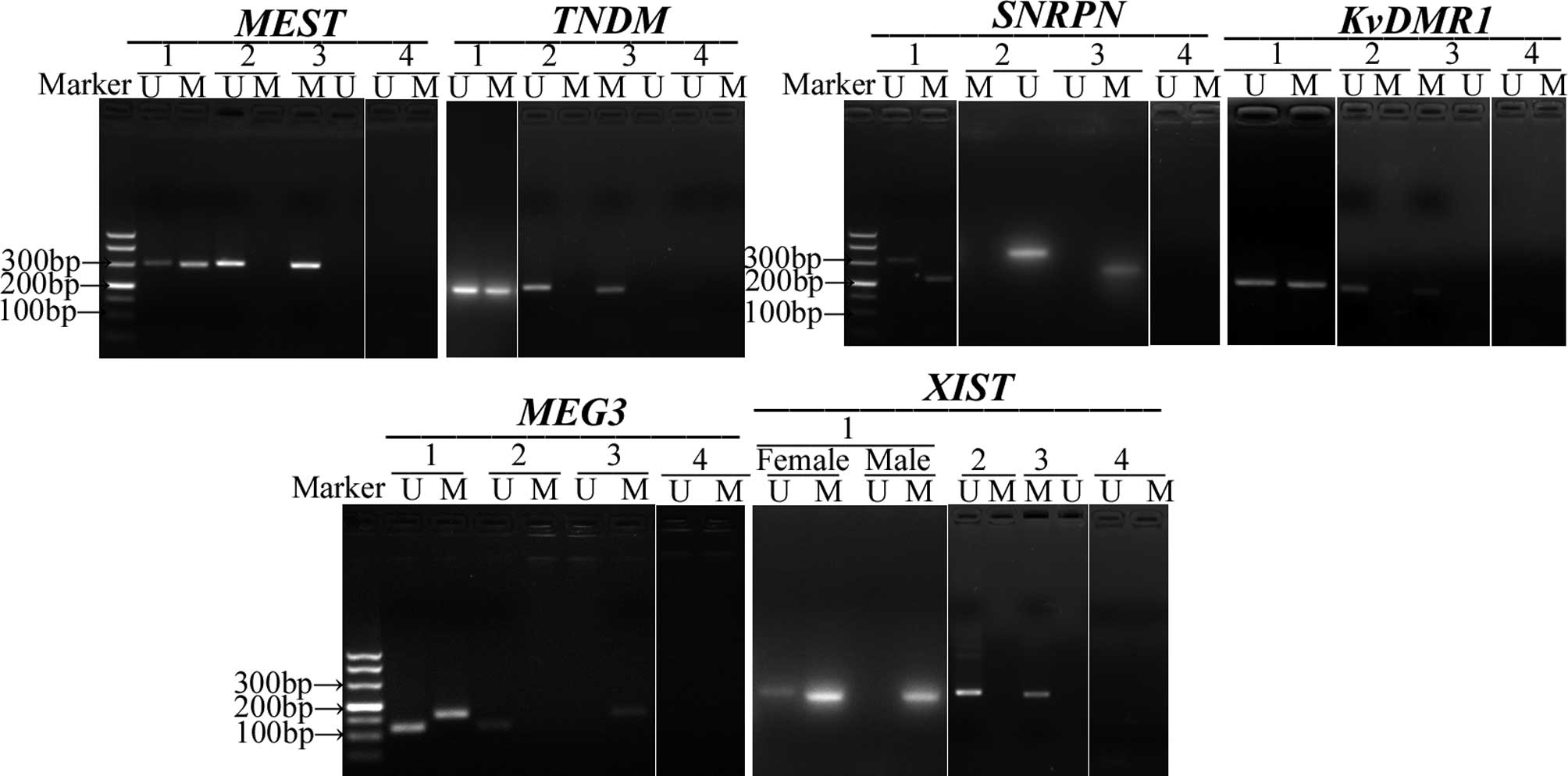 | Figure 1.Representative example of
methylation-specific PCR (MS-PCR) analysis of six DMRs
(MEST, TNDM, SNRPN, KvDMR1,
MEG3, XIST) in umbilical cord blood. Lane 1,
umbilical cord blood from neonates; lane 2, non-methylated DNA
standard was amplified for use as negative control; lane 3,
methylated DNA standard was amplified for use as positive control;
lane 4, untreated DNA. U, unmethylated-allele-specific products; M,
methylated-allele-specific products. |
Discussion
In the present study we found that DNA methylation
patterns in children born after ART were identical to the
methylation patterns in children conceived spontaneously. Our
research indicated a stable methylation status in umbilical cord
blood of children regardless of conception mode.
An important concern for infertile couples is to
produce a healthy baby; thus, the neonates were objectively and
representatively chosen for our study. During gametogenesis and
embryogenesis, existing imprints inherited from previous
generations are erased, and new imprints are established.
Consequently, many studies have focused on the alteration of DNA
methylation patterns in gametes and embryos, which are thought to
be linked to various steps in ART (6,28,29).
However, the number of human oocytes and embryos, to some extent,
were too small to be used to investigate the association between
offspring after ART and imprinting defects. It is not possible to
prove the normal fertilization ability of oocytes when they are
used in empirical studies. That is, normal embryos from patients
were suitable but not available for ethical reasons. It is
plausible to consider that the evaluation of offsprings conceived
by ART is the most direct way to evaluate the epigenetic safety of
ART compared with the study of human gametes and embryos.
In our study, the clinical data revealed that,
compared with the infants conceived spontaneously, infants
conceived by ART were more likely to have low birth weight and to
be born before term, which is mainly due to the high twinning rate
associated with IVF and ICSI (30). Pregnancies by ART have been
reported to be associated with an increased risk for adverse
perinatal outcomes, including low birth weight, preterm birth and
perinatal death. However, a recent study reported that subfertile
women conceiving without ART also appeared to be at an increased
risk of adverse outcomes, which indicates that these risks should
be considered when analyzing the adverse effects of ART (31). In addition, maternal age was higher
in the IVF group, which is mainly attributed to unsuccessful
attempts to conceive spontaneously. Patients who conceived by ICSI
may have begun their treatments earlier due to definite infertility
causes and thus had a similar maternal age compared with the
patients who conceived spontaneously. The advanced age of infertile
couples is also a risk factor for adverse perinatal outcomes
(32). Overall, these differences
between the groups do not indicate an association between ART and
adverse pregnancy outcomes. Moreover, the 3-year clinical follow-up
revealed that none of the studied children had clinical symptoms of
any imprinting disorders. Hansen et al suggested that most
major defects are likely to be detected by 1 year after birth
(33). Accordingly, a 3-year
follow-up was, to some extent, an adequate time period to provide
clinical evidence for evaluating the children studied.
Molecular studies regarding the association between
ART and imprinting errors in humans have been primarily carried out
using gametes and embryos. Thus, data on offsprings are limited,
and the results are inconclusive. Previous studies have revealed
that children conceived by ART do not show a higher degree of
imprint variability and do not have a higher risk for imprinting
disorders (17,34), which were consistent with our
results. Rossignol et al suggested that an epigenetic
imprinting defect of patients with BWS is not restricted to the
11p15 region (KCNQ1OT1 and H19), and the involvement
of other loci (PEG1/MEST, SNRPN) is also not
restricted to patients with BWS (14). Furthermore, several studies have
suggested an association between TNDM and disturbance of
TNDM DMR (26), human
tumors and imprinting defects of MEG3 and XIST
(25,35). Taking these data together, our
multi-gene study on children born after ART was necessary and
reliably assessed the safety of ART.
In the present study, MS-PCR was used to analyze the
methylation patterns of imprinted genes, which is a classic
strategy for investigating DNA methylation. The accuracy and
usefulness of MS-PCR indicates that it can be designed for many
DMRs in the genome (36). Our
results found normal methylation patterns in six DMRs of the
imprinted genes. One related problem of this method includes the
mis-amplification of the original untreated DNA with the primers
that we used. However, due to the high efficiency of DNA
modification with the bisulfite kit, the chance for this error was
negligibly small. Indeed, the untreated DNA was not amplified with
either primer set in our study (Fig.
1, lane 4). Moreover, Kobayashi et al suggested a
difference in the vulnerability of different DMRs to undergo
alteration (37). This difference
was not detected in our study. However, a slight alteration in the
degree of methylation cannot be excluded in our study. Tierling
et al observed small changes in methylation of several DMRs
in umbilical cord blood, but the authors suggested that the
statistical significant differences obtained should be interpreted
with caution (17). In contrast,
subtle methylation changes cannot be neglected since the
accumulation of epigenetic disturbances throughout generations
(38), above a certain threshold,
may lead to obvious aberrant phenotypes.
A study conducted by Doornbos et al found
that an increased incidence of imprinting diseases was associated
with increased fertility problems of the parents but not ART
(39). Particularly, the intrinsic
imprinting defects of spermatozoa appeared to be responsible for
the increased incidence of imprinting disorders (37). However, phenotype changes caused by
epigenetic disturbances may present at a later stage since the
disturbances can be tolerated during development. Meanwhile, an
animal study suggested that imprinting errors in fetal germ cells
can be transmitted to the next generation (40). Actually, the 32-year period
involved in the development of ART is not sufficient to assess the
long-term risks linked to epigenetic defects after ART. In additon,
the analyzed sample size in our study was far too small to
generally exclude rare imprinting disorders. Therefore, a complete
safety evaluation may require studies from a two-generation
perspective with a large population.
In conclusion, our results suggest that ART alone is
not associated with a significant increase in the methylation
variations in imprinted genes in children born after ART. As the
impact of epigenetic disturbances at a later stage in the lifespan
of humans born as a result of ART is not known, long-term clinical
follow-up studies as well as further molecular research of the
children born as a result of assisted reproduction are
recommended.
Acknowledgements
The authors thank all members of the
Research Center of Clinical Medicine of Nanfang Hospital for their
technical supports and valuable suggestions, and the nurse midwives
of Nanfang Hospital for collecting samples for our study. This
study was supported by the National Key Basic Research Development
Plan of China (973 Program) (2007CB948104).
References
|
1.
|
Manipalviratn S, DeCherney A and Segars J:
Imprinting disorders and assisted reproductive technology. Fertil
Steril. 91:305–315. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Odom LN and Segars J: Imprinting disorders
and assisted reproductive technology. Curr Opin Endocrinol Diabetes
Obes. 17:517–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33(Suppl): 245–254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ferguson-Smith AC and Surani MA:
Imprinting and the epigenetic asymmetry between parental genomes.
Science. 293:1086–1089. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Fauque P, Jouannet P, Lesaffre C, Ripoche
MA, Dandolo L, Vaiman D and Jammes H: Assisted Reproductive
Technology affects developmental kinetics, H19 Imprinting Control
Region methylation and H19 gene expression in individual mouse
embryos. BMC Dev Biol. 7:1162007. View Article : Google Scholar
|
|
6.
|
Sato A, Otsu E, Negishi H, Utsunomiya T
and Arima T: Aberrant DNA methylation of imprinted loci in
superovulated oocytes. Hum Reprod. 22:26–35. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shi W and Haaf T: Aberrant methylation
patterns at the two-cell stage as an indicator of early
developmental failure. Mol Reprod Dev. 63:329–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Li T, Vu TH, Ulaner GA, et al: IVF results
in de novo DNA methylation and histone methylation at an igf2-H19
imprinting epigenetic switch. Mol Hum Reprod. 11:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Cox GF, Burger J, Lip V, Mau UA, Sperling
K, Wu BL and Horsthemke B: Intracytoplasmic sperm injection may
increase the risk of imprinting defects. Am J Hum Genet.
71:162–164. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Allen C and Reardon W: Assisted
reproduction technology and defects of genomic imprinting. BJOG.
112:1589–1594. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
DeBaun MR, Niemitz EL and Feinberg AP:
Association of in vitro fertilization with Beckwith-Wiedemann
syndrome and epigenetic alterations of LIT1 and H19. Am J Hum
Genet. 72:156–160. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Maher ER, Brueton LA, Bowdin SC, et al:
Beckwith-Wiedemann syndrome and assisted reproduction technology
(ART). J Med Genet. 40:62–64. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Halliday J, Oke K, Breheny S, Algar E and
Amor DJ: Beckwith-Wiedemann syndrome and IVF: a case-control study.
Am J Hum Genet. 75:526–528. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Rossignol S, Steunou V, Chalas C, et al:
The epigenetic imprinting defect of patients with
Beckwith-Wiedemann syndrome born after assisted reproductive
technology is not restricted to the 11p15 region. J Med Genet.
43:902–907. 2006. View Article : Google Scholar
|
|
15.
|
Lidegaard O, Pinborg A and Andersen AN:
Imprinting diseases and IVF: Danish National IVF cohort study. Hum
Reprod. 20:950–954. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bowdin S, Allen C, Kirby G, et al: A
survey of assisted reproductive technology births and imprinting
disorders. Hum Reprod. 22:3237–3240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tierling S, Souren NY, Gries J, et al:
Assisted reproductive technologies do not enhance the variability
of DNA methylation imprints in human. J Med Genet. 47:371–376.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Gomes MV, Huber J, Ferriani RA, Amaral NA
and Ramos ES: Abnormal methylation at the KvDMR1 imprinting control
region in clinically normal children conceived by assisted
reproductive technologies. Mol Hum Reprod. 15:471–477. 2009.
View Article : Google Scholar
|
|
19.
|
Kubota T, Das S, Christian SL, Baylin SB,
Herman JG and Ledbetter DH: Methylation-specific PCR simplifies
imprinting analysis. Nat Genet. 16:16–17. 1997.PubMed/NCBI
|
|
20.
|
Zeschnigk M, Lich C, Buiting K, Doerfler W
and Horsthemke B: A single-tube PCR test for the diagnosis of
Angelman and Prader-Willi syndrome based on allelic methylation
differences at the SNRPN locus. Eur J Hum Genet. 5:94–98.
1997.PubMed/NCBI
|
|
21.
|
Gomes MV, Gomes CC, Pinto WJ and Ramos ES:
Methylation pattern at the KvDMR in a child with Beckwith-Wiedemann
syndrome conceived by ICSI. Am J Med Genet A. 143:625–629. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kosaki K, Kosaki R, Robinson WP, Craigen
WJ, Shaffer LG, Sato S and Matsuo N: Diagnosis of maternal
uniparental disomy of chromosome 7 with a methylation specific PCR
assay. J Med Genet. 37:E192000. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
White HE, Durston VJ, Harvey JF and Cross
NC: Quantitative analysis of SNRPN(correction of SRNPN) gene
methylation by pyrosequencing as a diagnostic test for Prader-Willi
syndrome and Angelman syndrome. Clin Chem. 52:1005–1013. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Murphy SK, Wylie AA, Coveler KJ, et al:
Epigenetic detection of human chromosome 14 uniparental disomy. Hum
Mutat. 22:92–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Kawakami T, Okamoto K, Ogawa O and Okada
Y: XIST unmethylated DNA fragments in male-derived plasma as a
tumour marker for testicular cancer. Lancet. 363:40–42. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Mackay DJ, Temple IK, Shield JP and
Robinson DO: Bisulphite sequencing of the transient neonatal
diabetes mellitus DMR facilitates a novel diagnostic test but
reveals no methylation anomalies in patients of unknown aetiology.
Hum Genet. 116:255–261. 2005. View Article : Google Scholar
|
|
27.
|
Al HS, Kupker W, Baschat AA, Sturm R,
Bauer O, Diedrich C and Diedrich K: Mini-swim-up: a new technique
of sperm preparation for intracytoplasmic sperm injection. J Assist
Reprod Genet. 12:428–433. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Borghol N, Lornage J, Blachere T, Sophie
GA and Lefevre A: Epigenetic status of the H19 locus in human
oocytes following in vitro maturation. Genomics. 87:417–426. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Chen SL, Shi XY, Zheng HY, Wu FR and Luo
C: Aberrant DNA methylation of imprinted H19 gene in human
preimplantation embryos. Fertil Steril. 94:2356–2358.e1. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ombelet W, De Sutter P, van der Elst J and
Martens G: Multiple gestation and infertility treatment:
registration, reflection and reaction–the Belgian project. Hum
Reprod Update. 11:3–14. 2005.PubMed/NCBI
|
|
31.
|
Jaques AM, Amor DJ, Baker HW, et al:
Adverse obstetric and perinatal outcomes in subfertile women
conceiving without assisted reproductive technologies. Fertil
Steril. 94:2674–2679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Carolan M and Frankowska D: Advanced
maternal age and adverse perinatal outcome: a review of the
evidence. Midwifery 2010 (E-pub ahead of print).
|
|
33.
|
Hansen M, Kurinczuk JJ, Bower C and Webb
S: The risk of major birth defects after intracytoplasmic sperm
injection and in vitro fertilization. N Engl J Med. 346:725–730.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Manning M, Lissens W, Bonduelle M, Camus
M, De Rijcke M, Liebaers I and van Steirteghem A: Study of
DNA-methylation patterns at chromosome 15q11-q13 in children born
after ICSI reveals no imprinting defects. Mol Hum Reprod.
6:1049–1053. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Benetatos L, Dasoula A, Hatzimichael E,
Georgiou I, Syrrou M and Bourantas KL: Promoter hypermethylation of
the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin
Lymphoma Myeloma. 8:171–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Kubota T, Nonoyama S, Tonoki H, et al: A
new assay for the analysis of X-chromosome inactivation based on
methylation-specific PCR. Hum Genet. 104:49–55. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Kobayashi H, Hiura H, John RM, et al: DNA
methylation errors at imprinted loci after assisted conception
originate in the parental sperm. Eur J Hum Genet. 17:1582–1591.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Daxinger L and Whitelaw E:
Transgenerational epigenetic inheritance: more questions than
answers. Genome Res. 20:1623–1628. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Doornbos ME, Maas SM, McDonnell J,
Vermeiden JP and Hennekam RC: Infertility, assisted reproduction
technologies and imprinting disturbances: a Dutch study. Hum
Reprod. 22:2476–2480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Anway MD, Cupp AS, Uzumcu M and Skinner
MK: Epigenetic transgenerational actions of endocrine disruptors
and male fertility. Science. 308:1466–1469. 2005. View Article : Google Scholar : PubMed/NCBI
|















