Introduction
Human sulfotransferases (SULTs) catalyze the
conjugation of sulfate groups to a variety of endogen and exogenous
substrates, including many drugs, neurotransmitters, thyroid and
steroid hormones and pro-carcinogenic agents (1,2).
SULTs are genetically polymorphic and are expressed in a wide
variety of tissues, such as the liver, lung, brain, kidney, and
platelets (3). To date, 13 human
cytosolic SULT isoforms have been identified and grouped as four
major families: SULT1, SULT2, SULT4 and SULT6 (4). The SULT1A1 gene mapped to chromosome
16p12.1-p11.2 encodes four different allozymes:
SULT1A1*1 (wild-type), SULT1A1*2,
SULT1A1*3 and SULT1A1*4. The SULT1A1 enzyme catalyzes
the sulfation of certain carcinogenic and mutagenic compounds
including heterocyclic and aromatic amines, and polycyclic aromatic
hydrocarbons (2). A genetic
polymorphism in exon 7 of the SULT1A1 gene at the nucleotide of 638
(codon 213), results in a substitution of histidine by arginine
(Arg213His). SULT1A1*2 (His213
allele) is associated with less enzymatic activity and thermal
stability compared with the wild-type allele (Arg213
allele) in platelets (5,6).
Prostate cancer, a serious health problem in the
Western world and Turkey, has shown an increasing incidence over
the last decade (7). Some reports
suggest that the risk of prostate cancer development is influenced
by both genetic and environmental factors, such as diet, hormone
levels, drinking habit, ethnicity and genetic background (8). It has been suggested by researchers
that the SULT1A1 Arg213His polymorphism may affect an
individual’s capacity in the metabolism of numerous endogenous and
exogenous compounds consequently resulting in the susceptibility of
an individual to cancer (2).
Studies have demonstrated the relationship between genetic
polymorphisms of SULT1A1 Arg213His and several cancer
types including prostate cancer (9–16).
On the other hand, the findings of these studies remain
controversial. This study investigated, for the first time, the
relationship between the SULT1A1 Arg213His polymorphism
and prostate cancer susceptibility in a Turkish population.
Materials and methods
Study population
The study population consisted of a total of 255
Turkish men (104 cases and 151 controls). The prostate cancer
patients were treated at the Urology Department, Cumhuriyet
University Hospital (Central Anatolia) during the year 2004. The
patients were newly diagnosed and histologically confirmed to have
prostate cancer and were previously untreated (by radiotherapy or
chemotherapy). The prostate cancer patients had elevated serum
levels of prostate-specific antigen (PSA). The controls were
selected randomly from healthy individuals without a history of
cancer and having serum levels of PSA <4 ng/ml. Members of the
study populations were informed in regards to the aim of this
study. During the study period, critical information, such as age
and smoking habit were collected from the members using a
standardized questionnaire. This study was approved by the Ethics
Committee of Cumhuriyet University.
SULT1A1 genotyping
Genomic DNA of the study populations was extracted
from blood leukocytes using the standard phenol-chloroform method
(17). SULT1A1
Arg213His genotypes were determined using polymerase
chain reaction (PCR)-based restriction fragment length polymorphism
(RFLP) assay. The PCR reaction was carried out in a total volume of
25 μl containing ∼100 ng genomic DNA, 200 mM deoxynucleo-tide
triphosphates (dATP, dCTP, dGTP and dTTP), 0.2 m of each SULT1A1
primer (forward, 5′-GGG TCT CTA GGA GAG GTG GC-3′; reverse, 5′-GCT
GTG GTC CAT GAA CTC CT-3′), 1X reaction buffer [75 mM Tris-HCI pH
8.8 at 25°C, 20 mM (NH4)2SO4,
0.01% Tween-20, MBI Fermentas], 1.5 mM MgCl2, and 2
units Taq polymerase (MBI Fermentas) in a thermal cycler (Techne,
UK). PCR conditions consisted of 94° for, 5 min, followed by 35
cycles of 30 sec at 94°C, 30 sec at 62°C, 30 sec at 72°C, with a
final extension step at 72°C for 5 min. Amplification products (333
bp) were observed in a 2% agarose gel. The PCR product (10 μl) was
digested with 10 units of HaeII restriction enzyme (New
England Biolabs, Beverly, MA) overnight. The fragments were
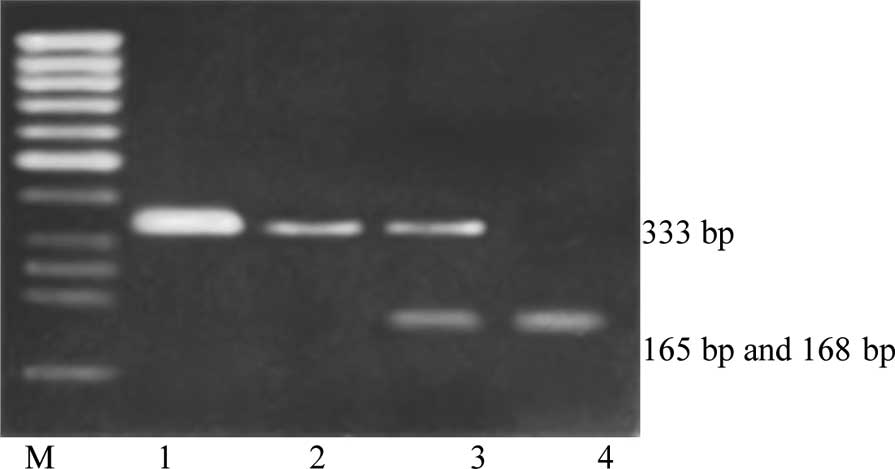
separated in a 2.5% agarose gel. The
SULT1A1*1/SULT1A1*1 (Arg/Arg213)
genotype yielded two distinct digestion products (168 and 165 bp),
the SULT1A1*1/SULT1A1*2
(Arg/His213) genotype yielded three distinct digestion
products (333, 168 and 165 bp), and the
SULT1A1*2/SULT1A1*2 (His/His213)
genotype yielded no digestion products (333 bp) (Fig. 1).
Statistical Package for the Social Sciences (SPSS)
release 10.0.1 software was used to perform the statistical
analyses. Hardy-Weinberg equilibrium, genotype frequencies and
allele frequencies were tested by the Pearson’s χ2 test.
The statistical significance of the differences in SULT1A1
Arg213His genotypes among the cases and controls was
determined by the χ2 test. Probability values <0.05
were regarded as statistically significant. Odds ratios and 95%
confidence intervals (CIs) for prostate cancer were calculated by
using a multivariate logistic regression analysis adjusting several
confounding variables such as age and smoking status.
Results
Demographic characteristics of the cases and
controls are summarized in Table
I. Mean ages of the cases and controls were 65.2±13.2 years
(range, 42–88) and 61±11.4 years (range, 41–82), respectively. No
significant relationship was found between the cases and controls
in terms of smoking status (P=0.83). Mean PSA levels were 3.4±0.5
and 30.6±11.2 ng/ml in the controls and patients, respectively.
 | Table ICharacteristics of the prostate cancer
patients and controls. |
Table I
Characteristics of the prostate cancer
patients and controls.
| Controls | | Patients |
|---|
| Sample size (n) | | | |
| Males | 151 | | 104 |
| Age (year) | | | |
| Range | 41–82 | | 42–88 |
| Mean ± SD | 61±11.4 | | 65.2±13.2 |
| Smoking history, n
(%) | | | |
| Smokers | 79 (52.3) | | 53 (50.9) |
| Non-smokers | 72 (47.7) | | 51 (49.1) |
| χ2 | | 0.045 | |
| P-valuea | | 0.830 | |
| PSA (ng/ml) | 3.4±0.5 | | 30.6±11.2 |
SULT1A1 Arg213His allele and genotype
frequencies are indicated in Table
II. The genotype and allele frequencies were found to be in
Hardy-Weinberg equilibrium. In the cases, the frequencies of the
homozygous wild-type genotypes (Arg/Arg213), the
heterozygous genotype (Arg/His213) and the homozygous
variant genotype (His/His213) were 52.8, 36.6 and 10.6%,
respectively; in the controls, these frequencies were 60.3, 35.8
and 3.9%, respectively.
 | Table IIGenotype and allele frequencies for
SULTA1 locus in cases and controls. |
Table II
Genotype and allele frequencies for
SULTA1 locus in cases and controls.
| Genotype | Controls (n=151) | Cancer patients
(n=104) | P-value | χ2 |
|---|
| Allele frequency, n
(%) | | | 0.072 | 3.23 |
| Arg allele | 236 (78.1) | 148 (71.1) | | |
| His allele | 66 (21.9) | 60 (38.9) | | |
| Genotype frequency, n
(%) | | | 0.099 | 4.62 |
| Arg/Arg | 91 (60.3) | 55 (52.8) | | |
| Arg/His | 54 (35.8) | 38 (36.6) | | |
| His/His | 6 (3.9) | 11 (10.6) | | |
The risk of prostate cancer in individuals carrying
the His213 allele was determined by combining
Arg/His213 and His/His213 genotypes. No
statistically significant difference was found between the cases
and controls in comparison of the genotype combination (P=0.24; OR,
1.36; 95% CI, 0.84–2.25). Concerning the smoker and non-smoker
populations, no significant relationship was evident between the
case and control groups regarding genotypic combinations (P=0.45;
OR, 1.32 and P=0.34; OR, 1.39, respectively) (Table III).
 | Table IIIRisk estimates for SULTA1 genotypes
categorized according to total cases and smoking status. |
Table III
Risk estimates for SULTA1 genotypes
categorized according to total cases and smoking status.
| Variable | Genotype
combinations | Controls n (%) | Cancer patients n
(%) | χ2 | P-value | aOR (95% CI) |
|---|
| Total | Arg/Arg | 91 (60.2) | 55 (52.8) | 1.37 | 0.24 | 1.36
(0.84–2.25) |
| Arg/His +
His/His | 60 (39.8) | 49 (47.2) | | | |
| Smoking status | | | | | | |
| Smokers | Arg/Arg | 44 (29.1) | 26 (25) | 0.56 | 0.45 | 1.32
(0.68–2.65) |
| Arg/His +
His/His | 35 (23.17) | 27 (25.9) | | | |
| Non-smokers | Arg/Arg | 47 (31.12) | 29 (27.8) | 0.89 | 0.34 | 1.39
(0.62–2.91) |
| Arg/His +
His/His | 25 (16.55) | 22 (21.1) | | | |
Discussion
The incidence of prostate cancer displays large
ethnic variations worldwide. While the lowest incidence rate of
prostate cancer is observed for Chinese men, African-American men
have the highest rate of incidence (7). It is believed that advanced age, an
intact androgen metabolism, ethnicity and genetic background are
risk factors for prostate cancer development (8). The majority of studies suggest that
genetic polymorphisms in xenobiotic metabolizing enzymes may play
an important role in the susceptibility of individuals to cancer
(18). In our previous study, we
found that the GSTM1 null genotype may play an important role as a
risk factor for prostate cancer development in the Turkish
population (14).
Allelic frequencies of the His213 allele
differ ranging from 5 to 32% among ethnic populations (5,19,20).
The frequency of the His213 allele was reported to be
18.5 and 22% in a study of a Turkish population (21,22).
In the present study, the frequency of this allele in the control
population was determined to be 21.9% which was higher than the
frequency in Chinese, Taiwanese and Koreans while lower than the
frequency reported for Caucasian and Nigerian populations (5,19,20).
Distributions of the SULT1A1 Arg213His genotypes and
alleles were also not significantly different between the cases and
controls in the present study.
In many studies, a significant relationship has been
demonstrated between the His213 allele and various
cancer types, including gastric, lung, colorectal, and breast
(9,10,13,21).
In addition, a statistical significant association was noted
between the His213 allele and primary brain tumor and
lung cancer incidence in our previous studies (21,22).
However, in the present study, no significant relationship was
determined between the His213 allele and prostate cancer
although His213 allele frequencies were higher in the
patients than in the controls. Another study which is in agreement
with ours was reported by Steiner et al (15). In contrast, Nowell et al
(11) found a positive association
between the Arg213 allele (rapid sulfation allele) and
prostate cancer risk. These controversial results may be due to the
dual role (bioactivation and detoxification) of SULT1A1 in the
metabolism of various carcinogens (23). Although SULT1A1 is considered as a
phase II enzyme, it has been demonstrated that this enzyme acts to
bioactivate various pro-carcinogens and pro-mutagens such as
dietary carcinogen 2-amino-1-methyl-6-phenyl-imidazo (4,5-b)
pyridine which induces prostate tumors in rats (24). Chou et al (25) and Ozawa et al (26) found that SULT1A1 catalyzes the
sulfation of N-hydroxy derivates of arylamines and heterocyclic
amines, to form more reactive DNA adduct-forming compounds. In this
context, differing environmental parameters may also influence the
function of a given allele. Thus, a different genetic background
and different carcinogen exposure may play an important role in the
different risk estimates associated with polymorphisms. In light of
this knowledge, we believe that the influence of the SULT1A1
polymorphism on cancer development risk may differ according to the
type of exposed carcinogen.
In the present study, there was no statistically
significant association between the SULT1A1 Arg213His
polymorphism and smoking status of the study population which is in
accordance with previously published studies (21,22).
In summary, our results suggest that the SULT1A1
Arg213His polymorphism does not play a role in prostate
cancer susceptibility in the Turkish population. However, since
this study is the first report carried out in a Turkish population,
it may contribute to the understanding of the relationship between
the SULT1A1 polymorphism and prostate cancer. In order to elucidate
the role of genetic polymorphisms in carcinogen metabolizing
enzymes in prostate cancer development more accurately,
environmental exposure to specific carcinogens must be investigated
in larger studies.
Acknowledgements
This study was supported by the
Research Council of Cumhuriyet University (CUBAP, project no.
F-217), Sivas, Turkey.
References
|
1
|
Falany CN: Enzymology of human cytosolic
sulfotransferases. FASEB J. 11:206–216. 1997.PubMed/NCBI
|
|
2
|
Glatt H, Engelke CE, Pabel U, Teubner W,
Jones AL, Coughtrie MW, Andrae U, Falany CN and Meinl W:
Sulfotransferases: genetics and role in toxicology. Toxicol Lett.
112:341–348. 2000. View Article : Google Scholar
|
|
3
|
Richard K, Hume R, Kaptein E, Stanley EL,
Visser TJ and Coughtrie MW: Sulfation of thyroid hormone and
dopamine during human development: ontogeny of phenol
sulfotransferases and arylsulfatase in liver, lung, and brain. J
Clin Endocrinol Metab. 86:2734–2742. 2001.
|
|
4
|
Blanchard RL, Freimuth RR, Buck J,
Weinshilboum RM and Coughtrie MW: A proposed nomenclature system
for the cytosolic sulfotransferase (SULT) superfamily.
Pharmacogenetics. 14:199–211. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Raftogianis RB, Wood TC, Otterness DM, Van
Loon JA and Weinshilboum RM: Phenol sulfotransferase
pharmacogenetics in human: association of common SULT1A1 alleles
with TS PST phenotype. Biochem Biophys Res Commun. 239:298–304.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nowell S, Ambrosone CB, Ozawa S, MacLeod
SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF and Lang NP:
Relationship of phenol sulfotransferase activity (SULT1A1) genotype
to sulfotransferase phenotype in platelet cytosol.
Pharmacogenetics. 10:789–797. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
National Cancer Institute: Annual Report
to the Nation Finds Declines in Cancer Incidence and Death Rates;
Special Feature Reveals Wide Variations in Lung Cancer Trends
across States. https://www.cancer.gov/newscenter/pressreleases/2008/reportnation2008release.
|
|
8
|
Dearnaley DP: Cancer of the prostate. BMJ.
308:780–784. 1994.Erratum in: BMJ 308: 975, 1994.
|
|
9
|
Bamber DE, Fryer AA, Strange RC, Elder JB,
Deakin M, Rajagopal R, Fawole A, Gilissen RA, Campbell FC and
Coughtrie MW: Phenol sulfotransferase SUL1A1*1 genotype
is associated with reduced risk of colorectal cancer.
Pharmacogenetics. 11:679–685. 2001.
|
|
10
|
Boccia S, Persiani R, La Torre G, Rausei
S, Arzani D, Gianfagna F, Romano-Spica V, D’Ugo D and Ricciardi G:
Sulfotransferase 1A1 polymorphism and gastric cancer risk: a pilot
case-control study. Cancer Lett. 229:235–243. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nowell S, Ratnasinghe DL, Ambrosone CB,
Williams S, Teague-Ross T, Trimble L, Runnels G, Carrol A, Green B,
Stone A, Johnson D, Greene G, Kadlubar FF and Lang NP: Association
of SULT1A1 phenotype and genotype with prostate cancer risk in
African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev.
13:270–276. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng CT, Chen JC, Yeh KT, Wang YF, Hou MF,
Lee TP, Shih MC, Chang JY and Chang JG: The relationship among the
polymorphisms of SULT1A1, 1A2 and different types of cancers in
Taiwanese. Int J Mol Med. 11:85–89. 2003.PubMed/NCBI
|
|
13
|
Seth P, Lunetta KL, Bell DW, Gray H,
Nasser SM, Rhei E, Kaelin CM, Iglehart DJ, Marks JR, Garber JE,
Haber DA and Polyak K: Phenol sulfotransferases: hormonal
regulation, polymorphism, and age of onset of breast cancer. Cancer
Res. 60:6859–6863. 2000.PubMed/NCBI
|
|
14
|
Silig Y, Pinarbası H, Güneş S, Ayan S,
Bagci H and Çetinkaya Ö: Polymorphisms of CYP1A1, GSTM1, GSTT1, and
prostate cancer risk in Turkish population. Cancer Invest.
24:41–45. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steiner M, Bastian M, Schulz WA, Pulte T,
Franke KH, Röhring A, Wolff JM, Seiter H and Schuff-Werner P:
Phenol sulphotransferase SULT1A1 polymorphism in prostate cancer:
lack of association. Arch Toxicol. 74:222–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng W, Xie D, Cerhan JR, Sellers TA, Wen
W and Folsom AR: Sulfotransferase 1A1 polymorphism, endogenous
estrogen exposure, well-done meat-intake, and breast cancer risk.
Cancer Epidemiol Biomarkers Prev. 10:89–94. 2001.PubMed/NCBI
|
|
17
|
Sambrook J, Fritsch E and Maniatis T:
Molecular Cloning A Laboratory Manual. 2nd edition. Cold Spring
Harbor Laboratory Press; Plainview: 1989
|
|
18
|
Wormhoudt LW, Commandeur JN and Vermeulen
NP: Genetic polymorphisms of human N-acetyltransferase, cytochrome
P450, glutathione-S-transferase, and epoxide hydrolase enzymes:
relevance to xenobiotic metabolism and toxicity. Crit Rev Toxicol.
29:59–124. 1999. View Article : Google Scholar
|
|
19
|
Coughtrie MW, Gilissen RA, Shek B, Strange
RC, Fryer AA, Jones PW and Bamber DE: Phenol sulphotransferase
SULT1A1 polymorphism: molecular diagnosis and allele frequencies in
Caucasian and African populations. Biochem J. 337:45–49. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arslan S, Siliğ Y and Pınarbaşı H: An
investigation of the relationship between SULT1A1
Arg213His polymorphism and lung cancer susceptibility in
a Turkish population. Cell Biochem Funct. 27:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim KA, Lee SY, Park PW, Ha JM and Park
JY: Genetic polymorphisms and linkage disequilibrium of
sulfotransferase SULT1A1 and SULT1A2 in a Korean population:
comparison of other ethnic groups. Eur J Clin Pharmacol.
61:743–747. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bardakci F, Arslan S, Bardakci S, Binatli
AO and Budak M: Sulfotransferase 1A1 (SULT1A1) polymorphism and
susceptibility to primary brain tumors. J Cancer Res Clin Oncol.
134:109–114. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Banoglu E: Current status of the cytosolic
sulfotransferases in the metabolic activation of promutagens and
procarcinogens. Curr Drug Metab. 1:1–30. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shirai T, Tamano S, Sano M, Masui T,
Hasegawa R and Ito N: Carcinogenicity of
2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP) in rats:
dose-response studies. Princess Takamatsu Symp. 23:232–239.
1995.
|
|
25
|
Chou HC, Lang NP and Kadlubar FF:
Metabolic activation of the N-hydroxy derivative of the carcinogen
4-aminobiphenyl by human tissue sulfotransferases. Carcinogenesis.
16:413–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ozawa S, Chou HC, Kadlubar FF, Nagata K,
Yamazoe Y and Kato R: Activation of
2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b] pyridine by
cDNA-expressed human and rat arylsulfotransferases. Jpn J Cancer
Res. 85:1220–1228. 1994.
|