Introduction
Large-cell neuroendocrine carcinomas (LCNEC) of the
lung are aggressive tumors exhibiting features of high-grade
neuroendocrine tumors, a poor clinical prognosis and a biological
behavior similar to that of small-cell lung carcinomas (SCLCs)
(1–3). However, the treatment of patients
with LCNEC has traditionally been based on that of non-small cell
lung carcinomas (NSCLCs). To improve the prognosis of patients with
LCNEC, understanding the clinicopathological features and the
effectiveness of treatment in patients with LCNEC is imperative.
Recent reports have revealed the effectiveness of
molecular-targeted therapy in patients with lung adenocarcinoma
(AC) (4,5). Epidermal growth factor receptor
(EGFR)-tyrosine kinase inhibitor (TKI) has been effective for
NSCLC, particularly in patients with lung AC and specific EGFR
mutations in exon 19 or exon 21, or Asian patients (4). The EGFR mutation status can be used
as a good predictor of the clinical benefit of EGFR-TKI (6). On the other hand, angiogenesis is
also one of the hallmarks of cancer (7). Vascular endothelial growth factor
(VEGF) is an important regulator of angiogenesis, and inhibition of
VEGF has demonstrated survival benefits for patients with
colorectal cancers (8). The
expression of VEGF may also be related to prognosis in NSCLC
(9), with monoclonal anti-VEGF
antibody therapy showing a potential survival benefit in these
patients (10).
Overexpression of the human epidermal growth factor
receptor 2 (HER2) tyrosine kinase, usually due to
over-amplification, has been linked to poor prognosis in breast
cancer (11). Treatment with
recombinant monoclonal anti-HER2 antibody (trastuzumab) displays
activity in patients with breast cancer, particularly with high
HER2 expression. A type III tyrosine kinase (KIT) is encoded by
proto-oncogene c-kit, and exhibits high expression rates in
gastrointestinal stromal tumors (12). The small-molecule tyrosine-kinase
inhibitor imatinib is active against the intracellular ABL kinase,
the chimeric BCR-ABL fusion oncoprotein of chronic myeloid
leukemia, the transmembrane receptor KIT and platelet-derived
growth factor receptors. Exposure of tumor cells dependent on the
KIT pathway to imatinib blocks the kinase activity of KIT, arrests
proliferation and causes apoptotic cell death (13). Imatinib has shown a high responsive
rate in gastrointestinal stromal tumors, a tumor otherwise
refractory to conventional chemoradiation therapy (13,14).
Little is currently known regarding expression of
these markers in LCNEC, or the use of their corresponding
target-specific therapies in patients with these tumors. In this
study, we report the gene expression and mutation analyses of
LCNEC, and discuss the possibility of molecular-targeted therapy in
the management of patients with LCNEC.
Patients and methods
We analyzed 13 tumors resected at the Kitasato
University Hospital and diagnosed as LCNEC according to the WHO
classification (1).
Immunohistochemical (IHC) staining was performed using a polyclonal
anti-chromogranin A antibody (Dako, Glostrup, Denmark), a
polyclonal anti-synaptophysin antibody (Dako) and an anti-neural
cell adhesion molecule (NCAM) antibody (Nippon Kayaku, Tokyo,
Japan). Neuroendocrine differentiation was identified by positive
IHC staining for chromogranin A, synaptophysin or NCAM (1).
In cases of combined LCNEC, the histological
components of LCNEC were selected according to microscopic findings
to perform analyses of IHC expression, gene expression or gene
mutation. Additionally, we analyzed 14 consecutive patients with
ACs as a comparative cohort. The Institutional Review Board of the
Kitasato University Hospital approved the protocols and procedures
(B08-32).
Expression and gene mutations of
molecular markers
IHC analyses. IHC analyses of c-KIT, HER2 and
VEGF were performed with anti-c-KIT (Dako, Tokyo, Japan), anti-HER2
(Dako) and anti-VEGF antibodies (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA). The expression of HER2 was evaluated
acccording to a previous study (15). Overexpression was indicated by a
score of 3+ or 2+, and weak expression by a score of 1+.
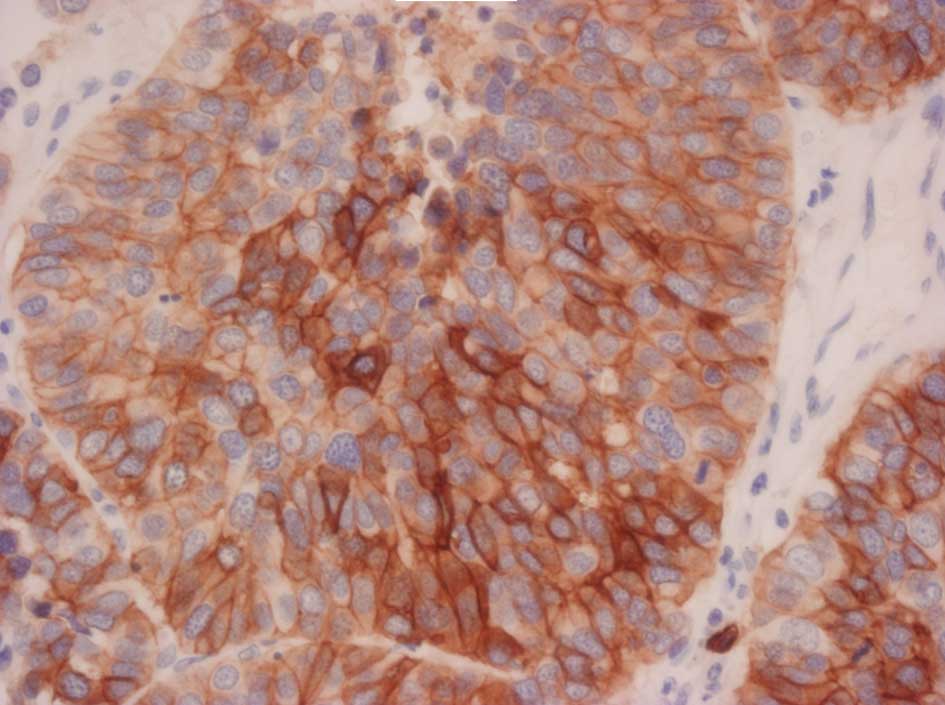
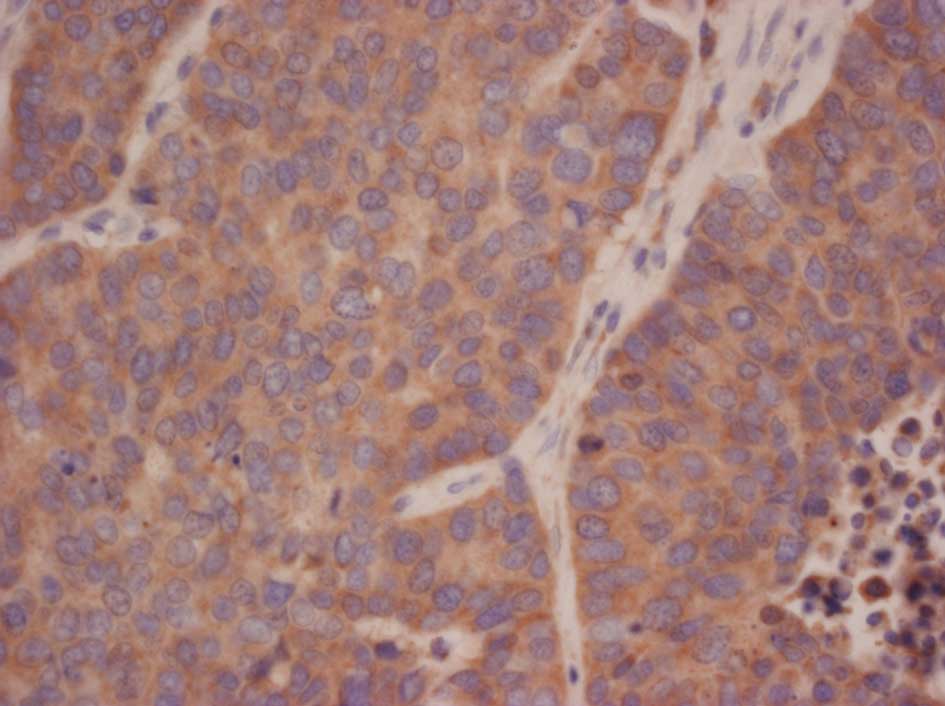
Representative stains are shown in Figs. 1–3.
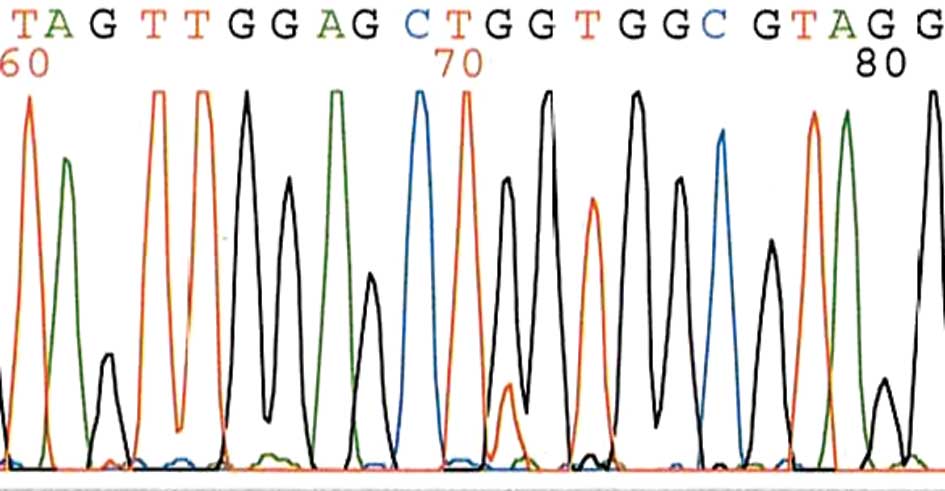
EGFR gene mutation analysis. H&E-stained
sections of formalin-fixed paraffin-embedded tissues were reviewed
to identify regions of tissue composed of tumor cells. Genomic DNA
was isolated using the QIAamp DNA Mini kit (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. DNA
extraction was identical for all mutation analyses. Exon sequences
for EGFR (kinase domain) were amplified with specific primers by
polymerase chain reaction (PCR). Molecularly pre-typed samples were
selected based on results from direct DNA sequencing of EGFR (exons
18, 19, 20 and 21) (Fig. 4).
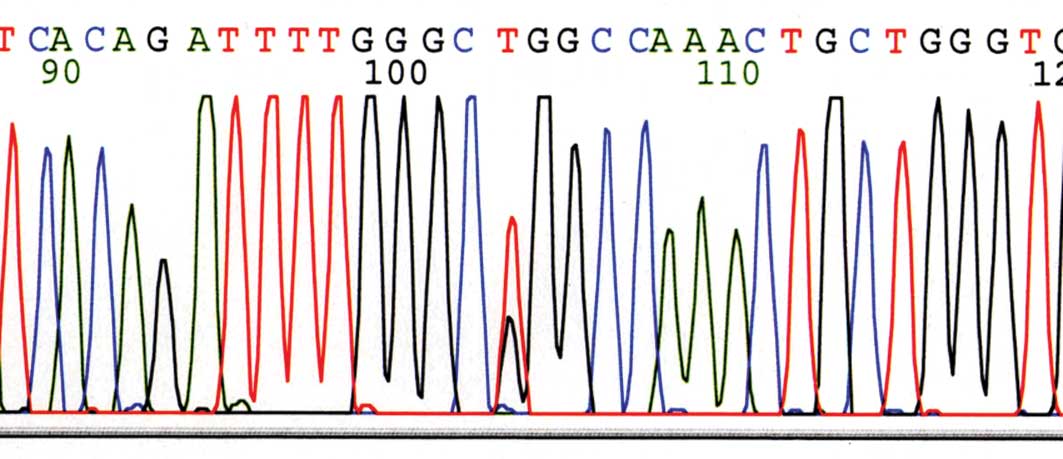
K-ras gene mutation analysis. After DNA
isolation, exon 2 of the K-ras gene was amplified by PCR using
TaKaRa Ex Taq Hot Start Version (Takara Bio Inc., Otsu, Japan) with
forward primer 5′-GTGTGACATGTTCTAATATAGTCA-3′ and reverse primer
5′-GTCCTGCACCAGTAATATGC-3′. The expected products of 209 base pairs
were sequenced bidirectionally using the BigDye Terminator Cycle
Sequencing kit (version 1.1; Applied Biosystems) and an ABI Genetic
Analyzer (model 3100; Applied Biosystems) (Fig. 5).
c-kit gene mutation analysis. Exons 9, 11, 13
and 17 of the c-kit gene were amplified by PCR using the following
oligonucleotide primer pairs: for exon 9, 5′-TCCTAGAGTAAGCCAGGG
CTT-3′/5′-TGGTAGACAGAGCCTAAACATCC-3′; for exon 11,
5′-GATCTATTTTTCCCTTTCTC-3′/5′-AGCCCC TGTTTCATACTGAC-3′; for exon
13, 5′-GCT TGA CAT CAG TTT GCC AG-3′/5′-AAA GGC AGC TTG GAC ACG GCT
TTA-3′; for exon 17, 5′-CTCCTCCAACCTAATAG
TGT-3′/5′-GTCAAGCAGAGAATGGGTAC-3′. The expected products were
sequenced bidirectionally using the BigDye Terminator Cycle
Sequencing kit (Applied Biosystems) and an ABI genetic analyzer
(model 3100; Applied Biosystems) (16).
EGFR gene copy number analysis by FISH. EGFR
gene copy number was evaluated by FISH using the LSI EGFR Dual
Color probe (Abbott Molecular, Des Plaines, IL, USA) hybridized to
the band region 7p12 in Spectrum Orange (EGFR DNA probe) and the
centromere of chromosome 7 (7p11.1-q11.1, D7Z1 locus) in Spectrum
Green (CEP7 DNA probe). We calculated >60 tumor cells, and
evaluated a gene-to-chromosome ratio per cell. The cut-off value
was 2.0, and we estimated that tumor samples had a high EGFR gene
copy number if a gene-to-chromosome ratio per cell was ≥2.0
(Fig. 6).
Statistical analysis
Fisher’s exact test was used to compare binomial
proportions. The χ2 test was used to assess differences
in gender, pathological stage, smoking status, IHC positivity for
c-KIT, HER2 and VEGF, gene mutations for K-ras and EGFR, and FISH
expression of the EGFR gene. The unpaired t-test was used to detect
significant differences in patient age. A p-value <0.05 was
considered statistically significant.
Results
There was no significant difference between patients
with LCNEC and AC in relation to age, gender and pathological stage
(Table I). There was a significant
difference in smoking status (Table
I).
 | Table I.Patients characteristics of 13 cases
of LCNEC and 14 cases of adenocarcinoma. |
Table I.
Patients characteristics of 13 cases
of LCNEC and 14 cases of adenocarcinoma.
| Factor | LCNEC (n=13) | Adenocarcinoma
(n=14) | p-value |
|---|
| Age | | | |
| Mean | 61.9 | 60.6 | 0.7356 |
| SD | 8.4 | 11.7 | |
| Gender | | | |
| Male | 11 (84.6%) | 7 (50.0%) | 0.1032 |
| Female | 2 (15.4%) | 7 (50.0%) | |
| Pathological
stage | | | |
| I | 7 (53.8%) | 8 (57.1%) | 0.2091 |
| II | 0 (0.0%) | 3 (21.4%) | |
| III | 5 (38.5%) | 3 (21.4%) | |
| IV | 1 (7.7%) | 0 (0.0%) | |
| Smoking | | | |
| Smoker | 13 (100%) | 7 (50.0%) | 0.0058 |
| Non-smoker | 0 (0.0%) | 7 (50.0%) | |
Differential expression of study markers between
LCNEC and AC are summarized in Table
II. Markers with statistically significant differences in
expression included c-KIT (p=0.0018) and HER2 (p=0.0037). The
frequency of EGFR gene mutations was higher in the AC group
(p=0.0128), with only a single EGFR mutation [(a silent mutation in
codon 725 (ACG to ACA) of exon 18)] identified in the LCNEC group.
Only 2 cases of LCNEC had overexpression of HER2, and no cases of
AC had overexpression of HER2.
 | Table II.Expression of molecular markers in all
patients. |
Table II.
Expression of molecular markers in all
patients.
| Molecular marker | Positive cases (%) of
LCNEC | Positive cases (%) of
adenocarcinoma | p-value |
|---|
| c-KIT | 10 (76.9) | 2 (14.3) | 0.0018 |
| HER2 | 2 (15.4)
overexpression/2 (15.4) weak | 0 (0)
overexpression/11 (78.6) weak | 0.0037 |
| VEGF | 13 (100.0) | 13 (92.9) | >0.9999 |
| EGFR mutation | 1 (7.7) | 8 (57.1) | 0.0128 |
| K-ras mutation | 0 (0.0) | 2 (14.3) | 0.4815 |
| EGFR FISH | 0 (0.0) | 2 (14.3) | 0.4815 |
For the LCNEC cases exhibiting expression of c-KIT,
we analyzed gene mutations of c-kit. However, no case with the
c-kit gene mutation was noted.
Discussion
Previous studies have reported poor prognosis for
patients with LCNEC, with 5-year survival rates ranging from 15 to
57% (2,3). Tumor recurrence is frequent and
problematic, and even patients with stage I disease show 5-year
survival rates of only 27–67%.
The role of adjuvant therapy in this disease is not
well understood. Studies of adjuvant therapy in LCNEC have been
few, with small series of patients, although some have shown
satisfactory response to chemotherapy (17).
Although molecular-targeted therapies for NSCLC,
especially lung AC, are increasingly investigated, there are no
current reports of such therapies in LCNEC. This may be partly due
to the fact that LCNEC was only recently officially recognized by
the World Health Organization in 1999, and because of the rarity of
this tumor. The purpose of the present study was to examine the
expression of some of these known molecular targets of therapy in
LCNEC.
EGFR mutations are frequently observed in lung AC.
In the present study, we confirmed EGFR mutations in 57.1% of ACs,
but in only 1 [a silent mutation in codon 725 (ACG to ACA) of exon
18] of the 13 examined cases of LCNEC. Similarly, although previous
reports have cited high EGFR copy number in ACs as predictive of
response to EGFR-TKI therapy, no EGFR copy number expansion
occurred in our LCNEC cohort as detected by FISH (18). Given these low rates of EGFR
mutations (noted in exon 18) and normal gene copy number, we
believe that EGFR-TKI is not likely to be an effective therapy for
patients with LCNEC.
Previous studies have shown a survival benefit with
the use of anti-angiogenic factors in NSCLC (10). In this study, LCNEC tumors
frequently exhibited VEGF expression when compared to AC,
suggesting a possible role for anti-VEGF therapy in the treatment
of LCNEC. Further studies are warranted to investigate anti-VEGF
therapies for this disease.
The effect of imatinib is largely predicted by the
presence of activating c-kit mutations (19). While we identified high expression
rates of c-KIT in patients with LCNEC compared to ACs, no c-kit
mutations were identified in our LCNEC cohort. Given that c-KIT
expression does not appear to correlate with the presence of an
activating c-kit mutation, further investigation is required to
determine what effectiveness KIT inhibitors may have in LCNEC.
In patients with breast carcinoma, the
overexpression of HER2 is an important predictor for the response
of trastuzumab (11). In this
study, lung ACs had a significantly higher rate of HER2 IHC
staining than LCNEC, with only 2 LCNEC cases showing overexpression
of HER2. Although the rates of overexpression of HER2 in LCNEC were
not high, potentially the small percentage of patients with tumors
showing strong HER2 expression may benefit by treatment with
trastuzumab.
In summary, we carried out a preliminary molecular
screening of putative targets of therapy for LCNEC. High expression
rates of VEGF, comparable to that observed in lung ACs, support the
further assessment of anti-VEGF therapies in these patients;
studies concerning anti-VEGF therapies in LCNEC patients have not
been reported to date. Likewise, strong expression of HER2 and
c-KIT in a subset of these patients suggests possible roles for
targeted therapies, such as trastuzumab and imatinib. Additional
analyses are warranted to better elucidate treatment responses to
imatinib in c-KIT-positive patients, even in the absence of a c-kit
mutation. Finally, the infrequent presence of an EGFR mutation and
increased gene copy number suggest a minimal role of EGFR-TKI
therapy for LCNEC patients.
Acknowledgements
The authors thank Kenji Nezu, Fumihiro
Ogawa, Yoshio Matsui and Naomi Kurouzu for the assistance in the
preparation of the manuscript. This study was supported in part by
a Grant-in-aid for Scientific research (C) 21591822 of the Japanese
Ministry of Education, Culture, Sports, Science, and
Technology.
References
|
1
|
Travis WD, Colby TV, Corrin B, Shimosato Y
and Brambilla E: Histological Typing of Lung and Pleural Tumours.
World Health Organization International Histological Classification
of Tumors, XIII; 3rd edition. Springer-Verlag; Berlin/Heidelberg:
1999, View Article : Google Scholar
|
|
2
|
Iyoda A, Hiroshima K, Nakatani Y and
Fujisawa T: Pulmonary large cell neuroendocrine carcinoma: its
place in the spectrum of pulmonary carcinoma. Ann Thorac Surg.
84:702–707. 2007.PubMed/NCBI
|
|
3
|
Iyoda A, Hiroshima K, Toyozaki T, Haga Y,
Fujisawa T and Ohwada H: Clinical characterization of pulmonary
large cell neuroendocrine carcinoma and large cell carcinoma with
neuroendocrine morphology. Cancer. 91:1992–2000. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takano T, Fukui T, Ohe Y, Tsuta K,
Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Furuta K
and Tamura T: EGFR mutations predict survival benefit from
gefitinib in patients with advanced lung adenocarcinoma: a
historical comparison of patients treated before and after
gefitinib approval in Japan. J Clin Oncol. 26:5589–5595. 2008.
View Article : Google Scholar
|
|
5
|
Zhu CQ, da Cunha Santos G, Ding K,
Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire
JA, Kamel-Reid S, Seymour L, Shepherd FA and Tsao MS; National
Cancer Institute of Canada Clinical Trials Group Study BR.21: Role
of KRAS and EGFR as biomarkers of response to erlotinib in National
Cancer Institute of Canada Clinical Trials Group Study BR.21. J
Clin Oncol. 26:4268–4275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitsudomi T, Kosaka T, Endoh H, Horio Y,
Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T and Yatabe Y:
Mutations of the epidermal growth factor receptor gene predict
prolonged survival after gefitinib treatment in patients with
non-small-cell lung cancer with postoperative recurrence. J Clin
Oncol. 23:2513–2520. 2005. View Article : Google Scholar
|
|
7
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
8
|
Hurwitz H, Fehrenbacher L, Novotny W,
Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S,
Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R and Kabbinavar F:
Bevacizumab plus irinotecan, fluorouracil, and leucovorin for
metastatic colorectal cancer. N Engl J Med. 350:2335–2342. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seto T, Higashiyama M, Funai H, Imamura F,
Uematsu K, Seki N, Eguchi K, Yamanaka T and Ichinose Y: Prognostic
value of expression of vascular endothelial growth factor and its
flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung
Cancer. 53:91–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sandler A, Gray R, Perry MC, Brahmer J,
Schiller JH, Dowlati A, Lilenbaum R and Johnson DH:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lennon S, Barton C, Banken L, Gianni L,
Marty M, Baselga J and Leyland-Jones B: Utility of serum HER2
extracellular domain assessment in clinical decision making: pooled
analysis of four trials of trastuzumab in metastatic breast cancer.
J Clin Oncol. 27:1685–1693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirota S, Isozaki K, Moriyama Y, Hashimoto
K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M,
Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y and Kitamura
Y: Gain-of-function mutations of c-kit in human gastrointestinal
stromal tumors. Science. 279:577–580. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Demetri GD, von Mehren M, Blanke CD, van
den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA,
Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL,
Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless
C, Fletcher CD and Joensuu H: Efficacy and safety of imatinib
mesylate in advanced gastrointestinal stromal tumors. N Engl J Med.
347:472–480. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Verweij J, Casali PG, Zalcberg J, LeCesne
A, Reichardt P, Blay JY, Issels R, van Oosterom A, Hogendoorn PC,
van Glabbeke M, Bertulli R and Judson I: Progression-free survival
in gastrointestinal stromal tumours with high-dose imatinib:
randomised trial. Lancet. 364:1127–1134. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsuda H, Akiyama F, Terasaki H, Hasegawa
T, Kurosumi M, Shimadzu M, Yamamori S and Sakamoto G: Detection of
HER-2/neu (c-erb B-2) DNA amplification in primary breast
carcinoma. Interobserver reproducibility and correlation with
immunohistochemical HER-2 overexpression. Cancer. 92:2965–2974.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirota S, Nishida T, Isozaki K, Taniguchi
M, Nakamura J, Okazaki T and Kitamura Y: Gain-of-function mutation
at the extracellular domain of KIT in gastrointestinal stromal
tumours. J Pathol. 193:505–510. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamazaki S, Sekine I, Matsuno Y, Takei H,
Yamamoto N, Kunitoh H, Ohe Y, Tamura T, Kodama T, Asamura H,
Tsuchiya R and Saijo N: Clinical responses of large cell
neuroendocrine carcinoma of the lung to cisplatin-based
chemotherapy. Lung Cancer. 49:217–223. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hirsch FR, Herbst RS, Olsen C, Chansky K,
Crowley J, Kelly K, Franklin WA, Bunn PA Jr, Varella-Garcia M and
Gandara DR: Increased EGFR gene copy number detected by fluorescent
in situ hybridization predicts outcome in non-small-cell lung
cancer patients treated with cetuximab and chemotherapy. J Clin
Oncol. 26:3351–3357. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heinrich MC, Owzar K, Corless CL, Hollis
D, Borden EC, Fletcher CD, Ryan CW, von Mehren M, Blanke CD, Rankin
C, Benjamin RS, Bramwell VH, Demetri GD, Bertagnolli MM and
Fletcher JA: Correlation of kinase genotype and clinical outcome in
the North American Intergroup Phase III Trial of imatinib mesylate
for treatment of advanced gastrointestinal stromal tumor: CALGB
150105 Study by Cancer and Leukemia Group B and Southwest Oncology
Group. J Clin Oncol. 26:5360–5367. 2008.
|