Introduction
Gastric cancer is one of the most common malignant
tumors of the digestive system, and its early diagnosis rate is
still low in China, where gastric cancer is typically diagnosed at
a more advanced stage; metastasis is typically present at the time
of diagnosis. However, conventional strategies based on radical
surgery for the treatment of gastric cancer are not yet
satisfactory. Therefore, investigation of the mechanisms of
metastasis and recurrence of gastric cancer, and exploration of the
methods for the effective prediction, prevention and treatment of
tumor metastasis are attracting increased attention in both basic
and clinical cancer research.
microRNAs (miRNAs), discovered in 1993, are a class
of non-coding RNAs 19–25 nt in length that regulate gene expression
at the post-transcriptional level (1,2).
These miRNAs regulate complicated cellular processes such as
differentiation, proliferation and apoptosis (3) and are widely involved in
tumorigenesis, tumor progression and metastasis (4,5).
Recent research suggests that miRNAs are involved in tumor invasion
and metastasis, in the context of breast cancer, colorectal cancer,
esophageal squamous cell carcinoma, and pancreatic endocrine tumors
(6,7). Few studies have reported the role of
miRNAs in gastric cancer owing to its relatively low incidence in
Europe and America. Therefore, profiling should be performed in
different types of gastric cancer to identify differentially
expressed miRNAs. In the present study, we investigated differences
in miRNA profiling between primary gastric cancer and paired lymph
node metastasis, and then detected the differentially expressed
miRNAs in 4 cell lines, 33 gastric cancer specimens and
corresponding adjacent gastric mucosa tissue, in an effort to
identify the miRNAs involved in gastric cancer lymph node
metastasis. Furthermore, our data provide evidence regarding the
effects of miRNAs on metastasis involved in the molecular
pathogenesis of gastric cancer.
Materials and methods
Specimens
Specimens used in the miRNA array
Cases of full-thickness gastric wall with invasion
and lymph nodes with tumor metastasis were assessed
intraoperatively; specimens from the primary tumor were retained.
The specimens were embedded in optimal cutting temperature compound
(OCT) and stored at −80°C until use. Experimental samples were
selected to pathologically confirm that invasive adenocarcinoma of
the stomach lesions was of full thickness and to verify the
existence of lymph node metastasis. In total, 5 pairs of specimens
(each of which included primary tumor and paired lymph node
metastasis tissues) were selected, and their clinicopathological
characteristics are summarized in Table I.
 | Table I.Data from the gastric cancer
patients. |
Table I.
Data from the gastric cancer
patients.
| No. | Gender | Age | Tumor location | Type |
Differentiation | Ta | Na | Others |
|---|
| 1 | Male | 67 | Lower 1/3 | Adenocarcinoma | II | 3 | 3 | Lymphatic
invasion |
| 2 | Male | 54 | Mid 1/3 | Adenocarcinoma | II–III | 3 | 1 | Lymphatic
invasion |
| 3 | Male | 62 | Lower 1/3 | Adenocarcinoma | II | 3 | 2 | Unknown |
| 4 | Male | 52 | Lower 1/3 | Adenocarcinoma | III | 3 | 1 | Neural
invasion |
| 5 | Male | 76 | Lower 1/3 | Adenocarcinoma | III | 3 | 3 | Neural
invasion |
Specimens used for validation
Surgical specimens were obtained from 33 randomly
selected patients with primary tumors among those who underwent D2
radical resection of gastric cancer at ZhongShan Hospital, Fudan
University (Shanghai, China) from May, 2006 to December, 2007. All
eligible cases had not received preoperative tumor-related
treatment, were pathologically confirmed as gastric adenocarcinoma,
and displayed full-thickness gastric wall invasion.
Clinicopathological statistical data are shown in Table IV. Another 10 cases were randomly
selected from the 33 cases mentioned above; normal gastric mucosa
tissues were taken from the tumor marginal zone (>5 cm from the
tumor). These samples were mixed in proper proportion after RNA
extraction and used as the control group (indicated below with an
N). Random sampling was performed with SPSS software. Ethical
approval was obtained from the ZhongShan Hospital Research Ethics
Committee.
 | Table IV.Clinicopathological characteristics
of the 33 cases of gastric cancer. |
Table IV.
Clinicopathological characteristics
of the 33 cases of gastric cancer.
| | P-value
|
|---|
| Variable | Cases, n (%) | miR-10a | miR-510 |
|---|
| Gender | | 0.861 | 0.918 |
| Male | 28 (84.8) | | |
| Female | 5 (15.2) | | |
| Age (year) | | 0.874 | 0.280 |
| <60 | 9 (27.3) | | |
| ≥60 | 24 (72.7) | | |
| Types | | 0.978 | 0.691 |
|
Adenocarcinoma | 23 (69.7) | | |
| Tubular
adenocarcinoma | 8 (24.2) | | |
| Signet ring cell
carcinoma | 2 (6.1) | | |
|
Differentiation | | 0.979 | 0.897 |
| Moderately
differentiated (II) | 10 (30.3) | | |
| Poorly
differentiated (III) | 21 (63.6) | | |
| No
differentiation (IV) | 2 (6.1) | | |
| Lymphatic
invasion | | 0.169 | 0.365 |
| Yes | 18 (54.4) | | |
| No | 15 (45.5) | | |
| Lymph node
metastasis | | 0.047 | 0.204 |
| Yes | 13 (39.4) | | |
| No | 20 (60.6) | | |
Gastric cancer cell lines
Human gastric cancer cell lines, AGS and SGC7901,
were purchased from the Cell Bank of the Chinese Academy of
Sciences. The human gastric mucosal cell line GES-1 and moderately
differentiated tubular adenocarcinoma cell line MKN-28 were a gift
from the Institute of Digestive Disease, Shanghai Jiaotong
University. All cells mentioned above were cultured in RPMI-1640
medium supplemented with 10% fetal bovine serum in 5%
CO2 at 37°C.
Methods
Laser capture microdissection
(LCM)
The collected tissues embedded in OCT were cut in
8-μm sections at −20°C, after being stained with hematoxylin and
eosin (H&E). The stained slides were placed into a Veritas
Microdissection Instrument (Veritas, Mountain View, CA). Areas that
were enriched in tumor cells (10 mm2/slides) were
selected and captured using ultraviolet laser cutting according to
the manufacturer’s recommended protocol. The tissue samples were
immediately placed in a microcentrifuge tube containing
lysis/binding buffer (Ambion, Inc., USA), which was vortex-mixed
and stored upside down at −20°C. RNA concentration and integrity
were determined by UV absorption and agarose gel electrophoresis,
respectively.
miRNA array
miRCURY LNA™ microRNA arrays (Agilent Technologies,
Santa Clara, CA), which contain more than 1,700 capture probes,
were used to compare the expression profiles of 5 paired primary
gastric cancer tissues and metastatic lymph nodes collected from
the same case. A difference in expression was defined as a change
of >1.5-fold in the corresponding level of miRNA detection
between paired specimens. The Student’s t-test was used to analyze
whether there were significant differences between paired
samples.
qPCR assessment of miRNA (8)
miRNA genes were selected from the Sanger Center
miRNA Registry (http://www.mirbase.org). The sequence-specific primers
for miRNAs and endogenous control U6 snRNA were synthesized by
Sangon (Shanghai, China) (Table
II). The reverse-transcription reaction was carried out with
miRNA-specific stem-loop RT primer and reverse transcriptase
(Fermantas, USA) according to the manufacturer’s instructions.
Real-time quantification polymerase chain reaction (qPCR) analysis
was carried out after cDNA was synthesized. Reaction conditions
were as follows: 95°C for 10 min; followed by 40 cycles of 95°C for
15 sec, 60°C for 15 sec, and 72°C for 45 sec for the amplification.
The threshold cycle (Ct) is defined as the fractional cycle number
at which the fluorescence passes the fixed threshold. The gene
expression Ct values of miRNAs from each sample were calculated by
normalization to the internal control U6 snRNA. All experiments
were repeated in triplicate.
 | Table II.Primer sequences used for miRNA
expression analysis with gene name, sequence (5′-3′). |
Table II.
Primer sequences used for miRNA
expression analysis with gene name, sequence (5′-3′).
| Primer name | Primer
sequence |
|---|
| miR-510-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTGATT-3′ |
| miR-10a-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAAA-3′ |
| U6-RT |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| U6-F |
5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| U6-R |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| miR-510-F |
5′-TACTCAGGAGAGTGGCAATCA-3′ |
| miR-10a-F |
5′-TACCCTGTAGATCCGAATTTG-3′ |
| Universal reverse
primer |
5′-GTGCAGGGTCCGAGGT-3′ |
Bioinformatic analysis of miRNA
miRGen (http://www.diana.pcbi.upenn.edu/miRGen/v3/miRGen.html)
was employed for the prediction of miR-10a target genes. Functional
analysis of the predicted targets was performed using MAS3.0
Bioinformatics Resources (http://bioinfo.capitalbio.com/mas3/).
Statistical analysis
miRNA expression in tumor samples was detected by
real-time quantitative PCR and then subjected to N normalization.
The relationship between changes in miRNA expression and
clinicopathologic characteristics was analyzed using one-way ANOVA.
Two-tailed P<0.05 was judged to be significant. All data and
statistics processing were performed with Excel 2007 (Microsoft,
USA) and SPSS 16.0 (SPSS, Chicago, IL).
Results
Results of the miRNA array
Considering the expression level of miRNAs in
primary gastric cancer as baseline, there were 76 upregulated and
106 downregulated miRNAs associated with lymph node metastasis,
among which four statistically significant miRNAs were included:
three downregulated (miR-24-1*, miR-510 and miR-1284)
and one upregulated miRNA (miR-10a) (Table III).
 | Table III.Differentially expressed miRNAs
detected by miRNA array. |
Table III.
Differentially expressed miRNAs
detected by miRNA array.
| Fold change | P-value | Normalization
|
|---|
| miRNA | LN/T | LN/T | T | LN |
|---|
|
hsa-miR-24-1* | 0.4831 | 0.0401 | 0.071 | 0.0343 |
| hsa-miR-510 | 0.3999 | 0.0069 | 0.0828 | 0.0331 |
| hsa-miR-1284 | 0.5716 | 0.0099 | 0.2366 | 0.1352 |
| hsa-miR-10a | 2.3929 | 0.0203 | 0.0401 | 0.096 |
Differentially expressed miRNAs in
primary gastric tumors
We examined the two most obviously changed miRNAs
(miR-10a and miR-510) in 33 cases of primary gastric cancer and
corresponding normal tissue (N). Each detected miRNA exhibited a
good amplification curve; the melting curve had a single peak. The
results suggested that the products of the real-time PCR were
specific, and the experimental results were reliable.
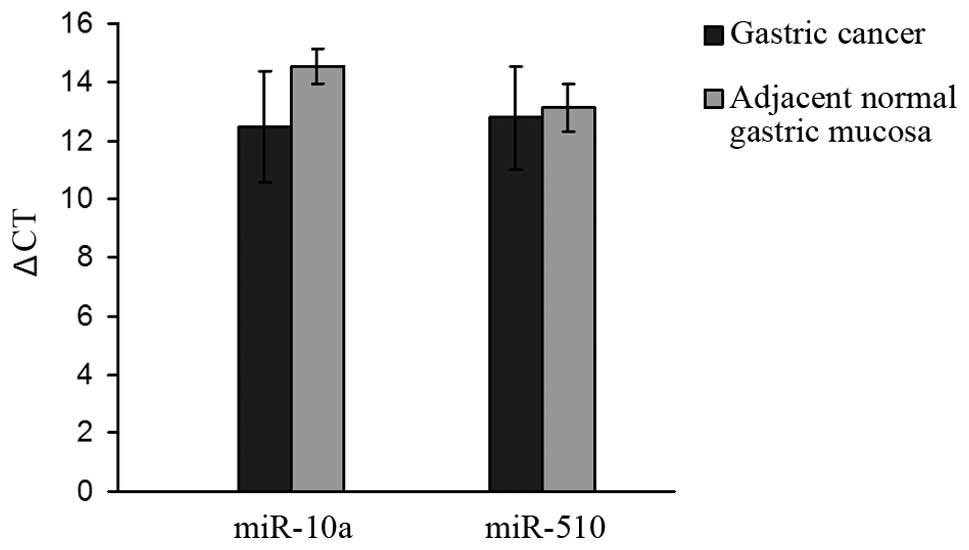
Expression levels of miR-10a and miR-510 were
measured in 33 cases of primary gastric cancer and its
corresponding normal tissue. The average ΔCt (9) values of the tumor tissue were
12.48±1.91 and 12.78±1.76, respectively. The average ΔCt values of
the corresponding adjacent normal tissue were 14.52±0.60 and
13.11±0.80, respectively. The expression of miR-10a was higher in
gastric cancer than that in the adjacent normal mucosa (P<0.05)
(Fig. 1).
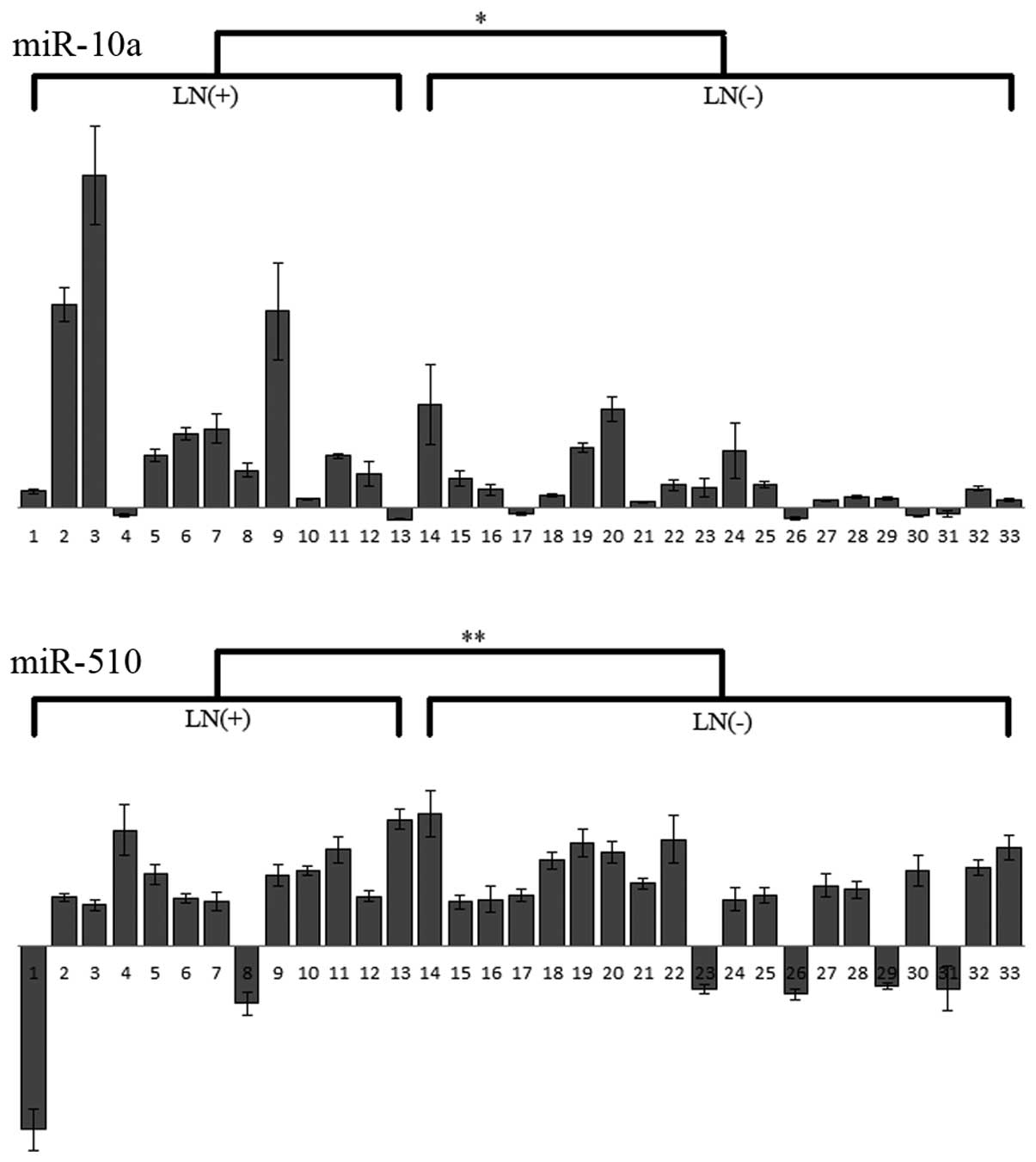
Clinicopathologic characteristics
We investigated the relationship between expression
of miR-10a and miR-510 and each of six clinicopathological
characteristics (patient age, gender, tumor differentiation, major
tumor histological classification, lymphatic invasion and lymph
node metastasis). The results showed that only miR-10a expression
was significantly related to node metastasis (P=0.047); this
measure was not related to the state of lymphatic invasion
(P=0.169) (Fig. 2 and Table IV).
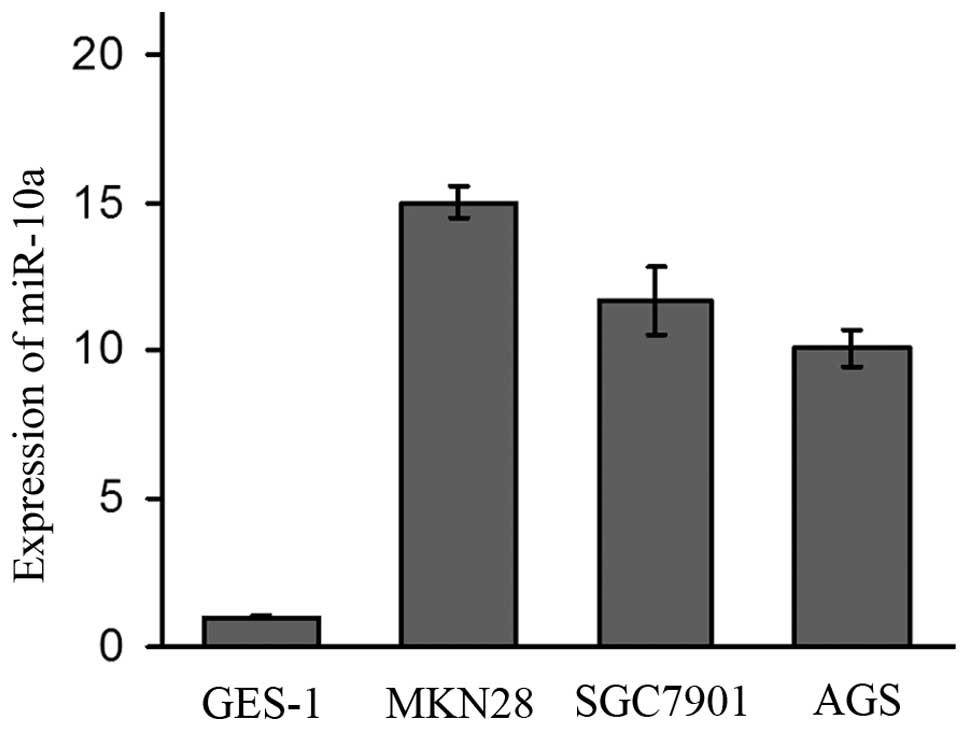
Expression of miR-10a in the cell
lines
The expression of miR-10a varied in all four cell
lines. The expression of miR-10a in the MKN28, SGC7901 and AGS cell
lines was 15-fold, 11.7-fold and 10.1-fold higher than that in the
GES-1 cell line (Fig. 3). As MKN28
and SGC7901 were established from lymph node metastasis, the
expression of miR-10a was higher in these two metastasis original
cell lines than that in the AGS cell line which was established
from a primary tumor.
Bioinformatic analysis of target genes
for miR-10a
Potential target genes of miR-10a were predicted
using miRGen; the corresponding results from the PicTar and
TargetScanS databases were chosen as candidate targets; 118
potential target genes were obtained. KEGG analysis showed that the
predicted target genes involved many signal pathways involved in
tumorigenesis and tumor metastasis, such as the Wnt signal pathway
and that initiated by cell adhesion molecules (CAMs) (Table V).
 | Table V.The most significant 5 pathways
identified in the functional analysis of the miR-10a target
genes. |
Table V.
The most significant 5 pathways
identified in the functional analysis of the miR-10a target
genes.
| Pathway | Gene | P-value |
|---|
| Wnt signaling
pathway | BTRC, NFAT5, DVL3,
CTNNBIP1, CAMK2G | 0.000013 |
| ErbB signaling
pathway | SHC1, CRK,
CAMK2G | 0.00069 |
| Focal adhesion | SHC1, ACTG1, CRK,
FLT1 | 0.00074 |
| Cell adhesion
molecules | NFASC, SDC1,
CADM1 | 0.0024 |
| Notch signaling
pathway | NCOR2, DVL3 | 0.0039 |
Discussion
Few research efforts to date have examined the lymph
node metastasis of gastric cancer using high-throughput methods
(10,11). These publications have focused
mainly on the genomic and proteomic differences between primary
gastric cancer patients with metastasis and those without
metastasis. Samples taken from different parts of the same person
may help to avoid false-positive results on the miRNA array (caused
by background differences among genes). In order to improve the
specificity of the chip-based results, we chose to investigate
primary gastric cancer and paired lymph node metastasis sites in
the same patient. As the ratio of lymph node metastasis increases
with the increased depth of invasion (12,13),
the specimens selected in the study all displayed full-thickness
invasion.
The results obtained identified four differentially
expressed miRNAs. miR-10a was upregulated while
miR-24-1*, miR-510 and miR-1284 were downregulated in
lymph node metastatic sites as compared to the primary gastric
cancer. We hypothesized that if some relationship exists between
the level of miRNA expression in the primary tumor sites and
metastasis, it is quite possible that miRNAs contribute to the
process of gastric cancer lymph node metastasis and may have the
ability to serve as indicators in lymph node metastasis detection.
In order to prove the hypothesis, we investigated the expression
level of miR-10a and miR-510, which were the most obviously changed
in the miRNA array, in the gastric cancer primary sites of 33
patients. The results showed that the expression level of miR-10a
was affected by the existence of lymphatic metastasis, while there
was no obvious relationship with the state of lymphatic invasion.
Furthermore, miR-10a was upregulated in gastric cancer at both the
tissue and cell levels; more obvious differences were noted at the
cell level. Noticeably, the expression of miR-10a in the two
metastasis original cell lines (MKN28 and SGC7910) was higher than
that of the AGS cell line which was established from a primary
tumor. This find also indicated that miR-10a was closely associated
with lymph node metastasis. To assess the possibility of using
miR-10a as a biomarker in lymph node metastasis, further research
with more samples is needed.
miR-10a belongs to the miR-10 family; the other
member is miR-10b. These two members differ from each other in only
one base. Recently, several reports found that miR-10 expression
was aberrantly increased in several types of cancers (14–17),
upregulated in glioma, liver cancer, colon cancer, melanoma and
breast cancer; downregulated in acute or chronic lymphocytic
leukemia and head and neck squamous-cell carcinoma. However,
studies concerning the function of miR-10 are still lacking.
Fortunately, miR-10 was proven to play a pivotal role in tumor
invasion and metastasis in these studies. Ma et al (18) found that miR-10b was significantly
correlated with breast tumor cell metastasis. They also
demonstrated the ability of grafted tumors in mice to invade
locally and transfer to distant sites and this was associated with
upregulated expression of miR-10b in breast cancer cells lacking
metastatic ability. In subsequent studies, the group effectively
inhibited the distant metastasis of grafted mouse tumors using
miR-10b antagomirs (chemically modified anti-miRNA
oligonucleotides) (19). These
results stimulated related research. Weiss et al (20) found that miR-10a was highly
expressed in pancreatic cancer cell lines with high metastatic
ability. Experimental manipulation of miR-10a expression altered
the ability of pancreatic cancer cells to metastasize and invade.
This pattern was also observed in zebrafish transplanted with
pancreatic cancer. In gastric cancer, research on miR-10 is still
limited to describing changes in miR-10 expression, with no
elucidation of the detailed mechanisms. Li et al (21) found a seven-miRNA signature in
gastric cancer (including miR-10b), which had a close relationship
with disease-free survival and overall survival rate. Another study
excluded a role for miR-10 family members (22). Although there is no report on
whether miR-10 contributes to the mechanism of gastric cancer
metastasis, the relationship of miR-10a to gastric cancer lymph
node metastasis demonstrated in our research provides promising
evidence for subsequent research.
miRNAs function post-transcriptionally during
biological processes; therefore, definitive identification of miRNA
target genes is essential. For this reason, we used bioinformatic
methods to predict miR-10a target genes, and then analyzed the
possible mechanisms of its role in gastric tumorigenesis and
metastasis. We chose the most commonly used target gene-predicting
databases, PicTar (23) and
TargetScanS (24), which are
available on the miRGen website (25). To narrow the scope of this hunt for
target genes and improve the specificity of the prediction, we
chose the results common to both databases and subjected these to
functional analysis. The analysis of KEGG (26) metabolic pathways showed that these
predicted targets took part in many pathways involved in
tumorigenesis, invasion and metastasis. For example, the Wnt
signaling pathway plays an important role in organism development
and gastrointestinal tumorigenesis (27), and cell adhesion molecules (CAMs)
localized at the cell surface take part in intercellular and
extracellular interactions.
These findings strongly suggest that miR-10a is
closely correlated with gastric tumorigenesis and metastasis. A
possible mechanism may be that it functions through interaction
with target genes involved in tumorigenesis- and metastasis-related
pathways.
Acknowledgements
This study is supported by the Key
Project of the Chinese Ministry of Education (no. 108015). We thank
other members of our department for their valuable comments and
help.
References
|
1.
|
B WightmanI HaG RuvkunPosttranscriptional
regulation of the heterochronic gene lin-14 by lin-4 mediates
temporal pattern formation in C.
elegansCell75855862199310.1016/0092-8674(93)90530-48252622
|
|
2.
|
RC LeeRL FeinbaumV AmbrosThe C.
elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14Cell758438541993
|
|
3.
|
DP BartelMicroRNAs: genomics, biogenesis,
mechanism, and
functionCell116281297200410.1016/S0092-8674(04)00045-514744438
|
|
4.
|
GA CalinCM CroceMicroRNA signatures in
human cancersNat Rev Cancer6857866200610.1038/nrc199717060945
|
|
5.
|
A Esquela-KerscherFJ
SlackOncomirs-microRNAs with a role in cancerNat Rev
Cancer6259269200610.1038/nrc1840
|
|
6.
|
Q HuangK GumireddyM SchrierThe microRNAs
miR-373 and miR-520c promote tumor invasion and metastasisNat Cell
Biol10202210200810.1038/ncb168118193036
|
|
7.
|
L MaRA WeinbergMicroRNAs in malignant
progressionCell Cycle7570572200810.4161/cc.7.5.5547
|
|
8.
|
C ChenDA RidzonAJ BroomerReal-time
quantification of microRNAs by stem-loop RT-PCRNucleic Acids
Res33e179200510.1093/nar/gni17816314309
|
|
9.
|
KJ LivakTD SchmittgenAnalysis of relative
gene expression data using real-time quantitative PCR and the
2[−Delta Delta C(T)] methodMethods254024082001
|
|
10.
|
L WangJS ZhuMQ SongComparison of gene
expression profiles between primary tumor and metastatic lesions in
gastric cancer patients using laser microdissection and cDNA
microarrayWorld J Gastroenterol12694969542006
|
|
11.
|
M MoriK MimoriY YoshikawaAnalysis of the
gene-expression profile regarding the progression of human gastric
carcinomaSurgery131Suppl
1S39S47200210.1067/msy.2002.11929211821786
|
|
12.
|
T UshijimaM SasakoFocus on gastric
cancerCancer Cell5121125200410.1016/S1535-6108(04)00033-9
|
|
13.
|
LF Onate-OcanaV Aiello-CrocifoglioR
Mondragon-SanchezSurvival benefit of D2 lympadenectomy in patients
with gastric adenocarcinomaAnn Surg
Oncol7210217200010.1007/BF0252365610791852
|
|
14.
|
X AgirreA Jimenez-VelascoE San
Jose-EnerizDown-regulation of hsa-miR-10a in chronic myeloid
leukemia CD34+ cells increase USF2-mediated cell
growthMol Cancer
Res618301840200810.1158/1541-7786.MCR-08-016719074828
|
|
15.
|
L ZhangJ HuangN YangMicroRNAs exhibit high
frequency genomic alterations in human cancerProc Natl Acad Sci
USA10391369141200610.1073/pnas.050888910316754881
|
|
16.
|
M Jongen-LavrencicSM SunMK
DijkstraMicroRNA expression profiling in relation to the genetic
heterogeneity of acute myeloid
leukemiaBlood11150785085200810.1182/blood-2008-01-13335518337557
|
|
17.
|
H VarnholtU DrebberF SchulzeMicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinomaHepatology4712231232200810.1002/hep.2215818307259
|
|
18.
|
L MaJ Teruya-FeldsteinRA WeinbergTumor
invasion and metastasis initiated by microRNA-10b in breast
cancerNature449682688200710.1038/nature0617417898713
|
|
19.
|
L MaF ReinhardtRA WeinbergTherapeutic
silencing of miR-10b inhibits metastasis in a mouse mammary tumor
modelNat Biotechnol28341347201010.1038/nbt.161820351690
|
|
20.
|
FU WeissIJ MarguesJM WolteringRetinoic
acid receptor antagonists inhibit miR-10a expression and block
metastatic behavior of pancreatic
cancerGastroenterology13721362145200910.1053/j.gastro.2009.08.06519747919
|
|
21.
|
X LiY ZhangD FanSurvival prediction of
gastric cancer by a seven-microRNA
signatureGut59579585201010.1136/gut.2008.17549719951901
|
|
22.
|
T UedaS VoliniaH OkumuraRelation between
microRNA expression and progression and prognosis of gastric
cancer: a microRNA expression analysisLancet
Oncol11136146201010.1016/S1470-2045(09)70343-220022810
|
|
23.
|
A KrekD GrunMN PoyCombinatorial microRNA
target predictionsNat Genet37495500200510.1038/ng1536
|
|
24.
|
BP LewisCB BurgeDP BartelConserved seed
pairing, often flanked by adenosines indicates that thousands of
human genes are microRNA
targetsCell1201520200510.1016/j.cell.2004.12.03515652477
|
|
25.
|
M MegrawP SethupathyB CordamiRGen: a
database for the study of animal microRNA genomic organization and
functionNucleic Acids
Res35D149D155200710.1093/nar/gkl90417108354
|
|
26.
|
M KanehisaS GotoKEGG: Kyoto encyclopedia
of genes and genomesNucleic Acids
Res282730200010.1093/nar/28.1.2710592173
|
|
27.
|
P PolakisWnt signaling and cancerGenes
Dev14183718512000
|