Introduction
MASPIN, a novel serine protease inhibitor (Serpin),
inhibits tumor invasion and metastasis of mammary carcinoma. The
gene encoding MASPIN has been initially identified in a search of
genes that display no or down-regulated expression in human breast
carcinoma cells (1). This 42-kDa
protein is known also as Serpin B5, protease inhibitor-5 or
proteinase inhibitor-5. As a tumor suppressor gene, MASPIN is
involved in tumor invasion, metastasis and angiogenesis. However,
the mechanisms of MASPIN are little known, particularly its signal
networks in lung cells.
MASPIN acts as a tumor suppressor gene by impairing
tumor growth, cell motility and metastasis. Shi et al
demonstrated that MASPIN reduced tumor growth as well as invasion
and metastasis through a combination of reduced angiogenesis and
increased apoptosis in a transgenic mouse model that overexpressed
MASPIN (2). Abraham et al
reported that MASPIN increased cell adhesion to the extracellular
matrix in prostate tumor cells and that it was capable of
decreasing the tumorigenic and metastatic potential of prostate
tumors (3). The decreased
expression of MASPIN in prostate cancer has also been reported to
inversely correlate with the development of local recurrence or
systemic tumor progression (4).
Furthermore, it has been reported that MASPIN is involved in the
processes of embryonic development, apoptosis and angiogenesis.
Schaefer and Zhang demonstrated that MASPIN played a significant
role in mammary development in a stage-dependent manner (5). The expression levels of MASPIN are
lower in virgin and early pregnancy mammary glands than in late
pregnancy and lactating mammary glands. Cher et al
demonstrated that MASPIN expression inhibited osteolysis, tumor
growth and angiogenesis in a model of prostate cancer bone
metastasis (6). Finally, the local
delivery of MASPIN to human prostate tumor cells in a mouse model
blocked tumor growth and dramatically reduced the density of
tumor-associated microvessels (5).
Over the past decade, with the expansion of studies
on MASPIN, novel protein-binding partners have been identified and
have provided insight into the molecular aspects on regulation and
divergent mechanisms. Naturally, the MASPIN gene contains a number
of potential transcription factor binding elements, which are
related to the regulation of its expression. These include
activator protein-1 (Ap-1), E26 transformation specific-1 (Ets),
hormonal responsive elements (HREs) and p53 binding sites (5). Recently, it has been shown that
MASPIN may be affected by promoter methylation levels and by
histone modification (7). These
data indicate the complexity of the MASPIN network. In this study,
we used comparative proteomics to systemically study the protein
profile changes in lung cell lines. The large-scale data and system
biology methodology may help us to illustrate the function of
MASPIN, its regulation pathways and networks in lung cancer.
Materials and methods
Cell culture
The human lung carcinoma cell line, LC5, was kindly
provided by the Cell Line Center of the Chinese Academy of Science.
The LC5 human lung carcinoma cell line was maintained in RPMI-1640
(Life Technologies, Gaithersburg, MD, USA) with 1 mM glutamate, 100
U/ ml penicillin, 100 ng/ml streptomycin and 10% fetal calf serum
(FCS). The cells were cultured at 37°C in a humidified atmosphere
of 5% CO2. Cell viability was evaluated by trypan blue
exclusion assay.
Porcine cytomegalovirus (pCMV)-maspin
construction
For construction of the pCMV-MASPIN plasmid,
full-length MASPIN complementary DNA (cDNA) was cloned into the
pCMV-Taq4C vector (Invitrogen, Carlsbad, CA, USA), using the
primers, 5′-atggatgccctgcaactagca-3′ and 5′-ttaaggagaac agaatttg-3′
(Sangon Co., Shanghai, China).
Transfection of the lung cancer
cells
The cells were transfected with small interfering
RNA (siRNA) or plasmids using the Effectene (Qiagen, Valencia, CA,
USA) or Amaxa electroporation system (Amaxa, Gaithersburg, MD,
USA), according to the manufacturer’s instructions. To develop cell
lines, which stably expressed MASPIN, following transfection, the
cells were then incubated in the medium with Geneticin 418 for 48
h, and selected for 2 weeks. We used a targeted SMART pool of
siRNAs from Dharmacon (Lafayette, CO, USA) to knockdown MASPIN
expression with the siLentFect lipid kit. In parallel, cells were
transfected with a fluorescein isothiocyanate-labeled, nonspecific
siRNA with a scrambled sequence.
Western blot analysis
Cells were harvested with ice-cold
phosphate-buffered saline and then lysed in lysis buffer containing
10 mM Tris (pH 7.5), 150 mM NaCl, 10 mM ethylenediaminetetraacetic
acid, 1% SDS, 1 mM sodium orthovanadate and a mixture of protease
inhibitors (Roche, Indianapolis, IN, USA). The lysates were
sonicated for 10 sec, centrifuged for 20 min at 20,000 × g, and
then stored at −70°C. Equal amounts (25 μg) of the cell lysates
were resolved by 12% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and were subjected to Western blot
analysis, using an enhanced chemiluminescence system (Amersham
Corp., Arlington Heights, IL, USA).
Two-dimensional gel electrophoresis (2DE)
and time-of-flight (TOF)/TOF mass spectometry (MS)
Proteins were collected from LC5 cells as described
previously. The extraction buffer included: 9 M urea, 0.5% SDS, 4%
CHAPS, 65 mM DTT, nucleases mixture (GE Healthcare, Piscataway, NJ,
USA) and protease inhibitor mixture (GE Healthcare). The crude
extraction was then treated with Clean-Up Kit (GE Healthcare). The
protein (100 μg) was loaded independently using 18 cm pH3–10 (L)
stripes by the GE Healthcare 2DE system. Second dimensional
SDS-PAGE was carried out, using a 12% gel and stained with the
PlusOne Silver Staining Kit (GE Healthcare). The gel images were
analyzed by the ImageMaster™ 2D Platinum v7.0 (GE Healthcare). The
spots were identified by 6200 Series Accurate-Mass TOF LC/MS
(Agilent, Santa Clara, CA, USA).
Results
Overexpression and knockdown of MASPIN in
LC5 human lung carcinoma cells
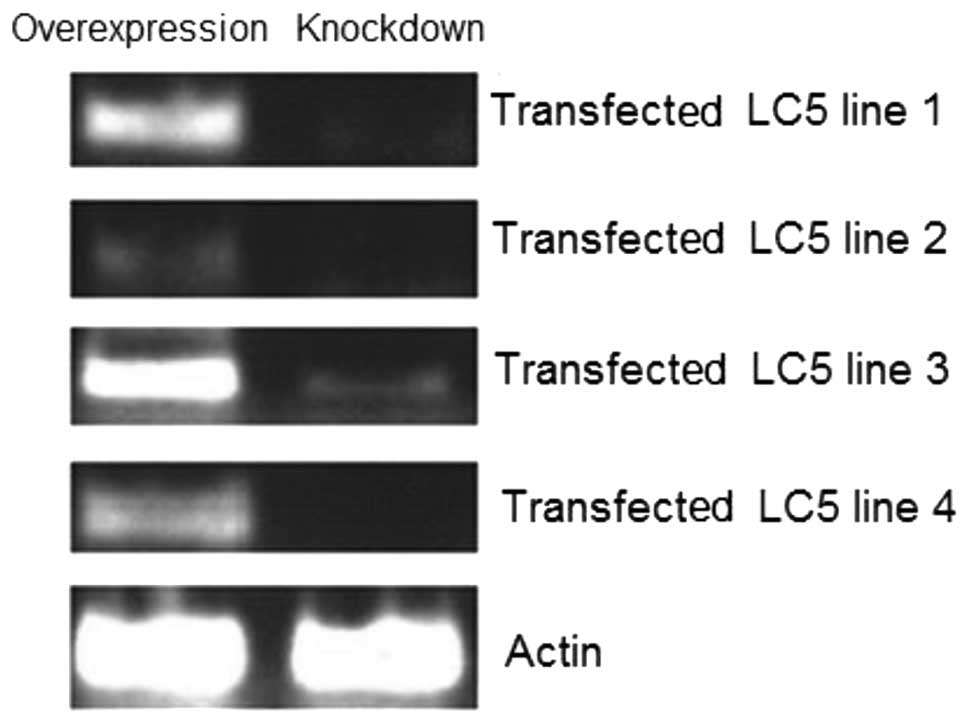
Total RNA was isolated from four exponentially
selected cell lines with overexpressed and knocked down MASPIN,
respectively. The results suggest that all eight cell lines were
successful. All of the transfected cells had a high expression of
MASPIN, while MASPIN expression was almost eliminated by
siRNA-MASPIN (Fig. 1).
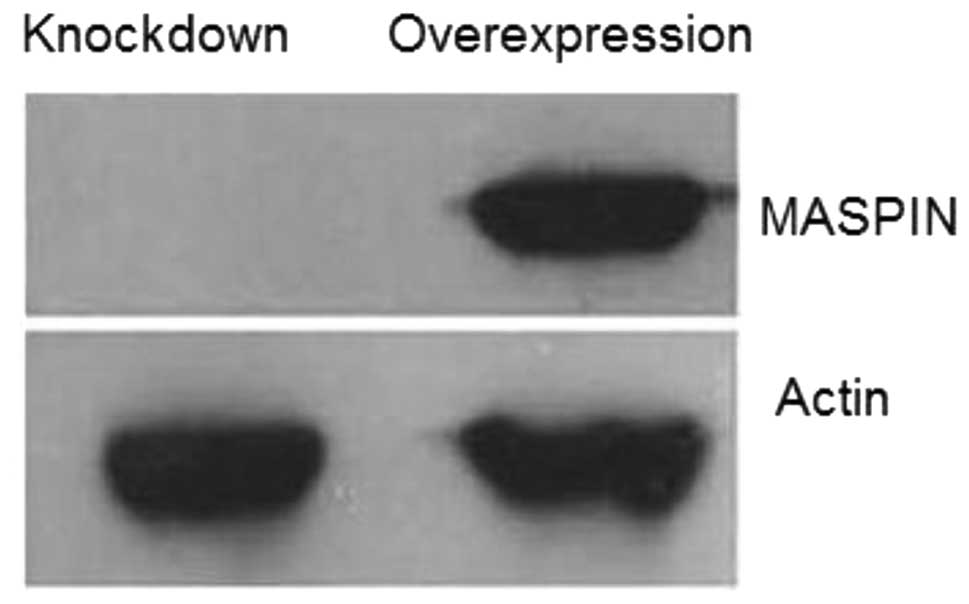
Furthermore, the expression of MASPIN protein was confirmed by
Western blot analysis. The abundance of MASPIN was detected in the
overexpressed cell lines by a specific anti-MASPIN antibody, but
was undetectable in the knocked down cell line (Fig. 2). The results indicated that the
overexpression or knockdown of MASPIN in LC5 cell lines was
successful.
Spot identification by TOF/TOF MS
The global view of a representative two-dimensional
gel is shown in Fig. 3. The MASPIN
overexpression and knockdown samples were loaded three times. Over
1,400 spots were represented in three duplicates. The excellent
reproduction in protein pattern and protein density between all
cell lines is evident in the comparison of four 2D gels to each
other. All gels were matched to the LC5 cells reference 2D map and
the relative density of each protein spot was quantified using
ImageMaster software. The data are shown in Table I.
 | Table I.Spots identification by TOF/TOF
MS. |
Table I.
Spots identification by TOF/TOF
MS.
| Protein name | Quantity in MASPIN
overexpression sample (V %) | Quantity in MASPIN
knockdown sample (V %) | Ratio
overexpression/knockdown |
|---|
| Sdccag8 | Unique | - | - |
| Ldoc1 | Unique | - | - |
| SCAI | Unique | - | - |
| SDCCAG3 | Unique | - | - |
| CT62 | Unique | - | - |
| Brms1 | Unique | - | - |
| CAGE1 | - | Unique | - |
| NEDD9 | - | Unique | - |
| TSP1 | 0.288 | 0.012 | 24.0 |
| RUNX | 0.294 | 0.017 | 17.3 |
| ORAOV1 | 0.335 | 0.022 | 15.2 |
| PTK2 | 0.396 | 0.024 | 16.5 |
| CASC5 | 0.415 | 0.065 | 6.38 |
| BCAR1 | 0.583 | 0.088 | 6.23 |
| GREB1L | 0.676 | 0.096 | 7.04 |
| BASE | 0.688 | 0.152 | 4.54 |
| CTAG2 | 0.707 | 0.169 | 4.18 |
| Blcap, | 0.077 | 0.835 | 0.09 |
| Casc5 | 0.098 | 0.799 | 0.12 |
| Casc3 | 0.104 | 0.761 | 0.13 |
| Hic2 | 0.112 | 0.733 | 0.15 |
| SNCG | 0.127 | 0.704 | 0.18 |
| HIC1 | 0.144 | 0.698 | 0.21 |
| DLC1 | 0.151 | 0.668 | 0.23 |
The results indicate that several tumor suppressor
genes are related to the expression of MASPIN. In this study, we
have found that at least six genes were unique in
MASPIN-overexpressed cell lines, in which most of them play crucial
roles in the invasion of cancer cells. For example, the suppressor
of cancer cell invasion (SCAI) acts as an inhibitor of cancer cell
invasion through the transcriptional control of β1-integrin
(8). The serologically defined
colon cancer antigen 3 (SDCCAG3) is necessary for the presentation
of TNF receptor 1 on the cell surface (9). Leucine zipper, down-regulated in
cancer 1 (LDOC1) inhibits NF-κB activation and sensitizes
pancreatic cancer cells to apoptosis (10). In addition, breast cancer
metastasis suppressor 1 (Brms1) functions as a co-repressor by
enhancing histone deacetylase 1-mediated deacetylation of RelA/p65
and promoting apoptosis, and inhibits gene expression by targeting
NF-κB activity (11).
However, two genes were discovered to be unique in
knockdown cell lines that are associated with the development and
progression of tumors. The expression of cancer-associated gene 1
(CAGE1) protein is associated with the progression of tumors and
has been used as a response criteria in several tumors during
chemotherapy (12). The neural
precursor cell expressed developmentally down-regulated protein 9
(NEDD9), as a melanoma metastasis gene, is regulated in human
neuroblastoma cells and in the embryonic hindbrain by all-trans
retinoic acid (13,14).
Other findings from this study were that nine genes
were extremely highly expressed in the MASPIN-overexpressed cell
lines. Most of these highly expressed genes are associated with
anti-thrombosis, inhibition of tumor cell mobility, metastasis and
angiogenesis. For example, soluble or local platelet-released
thrombospondin-1 (TSP-1) may protect unfolded endothelium-bound and
subendothelial von Willebrand factor (VWF) from degradation by
plasma ADAMTS13, thus securing platelet tethering and thrombus
adherence to the inflamed and injured endothelium, respectively
(15). Furthermore, runt-related
transcription factor 1 (RUNX1) plays a crucial role in the
transition of endothelial cells to hematopoietic cells, and in the
down-regulation of fetal liver kinase-1. By contrast, Runx1 is
weakly expressed in early erythroid cells, and its expression is
rapidly extinguished during later stages of erythropoiesis
(16,17). In addition, protein tyrosine kinase
2 (PTK2), or Focal adhesion kinase 1, plays a crucial role in
generating cell survival signals, as well as the cleavage of FAK
during caspase-mediated apoptosis (18). FAK catalytic activities are crucial
in promoting VEGF-associated tumor angiogenesis and
protease-associated tumor metastasis. Support is growing for the
theory that FAK and Src may be therapeutically relevant targets in
the inhibition of tumor progression (19).
Discussion
The findings of this study illustrate the importance
of the MASPIN gene and protein in regulating the signaling pathway
of tumor initiation, promotion and progression, which includes a
very complicated and intricate control system with multiple layers
that are connected to the expression of the MASPIN gene, including
transcription factors, transcription factor binding elements and
DNA methylation coalescence.
The expression level of MASPIN was used as an
indicator for tumor aggressiveness and metastatic potential. Breast
cancer progression from ductal carcinoma in situ, to locally
invasive cancer, and finally to lymph node metastasis has been
shown to correlate with a stepwise decrease in MASPIN expression
(20). Umekita et al
reported that the expression of MASPIN predicted poor prognosis in
breast cancer patients (21). The
down-regulation of the tumor suppressor gene, MASPIN, in breast
carcinoma was associated with a higher risk of distant metastasis
(20). In this study, we also
demonstrate that MASPIN may be an indicator of aggressiveness and
metastatic potential for lung cancer cells owing to its dual role
in the inhibition of tumor cells.
Previous studies on the regulatory mechanisms of
MASPIN have shown that hypermethylation and histone deacetylation
lead to silencing of the MASPIN gene in human breast cancer
(22,23). Bass et al demonstrated that
MASPIN was a non-inhibitory Serpin, which inhibited the migration
of tumor and vascular smooth muscle cells (24). Recombinant MASPIN specifically
inhibited activators of the cell surface-associated urokinase-type
plasminogen and fibrinogen-bound tissue-type plasminogen.
Endogenous MASPIN was also a potent inhibitor of the pericellular
urokinase-type plasminogen activator and may block tumor invasion
and motility by inhibiting localized pericellular proteolysis
(25).
Jiang et al reported that MASPIN sensitizes
breast carcinoma cells to induced apoptosis (26). Systemic delivery of the MASPIN gene
in a syngeneic tumor model inhibited breast tumor progression
(27). In this study, comparative
proteomic analysis was used to systemically study the protein
profile change in a lung cell line.
This study may help to illustrate the function of
MASPIN, as well as the regulatory pathway and functional network of
MASPIN in lung cancer. However, further studies are required to
investigate the functions of MASPIN in further detail.
References
|
1.
|
Z ZouC GaoAK Nagaichp53 regulates the
expression of the tumor suppressor gene maspinJ Biol
Chem27560516054200010.1074/jbc.275.9.605110692390
|
|
2.
|
HY ShiW ZhangR LiangModeling human breast
cancer metastasis in mice: maspin as a paradigmHistol
Histopathol18201206200312507299
|
|
3.
|
S AbrahamW ZhangN GreenbergM ZhangMaspin
functions as tumor suppressor by increasing cell adhesion to
extracellular matrix in prostate tumor cellsJ
Urol16911571161200310.1097/01.ju.0000040245.70349.3712576872
|
|
4.
|
S MachtensJ SerthC BokemeyerExpression of
the p53 and Maspin protein in primary prostate cancer: correlation
with clinical featuresInt J
Cancer95337342200110.1002/1097-0215(20010920)95:5%3C337::AID-IJC1059%3E3.0.CO;2-111494236
|
|
5.
|
JS SchaeferM ZhangRole of maspin in tumor
metastasis and angiogenesisCurr Mol
Med3653658200310.2174/156652403347951914601639
|
|
6.
|
ML CherHR Biliran JrS BhagatMaspin
expression inhibits osteolysis, tumor growth, and angiogenesis in a
model of prostate cancer bone metastasisProc Natl Acad Sci
USA10078477852200310.1073/pnas.133136010012788977
|
|
7.
|
A DokrasJ CoffinL Field6116172006
|
|
8.
|
DT BrandtC BaarlinkTM KitzingSCAI acts as
a suppressor of cancer cell invasion through the transcriptional
control of beta1-integrinNat Cell
Biol11557568200910.1038/ncb186219350017
|
|
9.
|
N NeznanovL NeznanovaB AngresAV
GudkovSerologically defined colon cancer antigen 3 is necessary for
the presentation of TNF receptor 1 on cell surfaceDNA Cell
Biol24777785200510.1089/dna.2005.24.77716332174
|
|
10.
|
K NagasakiC SchemC von
KaisenbergLeucine-zipper protein, LDOC1, inhibits NF-kappaB
activation and sensitizes pancreatic cancer cells to apoptosisInt J
Cancer105454458200310.1002/ijc.1112212712434
|
|
11.
|
M CicekR FukuyamaDR WelchN SizemoreG
CaseyBreast cancer metastasis suppressor 1 inhibits gene expression
by targeting nuclear factor-kappaB activityCancer
Res6535863595200510.1158/0008-5472.CAN-04-313915867352
|
|
12.
|
S ParkY LimD LeeIdentification and
characterization of a novel cancer/testis antigen gene
CAGE-1Biochim Biophys
Acta1625173182200310.1016/S0167-4781(02)00620-612531476
|
|
13.
|
M KimJD GansC NogueiraComparative
oncogenomics identifies NEDD9 as a melanoma metastasis
geneCell12512691281200610.1016/j.cell.2006.06.00816814714
|
|
14.
|
RA MerrillAW SeeML WertheimM
Clagett-DameCrk-associated substrate (Cas) family member, NEDD9, is
regulated in human neuroblastoma cells and in the embryonic
hindbrain by all-trans retinoic acidDev
Dyn231564575200410.1002/dvdy.20159
|
|
15.
|
A BonnefoyK DaenensHB FeysThrombospondin-1
controls vascular platelet recruitment and thrombus adherence in
mice by protecting (sub)endothelial VWF from cleavage by
ADAMTS13Blood107955964200610.1182/blood-2004-12-485616204318
|
|
16.
|
RB LorsbachJ MooreSO AngW SunN LennyJR
DowningRole of RUNX1 in adult hematopoiesis: analysis of
RUNX1-IRES-GFP knock-in mice reveals differential lineage
expressionBlood10325222529200410.1182/blood-2003-07-243914630789
|
|
17.
|
H HiraiIM SamokhvalovT FujimotoS
NishikawaJ ImanishiS NishikawaInvolvement of Runx1 in the
down-regulation of fetal liver kinase-1 expression during
transition of endothelial cells to hematopoietic
cellsBlood10619481955200510.1182/blood-2004-12-487215928041
|
|
18.
|
DD SchlaepferCR HauckDJ SiegSignaling
through focal adhesion kinaseProg Biophys Mol
Biol71435478199910.1016/S0079-6107(98)00052-2
|
|
19.
|
JA Bernard-TrifiloST LimS HouDD
SchlaepferD IlicAnalyzing FAK and Pyk2 in early integrin signaling
eventsCurr Protoc Cell Biol Chapter 14: Unit 1472006
|
|
20.
|
N MaassM TeffnerF RoselDecline in the
expression of the serine proteinase inhibitor maspin is associated
with tumour progression in ductal carcinomas of the breastJ
Pathol195321326200110.1002/path.94811673829
|
|
21.
|
Y UmekitaY OhiY SagaraH YoshidaExpression
of maspin predicts poor prognosis in breast-cancer patientsInt J
Cancer100452455200210.1002/ijc.1050012115529
|
|
22.
|
BW FutscherMM OshiroRJ WozniakRole for DNA
methylation in the control of cell type specific maspin
expressionNat Genet31175179200210.1038/ng88612021783
|
|
23.
|
N MaassM BiallekF RoselHypermethylation
and histone deacetylation lead to silencing of the maspin gene in
human breast cancerBiochem Biophys Res
Commun297125128200210.1016/S0006-291X(02)02136-812220518
|
|
24.
|
R BassAM FernandezV EllisMaspin inhibits
cell migration in the absence of protease inhibitory activityJ Biol
Chem2774684546848200210.1074/jbc.C20053220012384513
|
|
25.
|
H Biliran JrS ShengPleiotrophic inhibition
of pericellular urokinase-type plasminogen activator system by
endogenous tumor suppressive maspinCancer
Res6186768682200111751384
|
|
26.
|
N JiangY MengS ZhangE Mensah-OsmanS
ShengMaspin sensitizes breast carcinoma cells to induced
apoptosisOncogene2140894098200210.1038/sj.onc.120550712037665
|
|
27.
|
HY ShiR LiangNS TempletonM ZhangInhibition
of breast tumor progression by systemic delivery of the maspin gene
in a syngeneic tumor modelMol
Ther5755761200210.1006/mthe.2002.060212027560
|