Introduction
Hepatocellular carcinoma (HCC) is the fifth most
frequent type of cancer and the third leading cause of cancer
mortality worldwide, with over half a million cases of mortality
every year (1). HCC is also common
in China. According to the annual cancer incidence and mortality
report, the incidence and mortality rates of HCC in China over the
last decade were 300,000 and 306,000 cases, respectively (2,3).
This disease is strongly associated with several risk factors,
including chronic hepatitis B virus (HBV) and chronic hepatitis C
virus (HCV) infection, as well as alcohol abuse (4). Certain epidemic factors have been
identified as risk factors or outcome predictors for HCC (5–7);
however, the true mechanism of this cancer remains unknown. To
date, few studies have focused on the genetic factors associated
with age-at-onset of this cancer, although they have demonstrated
the genetic prevalence of this disease (8).
Hepatitis virus infection and alcohol abuse are
associated with increased oxidative stress in liver cells,
resulting in DNA changes including mitochondrial DNA (mtDNA)
instability (9,10). The human mitochondrial genome is 16
kb in length and is a closed-circular duplex molecule that contains
37 genes, including two ribosomal RNAs and complete sets of 22
transfer RNAs (tRNAs) (11). mtDNA
is believed to be more susceptible to DNA damage and acquires
mutations at a higher rate than nuclear DNA owing to high levels of
reactive oxygen species (ROS), lack of protective histones and
limited capacity for DNA repair in the mitochondria (12–14).
Thus, somatic mtDNA mutations occur in a wide variety of
degenerative diseases and cancers (15,16)
and may be homoplasmic by clonal expansion (17,18)
or heteroplasmic in tumor tissues (19,20).
In a number of cancers, including hepatitis virus-related HCC,
somatic mutations are frequently located in the mtDNA non-coding
region, termed the displacement loop (D-loop) (21,22).
This region is important for regulating the replication and
expression of the mitochondrial genome since it contains the
leading-strand origin of replication and is the main promoter for
transcription (23).
We sequenced the D-loop that contains a length of
1,122 bps (nucleotides 16024-16569 and 1–576; www.mitomap.org) in the blood from HCC patients and
identified 92 single nucleotide polymorphims (SNPs) in the D-loop.
We also identified cancer risk and outcome associated SNPs
(24,25). In the present study, we assess the
correlation between germline SNPs of the D-loop and age-at-onset in
HCC patients.
Materials and methods
Tissue specimens and DNA extraction
Blood samples were collected at the Fourth Hospital
of Hebei Medical University (China) from 60 HCC patients who
underwent HCC resection in the Department of Hepatobiliary Surgery
between 2007 and 2008. All patients originated from the Hebei
Province of China, a high-risk area for HCC. Whole blood was
obtained from corresponding HCC patients. Mitochondria isolation
and mtDNA extraction were carried out using the Blood Mitochondrial
DNA Extraction Kit (Genmed Scientific Inc., Shanghai, China). The
study was approved by the Human Tissue Research Committee of the
Fourth Hospital of Hebei Medical University. All patients provided
written informed consent for the collection of samples and
subsequent analysis.
Polymerase chain reaction (PCR)
amplification and sequence analysis
The forward primer, 5′-CCCCATGCTTACAAGCAA GT-3′
(nucleotide 16190–16209); and reverse primer, 5′-GCTTT
GAGGAGGTAAGCTAC-3′ (nucleotide 602-583) were used for the
amplification of a 982-bp product from the mtDNA D-loop region as
previously described (15). PCR
was performed according to the protocol of the PCR Master Mix Kit
(Promega, Madison, WI, USA) and purified prior to sequencing. Cycle
sequencing was carried out using the Dye Terminator Cycle
Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA,
USA) and the products were then separated on the ABI PRISM Genetic
Analyzer 3100 (Applied Biosystems). Polymorphisms were confirmed by
repeated analyses from the two strands. SNPs were identified
directly from blood mito-chondria.
Statistical analysis
The age-at-onset curve of the HCC patients was
calculated using the Kaplan-Meier method at each SNP site, and
compared using the log-rank test. Multivariate survival analysis
was performed using a Cox proportional hazards model. The
statistical analyses were carried out using the SPSS 11.5 software
package (SPSS Company, Chicago, IL, USA). A p-value of <0.05 was
considered to indicate statistically significant differences.
Results
A total of 60 patients, including 49 HBV-associated
and 11 alcohol-associated HCC patients, were enrolled in this
study. The age-at-onset distribution of HCC patients is listed in
Table I. Those analyzed included 6
patients aged <40 years, 12 patients aged 40–50, 30 patients
aged 51–60 and 12 patients aged >60. None of these patients had
received any adjuvant chemotherapy or radiation therapy following
HCC resection. The age-at-onset and clinical characteristics of the
HCC patients were analyzed using the Kaplan-Meier method and were
compared using the log-rank test. Gender, portal vein thrombosis,
child classification and tumor quantity were not associated with
age-at-onset according to the results of the log-rank test.
However, TNM classification and tumor size correlated with
age-at-onset at statistically significant levels (Table II).
 | Table I.Age-at-onset distribution in ESCC
patients. |
Table I.
Age-at-onset distribution in ESCC
patients.
| Age (years) | No. of cases
|
|---|
| Male | Female |
|---|
| ≤40 | 4 | 2 |
| 41–50 | 11 | 1 |
| 51–60 | 28 | 2 |
| >60 | 10 | 2 |
 | Table II.Clinical characteristics and their
association with age-at-onset in HCC patients. |
Table II.
Clinical characteristics and their
association with age-at-onset in HCC patients.
|
Characteristics | No. of cases | p-value |
|---|
| Gender | | 0.370 |
| Male | 53 | |
| Female | 7 | |
| TNM
classification | | 0.062 |
| I | 15 | |
| II | 45 | |
| Portal vein
thrombosis | | 0.405 |
| Yes | 10 | |
| No | 50 | |
| Child
classification | | 0.122 |
| A | 57 | |
| B | 3 | |
| Tumor size
(diameter) | | 0.024 |
| <5 cm | 10 | |
| ≥5 cm | 50 | |
| Tumor quantity | | 0.271 |
| Single | 50 | |
| Multiple | 10 | |
Subsequently, the correlation between mtDNA genotype
and age-at-onset was compared. We exluded the SNPs with a minor
allele frequency of <5% and obtained 21 SNPs for further
analysis. The HCC patients were divided into two groups on the
basis of their genotype at each SNP site, and the age-at-onset
curve was plotted using the Kaplan-Meier method for all HCC
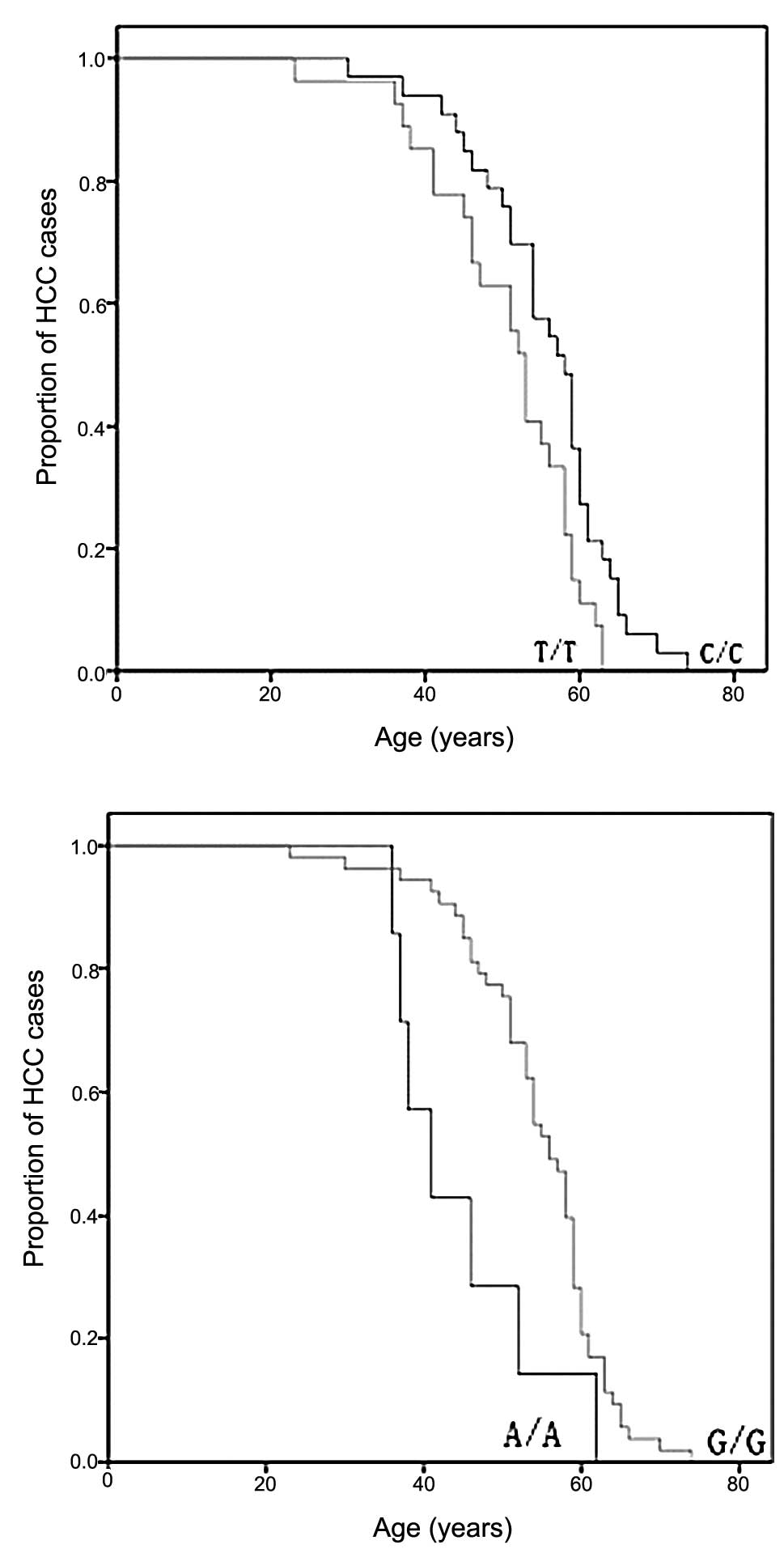
patients at these sites. A dramatic difference in age-at-onset
appeared at sites 489 and 235 as shown by the log-rank test
(Fig. 1). The 489C genotype, known
as mtDNA haplogroup M, was significantly associated with a late
age-at-onset of HCC when compared with halpogroup N (p=0.015)
(Fig. 1). Another
age-at-onset-associated SNP was identified at site 235 with
frequency allele 235G and was linked with the late onset of HCC
(p=0.009).
Using the Cox proportional hazards model, we
performed multivariate analysis of HCC age-at-onset predictors
including TNM classification, tumor size and the two SNPs. As shown
in Table III, haplogroup M (489C)
was identified as an independent predictive factor for age-at-onset
of HCC at borderline significant levels [relative risk, 1.736; 95%
confidence interval (CI), 0.967–3.115; p=0.065].
 | Table III.Multivariate analysis of predictive
factors associated with the age-at-onset of HCC. |
Table III.
Multivariate analysis of predictive
factors associated with the age-at-onset of HCC.
| Factors | Relative risk | 95% CI | p-value |
|---|
| Tumor size | 2.023 | 0.831–4.926 | 0.121 |
| TNM
classification | 1.179 | 0.573–2.427 | 0.654 |
| 235 (G/A) | 0.611 | 0.254–1.470 | 0.271 |
| 489 (C/T) | 1.736 | 0.967–3.115 | 0.065 |
Discussion
Selected SNPs in the D-loop region have previously
been examined for their ability to predict cancer risk and outcome
in many types of cancer (26–28).
The present study has extended these analyses to determine the
relationships between age-at-onset and germline SNPs in a
continuous sequence of mtDNA between nucleotides 16190 and 583 in
HCC patients. The SNPsm, 489C/T and 235G/A, were identified for
their association with age-at-onset at statistically significant
levels by the log-rank test. In an overall multivariate analysis,
haplogroup M (489C) was identified as an independent predictive
factor for age-at-onset of HCC at borderline significant levels.
The results from the log-rank test revealed that disease
advancement with a larger tumor size and a more serious clinical
stage was more prevalent in younger patients. However, this was not
shown in the multivariate analysis.
We have previously performed a number of studies on
D-loop SNPs as predictive factors for digestive tract cancers
(24,25,29).
In the present study, for the first time, we suggest that, other
than as predictors for cancer risk and outcome, SNPs in the D-loop
are also predictors for age-at-onset in HCC patients. All
non-African lineages belong to two founder clusters, namely
haplogroups M and N; 489C, defined as haplogroup M, and 489T,
defined as haplogroup N. According to the ‘out of Africa’ theory,
these are both derived from the L3 mtDNA African lineage (30). The functional significance of the
mtDNA haplo-groups and their association with human behaviors
requires further study, although certain haplogroups have been
identified as markers for special diseases (31,32).
The results from this study require validation in the form of
larger population sizes and laboratory-based functional
studies.
The D-loop region of mtDNA is crucial for the
regulation of mitochondrial genome replication and expression. SNPs
in this region might affect mtDNA replication and lead to the
alteration of the electron transport chain, which is responsible
for the release of highly reactive oxygen species (ROS) and could
contribute to nuclear genome damage as well as cancer initiation
and promotion (33–35). These two SNPs may alter the
transcription of the mitochondrial genome, and this may
subsequently enhance the production of ROS when the mitochondrial
transcription is altered (36).
These ROS-mediated mechanisms may thereby accelerate tumor
development.
In conclusion, SNPs in the D-loop have been found to
be biomarkers for the age-at-onset of HCC. The analysis of genetic
polymorphisms in the D-loop might help to identify patient
subgroups at high risk for an early onset, thereby helping to
refine therapeutic decisions in HCC cancers.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (No. 30801384). The
research was supported in part by the Natural Science Foundation of
Hebei Province (No. C2008000958).
References
|
1.
|
AI GomaaSA KhanMB ToledanoI WakedSD
Taylor-Robinsonhepatocellular carcinoma: Epidemiology, risk factors
and pathogenesisWorld J
Gastroenterol1443004308200810.3748/wjg.14.430018666317
|
|
2.
|
Z SunL MingX ZhuJ LuPrevetion and control
of hepatitis B in ChinaJ Med
Virol67447450200210.1002/jmv.1009412116043
|
|
3.
|
J FerlayF BrayP PisaniDM ParkinGlobocan
2000: Cancer Incidence, Mortality and Prevalence Worldwide Version
1.0IARC PressLyon2001
|
|
4.
|
S CaldwellSH ParkThe epidemiology of
hepatocellular cancer: from the perspectives of public health
problem to tumor biologyJ
Gastroenterol4496101200910.1007/s00535-008-2258-619148801
|
|
5.
|
A MakiH KonoM GuptaM AsakawaT SuzukiM
MatsudaH FujiiI RusynPredictive power of biomarkers of oxidative
stress and inflammation in patients with hepatitis C
virus-associated hepatocellular carcinomaAnal Surg
Oncol1411821190200710.1245/s10434-006-9049-117195915
|
|
6.
|
S OkadaK ShimadaJ YamamotoT TakayamaT
KosugeS YamasakiM SakamotoS HirohashiPredictive factors for
postoperative recurrence of hepatocellular
carcinomaGastroenterology1061618162419948194710
|
|
7.
|
M MinagawaM MakuuchiT TakayamaN
KokudoSelection criteria for repeat hepatectomy in patients with
recurrent hepatocellular carcinomaAnn
Surg238703710200310.1097/01.sla.0000094549.11754.e614578733
|
|
8.
|
N WongW YeoWL WongNL WongKY ChanFK MoJ
KohSL ChanAT ChanPB LaiTOP2A overexpression in hepatocellular
carcinoma correlates with early age onset, shorter patients
survival and chemoresistanceInt J
Cancer124644652200910.1002/ijc.2396819003983
|
|
9.
|
KB SchwarzOxidative stress during viral
infection: a reviewFree Radical Biol
Med21641649199610.1016/0891-5849(96)00131-18891667
|
|
10.
|
A MansouriB FromentyA BersonMA RobinS
GrimbertM BeaugrandS ErlingrD PessayreMultiple hepatic
mitochondrial DNA deletions suggest premature oxidative aging in
alcoholic patientsJ
Hepatol2796102199710.1016/S0168-8278(97)80286-3
|
|
11.
|
GS ShadelDA ClaytonMitochondrial DNA
maintenance in vertebratesAnnu Rev
Biochem66409435199710.1146/annurev.biochem.66.1.4099242913
|
|
12.
|
S DiMauroEA SchonMitochondrial DNA
mutations in human diseaseAm J Med
Genet1061826200110.1002/ajmg.139211579421
|
|
13.
|
MF BealMitochondia, free radicals, and
neurodegenerationCurr Opin
Neurobiol6661666199610.1016/S0959-4388(96)80100-08937831
|
|
14.
|
RN LightowlersPF ChinneryDM TurnbullN
HowellMammalian mitochondrial genetics: heredity, heteroplasmy and
diseaseTrends
Genet13450455199710.1016/S0168-9525(97)01266-39385842
|
|
15.
|
DC WallaceMouse models for mitochondrial
diseaseAm J Med Genet1067193200110.1002/ajmg.139311579427
|
|
16.
|
MS FlissH UsadelOL CaballeroL WuMR ButaSM
EleffJ JenD SidranskyFacile detection of mitochondrial DNA
mutations in tumors and bodily
fluidsScience28720172019200010.1126/science.287.5460.201710720328
|
|
17.
|
S NomotoK YamashitaK KoshikawaA NakaoD
SidranskyMitochondrial D-loop mutation as clonal markers in
multicentric hepatocellular carcimona and plasmaClin Cancer
Res8481487200211839667
|
|
18.
|
E MamboX GaoY CohenZ GuoP TalalayD
SidranskyElectrophile and oxidant damage of mitochondrial DNA
leading to rapid evolution of homoplasmic mutationsProc Natl Acad
Sci USA10018381843200310.1073/pnas.043791010012578990
|
|
19.
|
H YoneyamaT HaraY KatoT YamoriET MatsuuraK
KoikeNucleotide sequence variation is frequently in the
mitochondrial DNA displacement loop region of individual human
tumor cellsMol Cancer Res314202005
|
|
20.
|
JP JakupciakS MaraghME MarkowitzAK
GreenbergMO HoqueA MaitraPE BarkerPD WagnerWN RomS SrivastavaD
SidranskyCD O’ConnellPerformance of mitochondrial DNA mutations
detecting early stage cancerBMC
Cancer8285200810.1186/1471-2407-8-28518834532
|
|
21.
|
M NashikawaS NishiguchiS ShiomiA TamoriN
KohT TakedaS KuboK HirohashiH KinoshitaE SatoM InoueSomatic
mutation of mitochondrial DNA in cancerous and noncancerous liver
tissue in individuals with hepatocellular carcinomaCancer
Res6118431845200111280735
|
|
22.
|
M Sanchez-CespedesP ParrellaS NomotoD
CohenY XiaoM EstellerC JeronimoRC JordanT NicolWM KochM SchoenbergP
MazzarelliVM FazioD SidranskyIdentification of a mononucleotide
repeat as a major target for mitochondrial DNA alterations in human
tumorsCancer Res6170157019200111585726
|
|
23.
|
JW TaanmanThe mitochondrial genome:
structure, transcription, translation and replicationBiochim
Biophys Acta1410103123199910.1016/S0005-2728(98)00161-310076021
|
|
24.
|
R ZhangF ZhangC WangS WangY-H ShiaoZ
GuoIdentification of sequence polymorphism in the D-loop region of
mitochondria DNA as a risk factor for hepatocellular carcinoma with
distinct etiologyJ Exp Clin Cancer
Res29130201010.1186/1756-9966-29-13020849651
|
|
25.
|
C WangF ZhangH FanL PengR ZhangS LiuZ
GuoSequence polymorphisms of mitochondrial D-loop and
hepatocellular carcinoma outcomeBiochem Biophys Res
Commun406493496201110.1016/j.bbrc.2011.02.08821345333
|
|
26.
|
F NavagliaD BassoP FogarC SpertiE GrecoCF
ZambonA StrangesA FaldaS PizziA ParentiS PedrazzoliM
PlebaniMitochondrial DNA D-loop in pancreatic cancer: somatic
mutations are epiphenomena while the germline 16519 T variant
worsens metabolism and outcomeAm J Clin
Pathol126593601200610.1309/GQFCCJMH5KHNVX7316938655
|
|
27.
|
L WangWR BamletM de AndradeLA BoardmanJM
CunninghamSN ThibodeauGM PetersenMitochondrial genetic
polymorphisms and pancreatic cancer riskCancer Epidemiol Biomarkers
Prev1614551459200710.1158/1055-9965.EPI-07-011917627010
|
|
28.
|
L WangSK McDonnellSJ HebbringJM
CunninghamJ St SauverJR CerhanG IsayaDJ SchaidSN
ThibodeauPolymorphisms in mitochondrial genes and prostate cancer
riskCancer Epidemiol Biomarkers
Prev1735583566200810.1158/1055-9965.EPI-08-043419064571
|
|
29.
|
R ZhangR WangF ZhangC WuH FanY liC WangZ
GuoSingle nucleotide polymorphisms in the mitochondrial
displacement loop and outcome of esophageal squamous cell
carcinomaJ Exp Clin Cancer Res29155201010.1186/1756-9966-29-155
|
|
30.
|
AM GonzálezJM LarrugaKK Abu-AmeroY ShiJ
PestanoVM CabreraMitochondrial lineage M1 traces an early human
backflow to AfricaBMC Genomics8223200717620140
|
|
31.
|
V CarelliA AchilliML ValentinoC RengoO
SeminoM PalaA OlivieriM MattiazziF PallottiF CarraraHaplogroup
effects and recombination of mitochondrial DNA: novel clues from
the analysis of Leber hereditary optic neuropathy pedigreesAm J Hum
Genet78564574200610.1086/50123616532388
|
|
32.
|
N UdarSR AtilanoM MemarzadehDS BoyerM
ChwaS LuB MaguenJ LangbergP CoskunDC WallaceMitochondrial DNA
haplogroups associated with age-related macular degenerationInvest
Ophthalmol Vis Sci5019662974200910.1167/iovs.08-2646
|
|
33.
|
B BandyAJ DavisionMitochondrial mutations
may increase oxidative stress: implications for carcinogenesis and
aging?Free Radic Biol
Med8523539199010.1016/0891-5849(90)90152-92193852
|
|
34.
|
JJ GilleH JoenjeCell culture models for
oxidative stress: superoxide and hydrogen peroxide versus
normobaric heperoxiaMutat
Res275405414199210.1016/0921-8734(92)90043-O1383781
|
|
35.
|
MK ShigenagaTM HagenBN AmesOxidative
damage and mitochondrial decay in agingProc Natl Acad Sci
USA911077110778199410.1073/pnas.91.23.107717971961
|
|
36.
|
GA DementSC MaloneyR ReevesNuclear HMGA1
nonhistone chromatin proteins directly influence mitochondrial
transcription, maintenance, and functionExp Cell
Res3137787200710.1016/j.yexcr.2006.09.014
|