Introduction
Uterine carcinosarcoma (UCS), also known as
malignant mixed Müllerian tumor (MMMT), is a rare and aggressive
neoplasia characterized histologically by both
carcinomatous/epithelial and sarcomatous/mesenchymal elements
(1). These tumors are generally
thought to account for 3–7% of uterine cancers (2). In general, UCSs present in the late
seventh decade of life, are quite advanced and have a very poor
prognosis due to the aggressive nature of the tumors (3). An epidemiological study revealed that
UCS and endometrial carcinoma share a similar risk factor profile
including obesity and unopposed estrogen exposure (4). Moreover, the behavior of UCS more
closely resembles that of the highly aggressive endometrial
carcinomas (e.g. serous adenocarcinomas) than that of other uterine
sarcomas (5,6). For these reasons, the 2009 revised
FIGO staging for uterine sarcomas recommends that UCS be staged as
a carcinoma of the endometrium (1). Collectively, these clinical data
reflect a need to better understand the molecular characteristics
of UCS, particularly the similarities and differences with other
endometrial carcinomas.
The most contentious aspect of UCS in the past has
been the origin of the tumor itself. Certain investigators have
argued that UCS tumors are actually two independent tumors (the
collision model), while others contend that UCS begins as a single
tumor that differentiates into carcinomatous and sarcomatous
elements (the monoclonal model). While the collision model has not
been completely excluded, an increasing number of molecular studies
support the monoclonal model. For example, patterns of
X-inactivation in most UCSs are identical in the epithelial and
mesenchymal components (7,8). Similar findings have been reported
for loss of heterozygosity (LOH), the tumor suppressor TP53, the
oncogene KRAS and microsatellite instability (MSI) (8–11).
Thus, the emerging consensus is that the majority of UCSs are
monoclonal.
One aspect of UCS that has received little attention
to date is the pattern of microRNA (miRNA) expression in these
tumors compared either with benign endometrium or with other
endometrial cancers. miRNAs are small (21 to 23 nt) regulatory RNAs
whose known role in oncogenesis and cancer progression is rapidly
expanding (12,13). We determined the expression of 667
miRNAs in a panel of eight UCS tissue samples and compared the
expression patterns with a panel of five benign endometrium, four
endometrial endometrioid adenocarcinoma and four endometrial serous
adenocarcinoma tissue samples. We found that the expression of 114
miRNAs was significantly dysregulated (106 overexpressed and eight
underexpressed) as compared with benign endometrium, but only nine
of these miRNAs were found to overlap with those significantly
dysregulated in endometrial adenocarcinomas. We also found that,
among the 105 significantly dysregulated miRNAs that do not overlap
with the adenocarcinomas, nearly one-third (32 miRNAs) lie in the
250-kb imprinted region in chromosome 14q32. Moreover, all but
three of the 61 miRNAs in the region whose expression we measured
showed at least two-fold overexpression as compared with benign
endometrium. Global dysregulation of this region in UCS is further
evidenced by the altered expression of the reciprocally imprinted
δ-like homolog 1 (Drosophila) (DKL1)/non-coding RNA locus
maternally expressed 3 (GTL2) (also known as MEG3) locus that also
lies in the same chromosome 14q32 imprinted region (14,15).
The dysregulation of expression of such a large number of miRNAs in
such a small region suggests the possibility of the disruption of a
single regulatory factor, particularly in view of the evidence that
the entire region is imprinted.
Materials and methods
Carcinosarcomas and benign
endometrium
Primary carcinosarcoma tissues (n=8) and samples of
benign endometrium (n=5) were obtained from patients undergoing
surgery in the Gynecologic Oncology Division of the Department of
Obstetrics and Gynecology at the University of Iowa Hospitals and
Clinics. The study was approved by the Institutional Review Board
and informed consent was obtained from all patients. All carcinoma
patients presented as grade 3 in stages IB to IIIC. Benign
endometrium was obtained from post-menopausal patients who were
cancer-free. All tissue specimens were snap-frozen and stored at
−80°C following histological evaluation.
RNA purification
Tissue samples were transitioned from −80 to −20°C
following perfusion in RNAlater ICE (Ambion, Life Technologies)
according to the supplier's instructions. Total RNA was extracted
from homogenized tissue using the miRvana miRNA isolation kit
(Ambion, Life Technologies). RNA yield and quality were assessed on
a NanoDrop M-1000 spectrophotometer and an Agilent 2100
Bioanalyzer. An RNA quality cut-off value at RIN ≥7.0 was adhered
to and RNAs meeting that cut-off value were standardized to 200
ng/μl (NanoDrop confirmed) for all subsequent analyses.
qPCR assays
miRNA expression levels were determined on miRNA
TaqMan Low Density Arrays (TLDA; Applied Biosystems, Life
Technologies). The A-set (v. 2.0, 377 miRNAs) and the B-set (v.
2.0, 290 miRNAs) TLDA cards were used for these assays. Reverse
transcription of 600 ng of total RNA was carried out for each
tissue and TLDA card using the TaqMan miRNA reverse transcription
kit (Applied Biosystems, Life Technologies) and either the Human
A-pool (v. 2.1) or Human B-pool (v. 2.0) Megaplex RT primers
(Applied Biosystems, Life Technologies).
All 26 TLDA cards were processed on an Applied
Biosystems Model 7900 Genetic Analyzer and the resulting data
analyzed using Applied Biosystems StatMiner software. The
expression of all 667 miRNAs in all samples was normalized against
the expression of the RNU48 endogenous control RNA. Comparison of
individual, normalized miRNA expression levels was performed using
the ΔΔCt method where fold change between carcinosarcoma and benign
endometrium is given as 2−ΔΔCt for each miRNA (16). Statistical significance of the fold
change estimates were assessed using two-tailed t-tests with
unequal variances and potential false discovery was adjusted via
Bonferroni correction of the p-values. A p-value of ≤0.05 was
considered to indicate a statistically significant difference.
mRNA expression assays
Expression of the reciprocally imprinted DKL1/GTL2
locus was examined using 500 ng of RNA from each tissue using
Applied Biosystems (Life Technologies) gene-specific TaqMan Assays.
RNAs were reverse-transcribed with random hexamers using the
Applied Biosystems (Life Technologies) High Capacity cDNA Reverse
Transcription Kit. Expression differences between carcinosarcomas
and benign endometrium were calculated using the ΔΔCt method as
above. The expressions of DLK1 and GTL2 were normalized against 18S
rRNA prior to analysis.
Bisulfite sequencing
To assess methylation status, bisulfite sequencing
was performed on the differentially methylated region (DMR) lying
between DLK1 and GTL2 in the carcinosarcomas. DNA was purified from
tumor tissue and 2 μg aliquots were bisulfite converted using the
EZ DNA Methylation-Direct Kit (Zymo Research) according to the
manufacturer's instructions. The DKL1/GTL2 DMR was then amplified
from converted DNA using the following primer pair, DMRFOR:
5′-GTGGATTTGTGAGAAATGATTTTGT-3′ and DMRREV:
5′-CCATTATAACCAATTACAATACCAC-3′ as specified by Guens et al
(15). Amplicons were cloned into
pGEM T-EASY (Promega), multiple colonies were selected for each
tumor and DNA was prepared using the QIAprep Spin Miniprep Kit
(Qiagen). These DNAs were then sequenced in both directions on an
Applied Biosystems Model 3730xl DNA Sequencer using vector M13
primers.
Results
miRNA expression
The results of the TLDA array comparisons of the
eight UCS samples with the five benign endometrium samples are
presented in Table I. A total of
114 miRNAs were significantly altered in the UCSs versus benign
endometrium after the p-values were adjusted by Bonferroni
correction to reduce the false discovery rate (FDR) (17). The majority of these differences
are in the direction of overexpression among the UCSs (n=106) as
opposed to underexpression among the UCSs (n=8). A number of
miRNAs, including miR-431, miR-888 and
miR-206, are very highly expressed in UCS tissues as
compared with benign samples.
 | Table I.Fold change and adjusted p-values of
the 114 microRNAs significantly dysregulated in UCSs compared with
benign endometrium. |
Table I.
Fold change and adjusted p-values of
the 114 microRNAs significantly dysregulated in UCSs compared with
benign endometrium.
| MicroRNA | Fold change | Adjusted p-value | MicroRNA | Fold change | Adjusted p-value | MicroRNA | Fold change | Adjusted p-value |
|---|
| Underexpressed | | | | | | | | |
| miR-29c | −10.6 | 0.009 | miR-143 | −11.7 | 0.009 | miR-145 | −8.4 | 0.02 |
|
miR-195* | −7.1 | 0.03 | let-7c | −7.5 | 0.03 | miR-133α | −8.2 | 0.04 |
| let-7b | −6.2 | 0.04 | miR-29a | −6.0 | 0.04 | | | |
| Overexpressed | | | | | | | | |
| miR-431 | 1525.3 | 0.0001 | miR-888 | 132.7 | 0.0001 | miR-216b | 91.5 | 0.0001 |
| miR-543 | 65.8 | 0.0002 | miR-206 | 135.1 | 0.0003 | miR-330-5p | 46.9 | 0.0003 |
|
miR-15b* | 56.4 | 0.0003 | miR-153 | 30.1 | 0.0004 | miR-183 | 43.7 | 0.0004 |
| miR-33b | 30.2 | 0.0004 |
let-7f-1* | 79.4 | 0.0004 | miR-432 | 70.2 | 0.0007 |
|
miR-541 | 32.9 | 0.0008 |
miR-369-3p | 67.9 | 0.0008 |
miR-20b* | 92.9 | 0.001 |
| miR-622 | 20.9 | 0.001 | miR-205 | 26.4 | 0.002 | miR-769-3p | 86.7 | 0.002 |
|
miR-433 | 36.1 | 0.002 | miR-182 | 23.3 | 0.003 |
miR-183* | 26.4 | 0.003 |
|
miR-656 | 20.3 | 0.004 | miR-605 | 23.0 | 0.004 |
miR-493* | 31.2 | 0.004 |
|
miR-376a* | 33.0 | 0.004 |
miR-516α-3p | 17.1 | 0.005 | miR-296-3p | 18.1 | 0.005 |
| miR-615-3p | 53.8 | 0.006 | miR-551a | 43.7 | 0.006 |
miR-192* | 17.5 | 0.006 |
|
miR-376b | 32.6 | 0.006 | miR-892a | 58.3 | 0.006 |
miR-130b* | 15.8 | 0.006 |
|
miR-488* | 38.7 | 0.006 |
miR-154* | 32.9 | 0.006 | miR-377 | 46.8 | 0.006 |
|
miR-889 | 15.4 | 0.007 | miR-496 | 40.9 | 0.007 | miR-668 | 32.8 | 0.007 |
|
miR-541* | 35.7 | 0.007 |
miR-18a* | 12.4 | 0.008 | miR-493 | 15.4 | 0.008 |
| miR-518d-3p | 14.2 | 0.008 |
miR-323-3p | 13.4 | 0.009 |
miR-155* | 28.9 | 0.009 |
| miR-7 | 13.3 | 0.009 | miR-641 | 25.8 | 0.01 |
miR-135b* | 17.5 | 0.01 |
|
miR-223* | 12.6 | 0.01 |
miR-654-5p | 17.3 | 0.01 | miR-922 | 26.7 | 0.01 |
| miR-217 | 14.1 | 0.01 |
miR-551b* | 9.7 | 0.01 | miR-539 | 13.3 | 0.01 |
| miR-147b | 12.8 | 0.01 | miR-499-5p | 13.7 | 0.01 | miR-382 | 18.4 | 0.01 |
| miR-887 | 10.7 | 0.01 |
miR-195* | 21.1 | 0.02 | miR-639 | 11.0 | 0.02 |
| miR-373 | 22.5 | 0.02 | miR-107 | 10.0 | 0.02 | miR-582-3p | 10.2 | 0.02 |
| miR-891a | 14.6 | 0.02 |
miR-409-5p | 10.5 | 0.02 |
miR-138-1* | 9.6 | 0.02 |
|
miR-210 | 8.1 | 0.02 |
let-7f-2* | 10.1 | 0.02 |
let-7a* | 13.5 | 0.02 |
| miR-651 | 10.7 | 0.02 | miR-198 | 9.8 | 0.02 | miR-346 | 8.2 | 0.02 |
|
miR-146a* | 17.5 | 0.02 | miR-937 | 12.0 | 0.02 | miR-450b-3p | 16.6 | 0.02 |
| miR-505 | 9.4 | 0.02 | miR-491-3p | 18.0 | 0.02 | miR-501-3p | 13.4 | 0.02 |
|
miR-369-5p | 8.5 | 0.02 | miR-630 | 7.6 | 0.02 |
miR-181c* | 8.3 | 0.03 |
| miR-18b | 8.4 | 0.03 | miR-643 | 7.6 | 0.03 | miR-506 | 15.7 | 0.03 |
| miR-943 | 16.1 | 0.03 | miR-570 | 7.7 | 0.03 | miR-188-3p | 14.0 | 0.03 |
|
miR-299-3p | 8.7 | 0.03 | miR-941 | 8.7 | 0.03 | miR-584 | 8.5 | 0.03 |
|
miR-545* | 7.9 | 0.03 | miR-638 | 8.0 | 0.04 |
miR-409-3p | 9.6 | 0.04 |
| miR-875-5p | 7.5 | 0.04 |
miR-485-3p | 8.9 | 0.04 |
miR-379* | 7.5 | 0.04 |
| miR-561 | 7.2 | 0.04 | miR-661 | 7.2 | 0.04 |
miR-432* | 7.1 | 0.04 |
| miR-223 | 7.0 | 0.04 |
miR-487b | 8.6 | 0.05 | miR-301b | 8.2 | 0.05 |
|
miR-127-5p | 6.7 | 0.05 |
miR-654-3p | 7.4 | 0.05 |
miR-376a | 7.2 | 0.05 |
| miR-372 | 6.8 | 0.05 | | | | | | |
When compared with the 71 miRNAs found to be
significantly dysregulated among endometrial adenocarcinomas
(18), only nine were found to
overlap with altered UCS miRNAs (Table
I). These nine miRNAs are the underexpressed miR-133a
and let-7c and the overexpressed miR-107,
miR-181c*, miR-205, miR-210,
miR-516a-3p, miR-888 and
let-7f-2*.
Among the twenty most highly expressed miRNAs are
miR-431, miR-432, miR-541, miR-543 and
miR-369-3p. These five miRNAs are all from the cluster of
miRNAs found in the imprinted region of chromosome 14q32,
demonstrating that a quarter of the most highly expressed miRNAs in
UCS are found in this small (250 kb) region. A complete scan of the
A-set and B-set miRNAs on the TLDA arrays revealed that 43 miRNA
loci in this chromosome 14q32 region, accounting for a total of 61
miRNAs (9.2%), were represented. The current miRNA content of the
chromosome 14q32 cluster is 57 loci accounting for 71 miRNAs
(miRBase, release 17 April 2011). Of the 61 chromosome 14q32
cluster miRNAs represented on the TLDA cards, all but three
(miR-154, miR-329 and miR-412) were
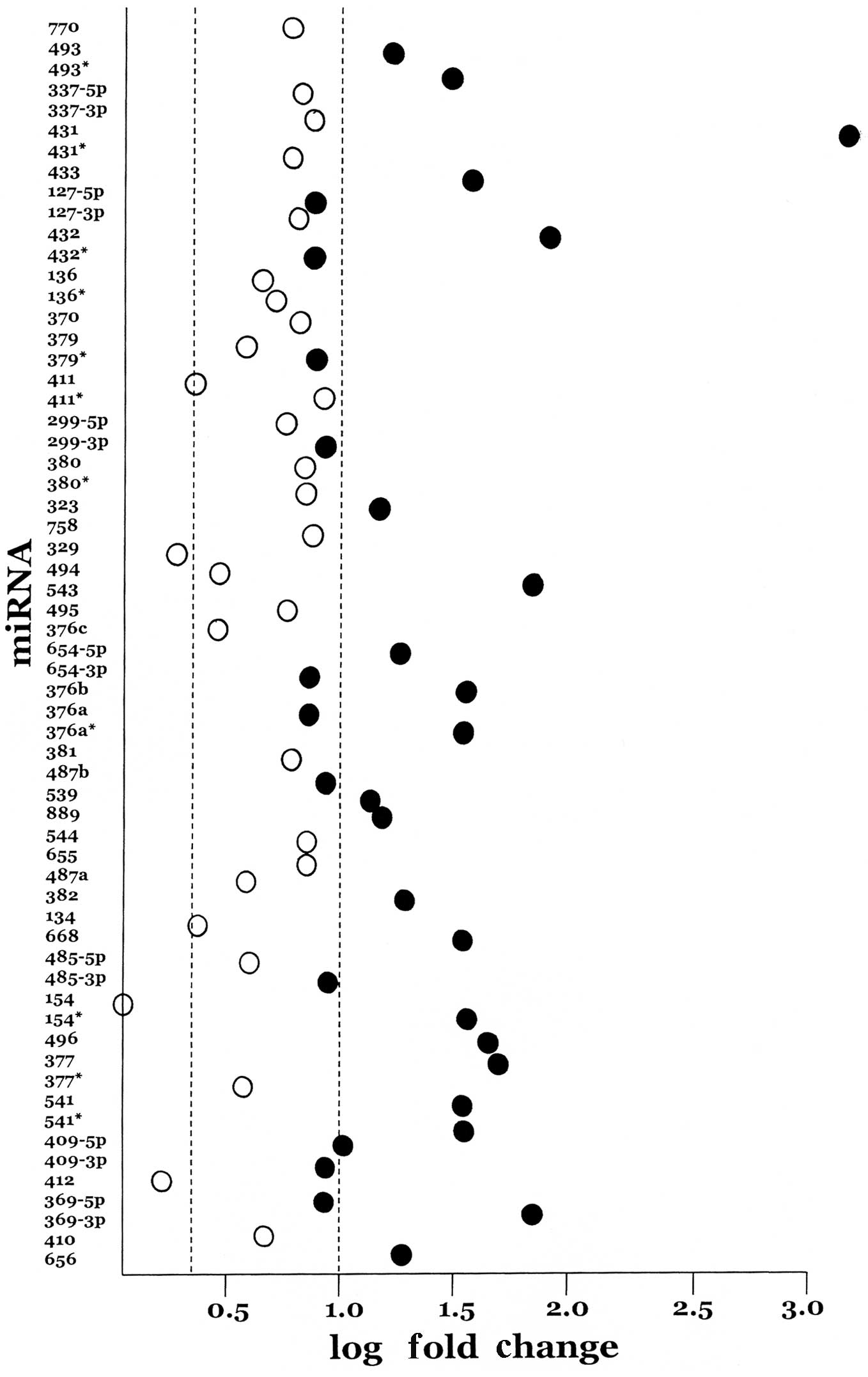
overexpressed in the UCSs by at least two-fold and, of these, 32
achieved statistical significance (Fig. 1). Thus, while chromosome 14q32
miRNAs represent 9.2% of the miRNAs present on the TLDA arrays,
they account for a statistically significant 30.5%
(χ2=56.9, p<0.001) proportion of the significantly
overexpressed miRNAs. Moreover, among the 18 miRNAs in which mature
sequences from both sides of the hairpin are expressed, that is,
both the 5p and the 3p arms, eight show nearly equal
expression.
DLK1/GTL2 expression in UCS
On the centromeric flank of the chromosome 14q32
miRNA cluster is the reciprocally imprinted locus composed of DLK1
and GTL2. In order to discover whether the global dysregulation of
the miRNAs in chromosome 14q32 extends to other genes, the
expression of both DKL1 and GTL2 was assessed in the UCS tumor
panel as well as the benign endometrium group. DKL1/GTL2 expression
in UCSs and benign endometrium, normalized against 18S rRNA
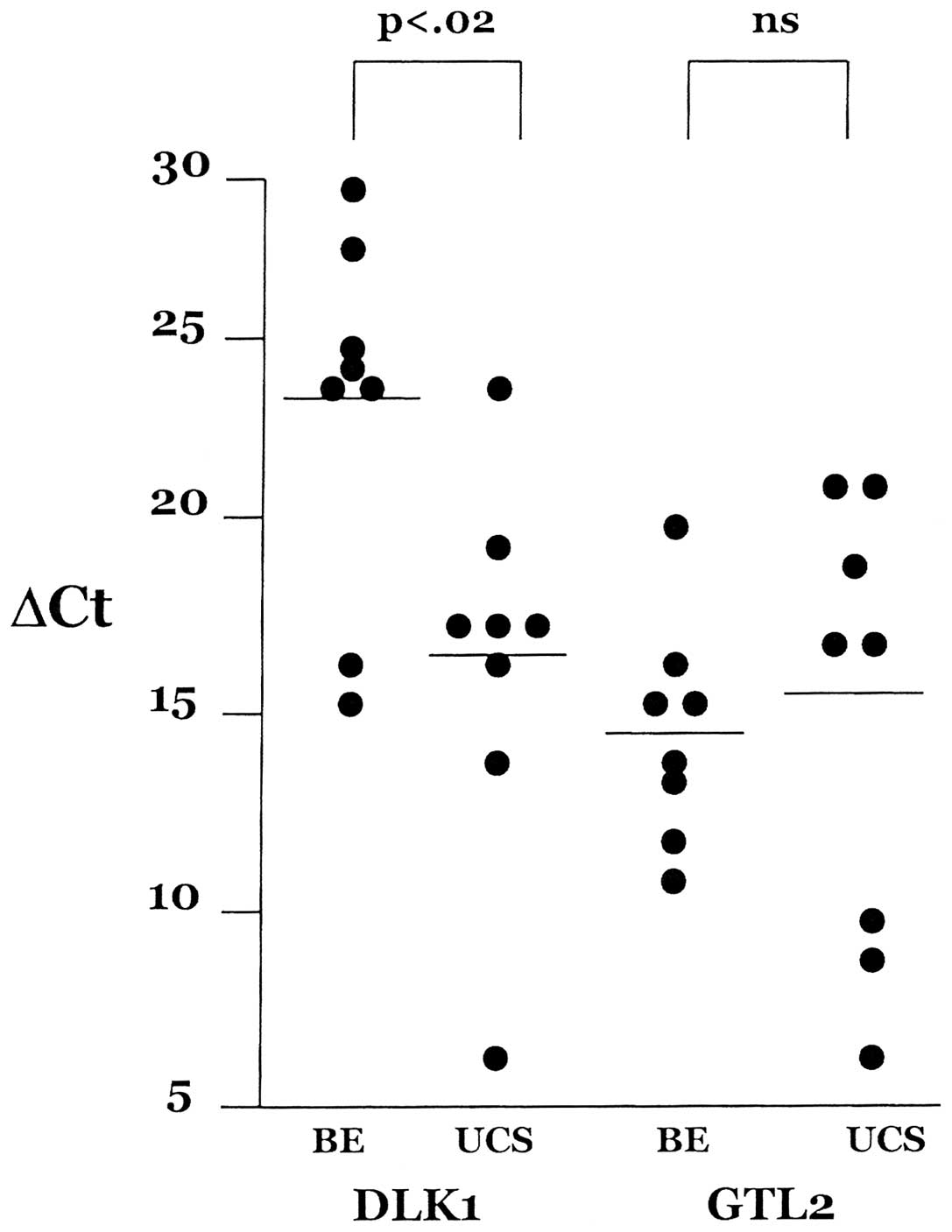
transcription, is shown in Fig. 2.
These data show that GTL2 expression is higher (lower ΔCt) than
DLK1 expression (higher ΔCt) in the benign endometrium panel
whereas levels are nearly equal in the UCS panel. In a reciprocally
imprinted locus a modest negative correlation of expression levels
of the two transcripts is anticipated. In the benign endometrium
panel, the correlation between DLK1 and GTL2 is r=−0.21 [not
significant (ns)] whereas we see a positive correlation in the UCS
panel, r=0.52 (ns). While GTL2 expression levels were not altered
in the UCSs as compared to benign endometrium, the DLK1 expression
level increased 92-fold (p<0.02) in the UCSs. This result
suggests that paternal chromosome expression (DLK1) has become
uncoupled from maternal chromosome expression (GTL2).
Previous studies have shown that reciprocal
expression of DKL1 and GTL2 is regulated in part by the methylation
status of a DMR located between the two loci (14). Bisulfite sequencing of the
DKL1/GTL2 DMR in the eight carcinosarcomas and five benign
endometrium samples revealed no change in methylation patterns.
This implies that the change in expression of DLK1 in UCS tissues
versus benign endometrium is not attributable to altered
methylation.
Discussion
UCSs display a unique miRNA expression
signature
A recent TLDA-based miRNA expression survey of
uterine cancers including a small number of UCSs concluded that
these tumors display a unique miRNA dysregulation signature
compared with other uterine cancers, specifically endometrial
adenocarcinomas (19). Our data
reinforce and expand upon this observation. Specifically, we show
from our larger panel of miRNAs that, among the same 667 miRNAs
surveyed by us in both endometrial adenocarcinoma (18) and UCS, only nine of the 71 miRNAs
significantly dysregulated in endometrial adenocarcinoma compared
with benign endometrium overlap with the 114 miRNAs significantly
dysregulated in UCS compared with benign endometrium. Apart from
these few miRNAs, it is the unique miRNA signature of UCSs that
dominates the data presented here. A recent TLDA-based miRNA study
of UCSs focused upon the biphasic nature of these tumors (20). Using the A-set miRNAs, the present
study separated the epithelial from the mesenchymal components of
23 tumors and reported on a modest differential miRNA expression
signature observed in the two components. Not surprisingly, a
number of the miRNAs reported in that study also appear in the
present study. Among these is miR-29c which is more highly
expressed in the epithelial component but significantly
underexpressed overall compared with benign endometrium. This miRNA
has been shown to be involved in the regulation of the
extracellular matrix marker SPARC and other markers associated with
invasion and metastasis (21).
Another miRNA shown to be differentially expressed in UCSs and
underexpressed overall in UCSs compared with benign endometrium is
miR-23b which is also negatively associated with the
proliferation of cancer cells (22). One notable finding is that none of
the five members of the miR-200 family, known to be
intimately involved in epithelial to mesenchymal transition
(19,23–25)
and significantly overexpressed in the epithelial component of UCSs
(20) as well as in endometrial
adenocarcinomas (18), achieved
statistical significance in our data, although all five
miR-200 family members were more highly expressed in UCSs
than in benign endometrium.
miRNAs in the chromosome 14q32 imprinted
region are globally dysregulated
In the above-mentioned study on miRNA expression in
the epithelial and mesenchymal components of UCSs (20), 24 miRNAs were shown to be more
highly expressed in the mesenchymal component than in the
epithelial component. Of these, seven (29.2%) are from the
imprinted region at chromosome 14q32. This is a statistically
significant over-representation of the chromosome 14q32 imprinted
region miRNAs present on the A-set TLDA card (χ2=7.7,
p<0.05) and is consistent with the statistically significant
overrepresentation of miRNAs from this region in UCSs compared with
benign endometrium that we report here. Our data also demonstrate
that the dysregulation of miRNA expression among the UCSs is
distributed across the entire 251 kb breadth of the region.
However, the extent of this phenomenon is clearly contained within
the boundaries of the imprinted region as three miRNAs at the 5′
boundary, miR-342-5p, miR-342-3p and miR-345,
and the miRNA closest to the 3′ boundary, miR-203, are all
expressed at the same level in the UCSs as in the benign
endometrium. Future studies are necessary to understand the
mechanisms by which miRNA expression in this region is coordinately
regulated and dysregulated.
The correlated expression of miRNAs in the
chromosome 14q32 imprinted region has previously been reported. For
example, correlated overexpression was observed in acute myeloid
leukemia (26) and in
gastrointestinal stromal tumors without loss of chromosome 14q
(27). Cytogenetic effects, either
amplifications or deletions, are not at issue here as no such
events have ever been reported on chromosome 14 in uterine cancers
(28,29).
miRNAs in the chromosome 14q32 imprinted cluster
show uncharacteristically low complementarity with cellular mRNA
targets, suggesting that they are involved in a shared mechanism of
action involving general inhibition of translation rather than
degradation of specific mRNA targets (27). In mice, coordinated expression of
the entire cluster has been linked with a DMR between the imprinted
non-miRNA loci DLK1/GTL2 (30).
Our examination of the methylation status of this DMR in relation
to DLK1/GTL2 expression in our UCSs and benign endometrium clearly
shows that this cis-acting regulation is disrupted, and it
is likely that this regulatory disruption extends throughout the
region and results in the observed miRNA expression patterns as
well.
We have carried out an miRNA expression survey of
UCSs which confirms that these tumors display a largely unique
miRNA signature. Most striking is the observation that the entire
imprinted region at chromosome 14q32 is dysregulated with all but
one of the 61 miRNAs assayed overexpressed in UCSs compared with
benign endometrium and more than half (32 of 60) reaching
statistical significance. We find that this dysregulation includes
the reciprocally imprinted gene pair DLK1/GTL2 lying at the
upstream boundary of the region where, in spite of a ‘normal’
methylation pattern in the intergenic DMR, both members of the pair
are highly expressed. It is possible that this DMR regulates
expression of the entire region in cis (30). Coordinated
overexpression of such a large block of miRNAs may contribute to
the unique nature and aggressive behavior of these tumors. In
addition, the significant increase in DLK1 expression may also play
a role, as DLK1 has been shown to inhibit Notch signaling in
mesenchymal multipotent cells (31). Whatever the effect of increased
DLK1 expression coupled with the global overexpression of the
chromosome 14q32 miRNA cluster may turn out to be, understanding
the mechanism that leads to the dysregulation of multiple miRNAs at
once may present a unique opportunity to identify a therapeutic
target whose effects are widespread.
Acknowledgements
This study was supported by NIH Grant
RO1CA99908 to K.K.L. and in part by the Department of Obstetrics
and Gynecology Research Development Fund. We thank the University
of Iowa Carver College of Medicine DNA Core Facility, particularly
Garry Hauser and Mary Boes, for their invaluable assistance and Dr
Kristina W. Thiel for assistance in manuscript preparation.
References
|
1.
|
D'Angelo E and Prat J: Uterine sarcomas: A
review. Gynecol Oncol. 116:131–139. 2010.
|
|
2.
|
Evans HL, Chawla SP, Simpson C and Finn
KP: Smooth muscle neoplasms of the uterus other than ordinary
leiomyoma. A study of 46 cases with emphasis on diagnostic criteria
and prognostic factors. Cancer. 62:2239–2247. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kernochan LE and Garcia RL:
Carcinosarcomas (malignant mixed Müllerian tumor) of the uterus:
Advances in elucidation of biologic and clinical characteristics. J
Natl Compr Canc Netw. 7:550–557. 2009.
|
|
4.
|
Zelmanowicz A, Hildesheim A, Sherman ME,
et al: Evidence for a common etiology for endometrial carcinomas
and malignant mixed mullerian tumors. Gynecol Oncol. 69:253–257.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Silverberg SG, Major FJ, Blessing JA, et
al: Carcinosarcoma (malignant mixed mesodermal tumor) of the
uterus. A Gynecologic Oncology Group pathologic study of 203 cases.
Int J Gynecol Pathol. 9:1–19. 1990. View Article : Google Scholar
|
|
6.
|
Sreenan JJ and Hart WR: A pathologic study
of 29 metastatic tumors: Further evidence for the dominant role of
the epithelial component and the conversion theory of histogenesis.
Am J Surg Pathol. 19:666–674. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Jin Z, Ogata S, Tamura G, et al:
Carcinosarcomas (malignant mullerian mixed tumors) of the uterus
and ovary: A genetic study with special reference to histogenesis.
Int J Gynecol Pathol. 22:368–373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Wada H, Enomoto T, Fujita M, et al:
Molecular evidence that most but not all carcinosarcomas of the
uterus are combination tumors. Cancer Res. 57:5379–5385.
1997.PubMed/NCBI
|
|
9.
|
Abeln EC, Smit VT, Wessels JW, et al:
Molecular genetic evidence for the conversion hypothesis of the
origin of malignant mixed Müllerian tumors. J Pathol. 183:424–431.
1997.PubMed/NCBI
|
|
10.
|
Fujii H, Yoshida M, Gong ZX, et al:
Frequent genetic heterogeneity in the clonal evolution of
gynecological carcinosarcoma and its influence on phenotypic
diversity. Cancer Res. 60:114–120. 2000.PubMed/NCBI
|
|
11.
|
Taylor NP, Zighelboim I, Huettner PC, et
al: DNA mismatch repair and TP53 defects are early events in
uterine carcinosarcoma tumorigenesis. Mod Pathol. 19:1333–1338.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Melo SA and Esteller M: Dysregulation of
microRNAs in cancer: Playing with fire. FEBS Lett. 585:2087–2099.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Rovira C, Güida MC and Cayota A: MicroRNAs
and other small silencing RNAs in cancer. IUBMB Life. 62:859–868.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kircher M, Bock C and Paulsen M:
Structural conservation versus functional divergence of maternally
expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC
Genomics. 9:3462008. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Guens E, De Temmerman N, Hilven P, et al:
Methylation analysis of the intergenic differentially methylated
region of DLK1-GTL2 in human. Eur J Hum Genet. 15:352–361. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Guo W, Sarkar SK and Peddada SD:
Controlling false discoveries in multidimensional directional
decisions, with applications to gene expression data on ordered
categories. Biometrics. 66:485–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Devor EJ, Hovey AM, Goodheart MJ,
Ramachandran S and Leslie KK: microRNA expression profiling of
endometrial endometrioid adenocarcinomas and serous adenocarcinomas
reveals profiles containing shared, unique and differentiating
groups of microRNAs. Oncol Rep. 26:995–1002. 2011.
|
|
19.
|
Ratner ES, Tuck D, Richter C, et al:
MicroRNA signatures differentiate uterine cancer tumor subtypes.
Gynecol Oncol. 118:251–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Castilla MA, Moreno-Bueno G, Romero-Perez
L, et al: Micro-RNA signature of the epithelial-mesenchymal
transition in endometrial carcinosarcoma. J Pathol. 223:72–80.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Sengupta S, den Boon JA, Chen IH, et al:
MicroRNA 29c is down-regulated in nasalpharyngeal carcinomas,
up-regulating mRNAs encoding extracellular matrix proteins. Proc
Natl Acad Sci USA. 105:5874–5878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Salvi A, Sabelli C, Moncini S, et al:
MicroRNA-23b mediates urokinase and c-met downmodulation and a
decreased migration of human hepatocellular carcinoma cells. FEBS
J. 276:2966–2982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Cochrane DR, Spoelstra NS, Howe EN,
Nordeen SK and Richer JK: MicroRNA-200c mitigates invasiveness and
restores sensitivity to microtubule-targeting chemotherapeutic
agents. Mol Cancer Ther. 8:1055–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Gregory P, Bert A, Paterson E, et al: The
miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Vrba L, Jensen TJ and Garbe JC: Role for
DNA methylation in the regulation of miR-200c and miR-141
expression in normal and cancer cells. PLoS ONE. 5:e86972010.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Dixon-McIver A, East P, Mein CA, et al:
Distinctive patterns of microRNA expression associated with
karyotype in acute myeloid leulaemia. PLoS ONE. 3:e21412008.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Haller F, von Heydebreck A, Zhang JD, et
al: Localization- and mutation-dependent microRNA (miRNA)
expression signatures in gastrointestinal stromal tumours (GISTs),
with a cluster of co-expressed miRNAs located at 14q32.31. J
Pathol. 220:71–86. 2010. View Article : Google Scholar
|
|
28.
|
Micci F, Tiexeira MR, Haugom L, Abeler VM
and Heim S: Genomic aberrations in carcinomas of the uterine
corpus. Genes Chrom Cancer. 40:229–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Salvesen HB, Carter SL, Mannelqvist M, et
al: Integrated genomic profiling of endometrial carcinoma
associates aggressive tumors with indicators of PI3 kinase
activation. Proc Natl Acad Sci USA. 106:4834–4839. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Seitz H, Royo H, Bortolin M-L, et al: A
large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2
domain. Genome Res. 14:1741–1748. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Sánchez-Solana B, Nueda ML, Ruvira MD, et
al: The EGF-like proteins DLK1 and DLK2 function as inhibitory
non-canonical ligands of NOTCH1 receptor that modulate each other's
activities. Biochim Biophys Acta. 1813:1153–1164. 2011.PubMed/NCBI
|