Introduction
MicroRNAs (miRNAs) are endogenous, single-stranded
non-coding RNAs that consist of approximately 22 nucleotides. They
play important regulatory roles at the post-transcriptional level
by binding to their targeted mRNAs. They either block their
translation or initiate their degradation, according to the degree
of complementarity with their targets (1). In addition to controling
hematopoiesis, miRNAs play key regulatory roles in a diverse range
of pathways, including developmental timing, cell differentiation,
apoptosis, cell proliferation and organ development (2). Direct involvement of miRNAs in cancer
has been suggested by studies demonstrating that several miRNAs are
localized in genomic regions associated with cancer, such as
breakpoint regions in chromosome aberrations involving oncogenes or
tumor-suppressor genes, minimal regions of loss of heterozygosity,
minimal regions of amplification, and at loci close to fragile
sites and integration sites of the human papillomavirus (3). miRNAs are proposed to play a direct
role in oncogenesis, as they can function as both oncogenes and
tumor-suppressor molecules. Currently, at least three mechanisms
are understood whereby miRNAs are deregulated in cancer: i)
chromosomal lesions at regions encoding microRNAs, ii) defects in
the miRNA biosynthetic pathway machinery, and iii) epigenetic
regulation (4). miRNAs also have
been demonstrated to have diagnostic, prognostic and therapeutic
potential in cancer (5–7).
Diffuse large B-cell lymphoma (DLBCL) is the most
common adult lymphoma accounting for nearly 40% of all lymphoid
tumors (8). Gene expression and
immunohistochemical studies of this clinically heterogeneous
disease have revealed the presence of at least two distinct
subtypes of DLBCL: germinal centre B cell-like (GCB) and
non-germinal centre-like (non-GCB) (9,10). A
hallmark of non-GCB DLBCL biology is the constitutive activation of
the NF-κB pathway, which promotes proliferation and differentiation
and suppresses apoptosis (11).
miR-155 and miR-146a are located on chromosome 21q21 and 5q33
dividedly. These two miRNAs are both thought to be regulated by
NF-κB (12–14).
Formalin-fixed/paraffin-embedded (FFPE) tissue
samples are the most readily available archival material. They
represent an invaluable source for the study of human disease. In
contrast to traditional gene expression studies that require
difficult to obtain freshly frozen clinical material, miRNAs can be
successfully isolated from routinely processed formalin-fixed
material, because of their relative resistance to RNase degradation
and their small size. Such samples, which have been matched to
frozen tissue, give remarkably similar results by real-time
quantitive PCR (RTQ-PCR) and microarray analysis (15,16).
Despite new drugs, such as rituximab, that have been
used in treatment regimes, the majority of patients succumb to this
aggressive disease, especially those with the non-GCB subtype
(17). Thus, it is necessary to
discover new potential diagnostic and prognostic markers for
clinical use. In this study, we investigated the expression levels
of miR-155 and miR-146a in FFPE tissue samples of DLBCL patients. A
correlation between miR-155 and miR-146a expression levels and
patient outcomes (complete remission rate, overall response rate
and prognosis-free survival) with DLBCL subtypes was found by
multivariate analysis.
Materials and methods
Patient samples
FFFPE biopsy samples of 90 of de novo DLBCL
cases and 31 reactive hyperplasia lymphoid node cases (16 males, 15
females; median age 45.3 years) were obtained from the Pathology
Department of Renji Hospital. All samples were collected at the
time of initial diagnosis (prior to treatment). A summary of DLBCL
de novo patient details are provided in Table I. Informed consent was provided
according to the Declaration of Helsinki. The study protocol was
approved by the Medical Ethics Committee of Renji Hospital.
 | Table I.Patient characteristics at initial
diagnosis (n=90). |
Table I.
Patient characteristics at initial
diagnosis (n=90).
| Clinical
feature | No. | GCB n (%) | non-GCB n (%) | P-value |
|---|
| Age (years) | | | | 0.222 |
| <60 | 54 | 15 (71.4) | 39 (56.5) | |
| ≥60 | 36 | 6 (28.6) | 30 (43.5) | |
| Gender | | | | 0.689 |
| Female | 48 | 12 (57.1) | 36 (52.2) | |
| Male | 42 | 9 (42.9) | 33 (47.8) | |
| Stage | | | | 0.363 |
| I | 9 | 4 (19.0) | 5 (7.2) | |
| II | 31 | 8 (38.1) | 23 (33.3) | |
| III | 24 | 4 (19.0) | 20 (29.0) | |
| IV | 26 | 5 (23.8) | 21 (30.4) | |
| ECOG PS | | | | 0.098 |
| 0–1 | 68 | 19 (90.5) | 49 (73.1) | |
| ≥2 | 22 | 3 (9.5) | 19 (26.9) | |
| LDH | | | | 0.346 |
| Normal | 61 | 16 (76.2) | 45 (65.2) | |
| >Normal | 29 | 5 (23.8) | 24 (34.8) | |
| Extranodal
involvement | | | | 0.005 |
| 0–1 | 65 | 20 (95.2) | 45 (63.1) | |
| ≥2 | 25 | 1 (4.8) | 24 (36.9) | |
| B symptom | | | | 0.606 |
| No | 47 | 12 (57.1) | 35 (50.7) | |
| Yes | 43 | 9 (42.9) | 34 (49.3) | |
| IPI status | | | | 0.004 |
| 0–1 | 44 | 16 (76.2) | 28 (40.6) | |
| ≥2 | 46 | 5 (23.8) | 41 (59.4) | |
|
β2-MG | | | | 0.233 |
| Normal | 53 | 13 (68.4) | 40 (53.0) | |
| >Normal | 37 | 6 (31.6) | 31 (47.0) | |
| Treatment
protocol | | | | 0.750 |
| R-CHOP | 49 | 14 (66.7) | 35 (50.7) | |
| CHOP | 41 | 7 (33.3) | 34 (49.3) | |
Immunohistochemical staining
DLBCL de novo cases were classified
immunohistochemically as GCB- or non-GCB-type using monoclonal
antibodies against CD10, BCL6 and melanoma-associated antigen
(mutated) 1 (MUM1) as previously described (15). Monoclonal antibody against c-myc
(Shanghai Long Island Biotech, Shanghai, China) was also used to
examine the protein expression. Relevant ethical permission was
obtained for the use of all samples.
RNA purification and RTQ-PCR
Total RNA was purified from 4×20 μm FFPE
sections using the Recoverall kit from Ambion (Huntingdon, UK) in
accordance with the manufacturer’s instructions. TaqMan miRNA
assays were used to detect and quantify mature miR-155 and miR-146a
as previously described (18)
using TaqMan® MicroRNA Reverse Transcription kit
(4366597; Applied Biosystems, Foster City, CA, USA) and PCR 9700
sequence detection system (Applied Biosystems). Normalization was
performed with U48. RTQ-PCR was performed in triplicate, including
no-template controls. The 2−ΔCt method was used in the
analysis of PCR data [ΔCt = mean Ct (microRNA of interest) - mean
Ct (U48)].
Chemotherapy protocol
De novo patients with DLBCL were treated with
the CHOP (cyclophosphamide 750 mg/m2 on day 1,
vincristine 1.4 mg/m2 on day 1, epirubicin 60
mg/m2 on day 1 and prednisone 60 mg/m2 on
days 1–5) or R-CHOP (rituximab 375 mg/m2 on day 0,
cyclophosphamide 600 mg/m2 on day 1, vincristine 1.4
mg/m2 on day 1, epirubicin 60 mg/m2 on day 1
and prednisone 60 mg/m2 on days 1–5) regimen.
Statistical and survival analysis
miRNA expression levels and clinicopathological
parameters were analyzed using SPSS 16.0 (SPSS Inc., Chicago, IL,
USA). The χ2 test, the Mann-Whitney U test, Kaplan-Meier
survival analysis and multivariate Cox regression analysis were
carried out to assess progression-free survival (PFS) times and
overall survival (OS) time of the de novo DLBCL cases.
P-value <0.05 was considered to denote statistical significance.
Receiver operating characteristic (ROC) analysis was performed to
determine an optimal cutoff value of miRNA expression
(2−ΔCt) to split the patients into a low-expression
group and a high-expression group. PFS was calculated as the time
of diagnosis to the date of clinical relapse, death or last
contact. OS was defined as the date of diagnosis to death from any
cause. Data were censored if the patients were alive at last
follow-up. Mean follow-up time was 26.3 months (range 6–83). Curves
were compared by univariate (log-rank) analysis using GRAPHPAD
Prism version 4.00 (La Jolla, CA, USA).
Results
miR-155 and miR-146a expression levels of
DLBCL patients are distinct from reactive hyperplasia lymphoid
nodes and differ between GCB and non-GCB subtypes of DLBCL
The expression levels of both tumor-associated
miRNAs (miR-155 and miR-146a) in biopsy samples from DLBCL patients
(n=90) were compared to those of reactive hyperplasia lymphoid
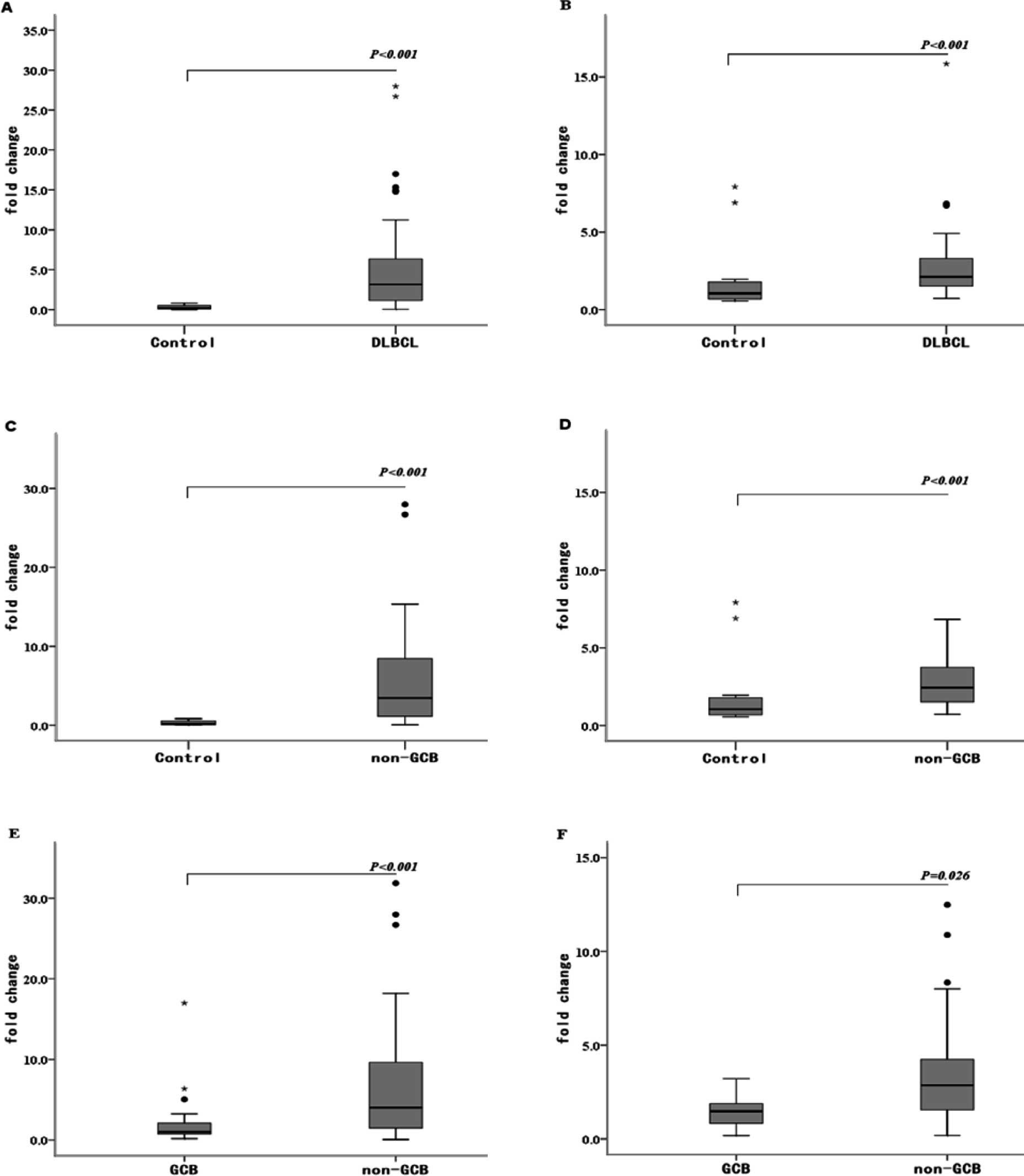
nodes (n=31) by RTQ-PCR. The levels of both miRNAs were
up-regulated in the DLBCL patients (P<0.001) (Fig. 1A and B), corresponding to an
average fold-change of 18.97 and 2.46, respectively.
To ascertain whether the expression of miR-155 and
miR-146a is DLBCL immunophenotype-specific, we measured the
expression levels of these miRNAs in 21 patients with GCB subtype
DLBCL and in 69 patients with non-GCB subtype by RTQ-PCR. The
expression levels of miR-155 and miR-146a were higher in the
non-GCB DLBCL patients than in reactive hyperplasia lymphoid nodes
(P<0.001) (Fig. 1C and D),
corresponding to an average fold-change of 20.80 and 2.13,
respectively. We also found that the expression levels of miR-155
and miR-146a were higher in the non-GCB DLBCL patients compared to
the GCB DLBCL patients, corresponding to an average fold-change of
1.58 and 1.69, respectively (P<0.001 and P=0.026, respectively)
(Fig. 1E and F).
miR-146a expression levels are associated
with miR-155 expression levels and clinical characteristics in the
de novo DLBCL patients
Clinical data, including lactate dehydrogenase
(LDH), β 2 microglobulin (β2-MG), c-myc, International
Prognostic Index (IPI) status and Eastern Cooperative Oncology
Group physical score (ECOG PS), were available for 90 cases of the
de novo DLBCL patients. The cases were split into two groups
according to low IPI scores (0 and 1) or high IPI scores (2, 3 and
4), and low ECOG PS (0 and 1) or high scores (2, 3 and 4). We found
that miR-146a expression levels were associated with miR-155
expression levels (r2=0.323, P<0.001), LDH
(r2=0.215, P<0.001), β2-MG
(r2=0.118, P=0.001), c-myc (r2=0.175,
P<0.001), IPI (r2=0.337, P<0.001) and ECOG PS
(r2=0.227, P=0.001) in the same patients (Fig. 2A–F). miR-146a expression levels
showed no association with gender, age, clinical stage, B symptom
and extranodal involvement. Meanwhile, there was no association
between miR-155 expression levels and all the clinical
characteristics mentioned above.
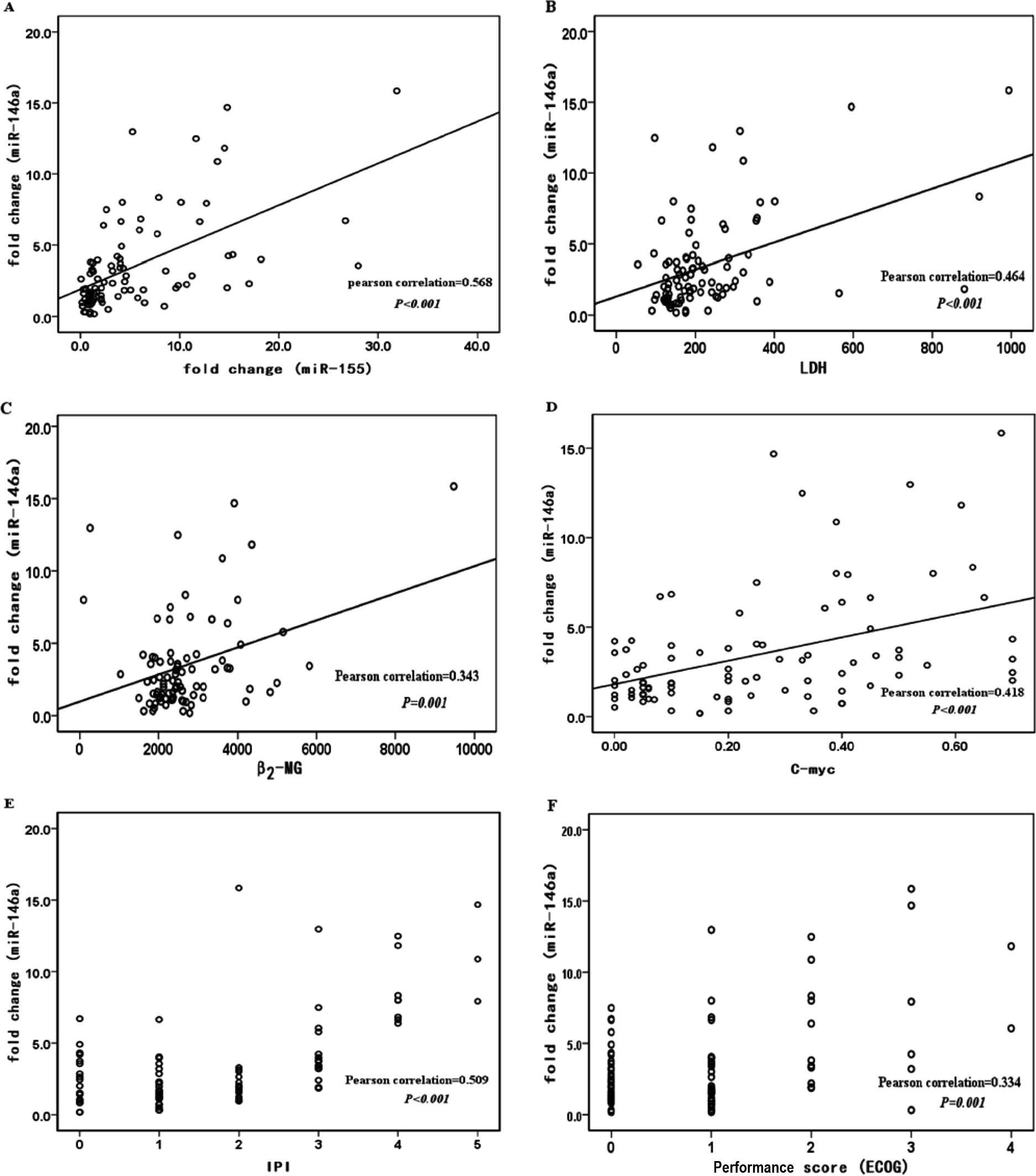 | Figure 2.Expression levels of miR-146a are
correlated with miR-155 and clinical patient characteristics in
formalin-fixed/paraffin-embedded samples of DLBCL patients. Linear
regression lines are shown. (A) miR-146a and miR-155,
r2=0.323, P<0.001; (B) miR-146a and LDH,
r2=0.215, P<0.001; (C) miR-146a and β2-MG,
r2=0.118, P=0.001; (D) miR-146a and c-myc,
r2=0.175, P<0.001; (E) miR-146a and ECOG performance
score, r2=0.227, P=0.001; (F) miR-146a and IPI status,
r2=0.337, P<0.0018. |
miR-155 and miR-146a expression is
associated with clinical outcome in de novo DLBCL patients
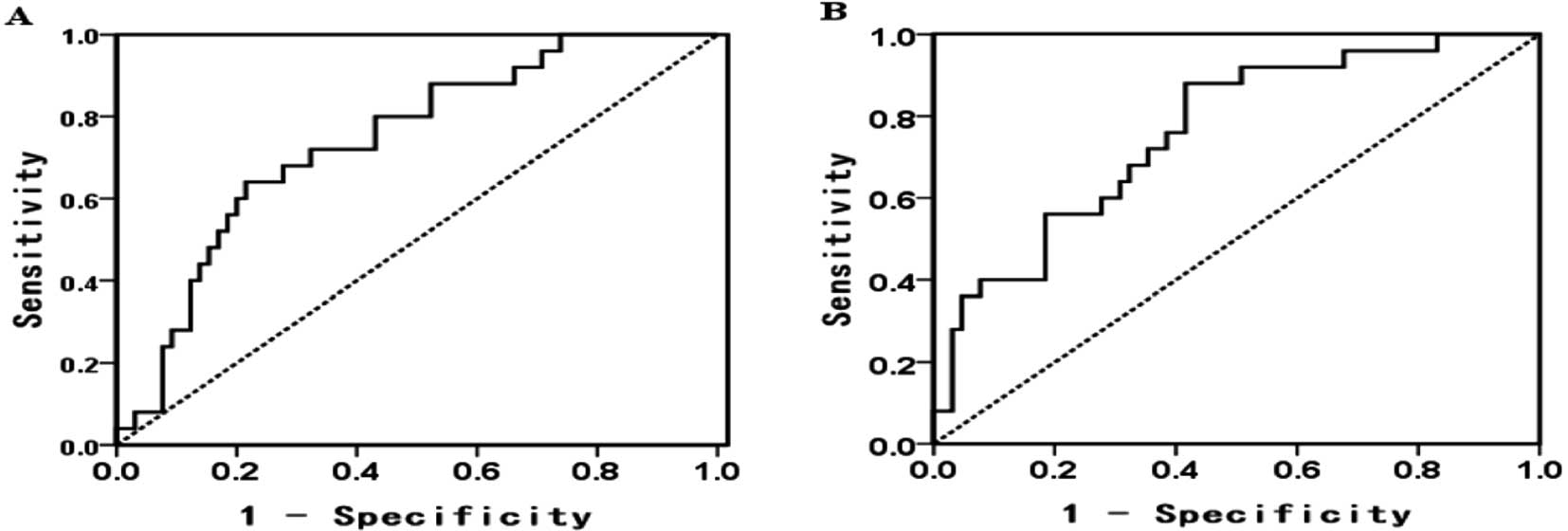
The cutoff value, sensitivity and specificity of
miR-155/miR-146a were 3.9840/2.0196, 80/88% and 58.5/56.9%,
respectively (Fig. 3A and B).
De novo DLBCL patients with low expression of miR-155 and
miR-146a were found to be associated with a high complete remission
(CR) rate (miR-155 76.5 vs. 43.8%, P=0.032; miR-146a 83.3 vs. 50%,
P=0.013) and a high overall response (OR) rate (miR-155 85.3 vs.
50%, P=0.008; miR-146a 87.5 vs. 61.5%, P=0.037).
To assess the potential prognostic impact of these
two miRNAs in de novo DLBCL cases, we performed a
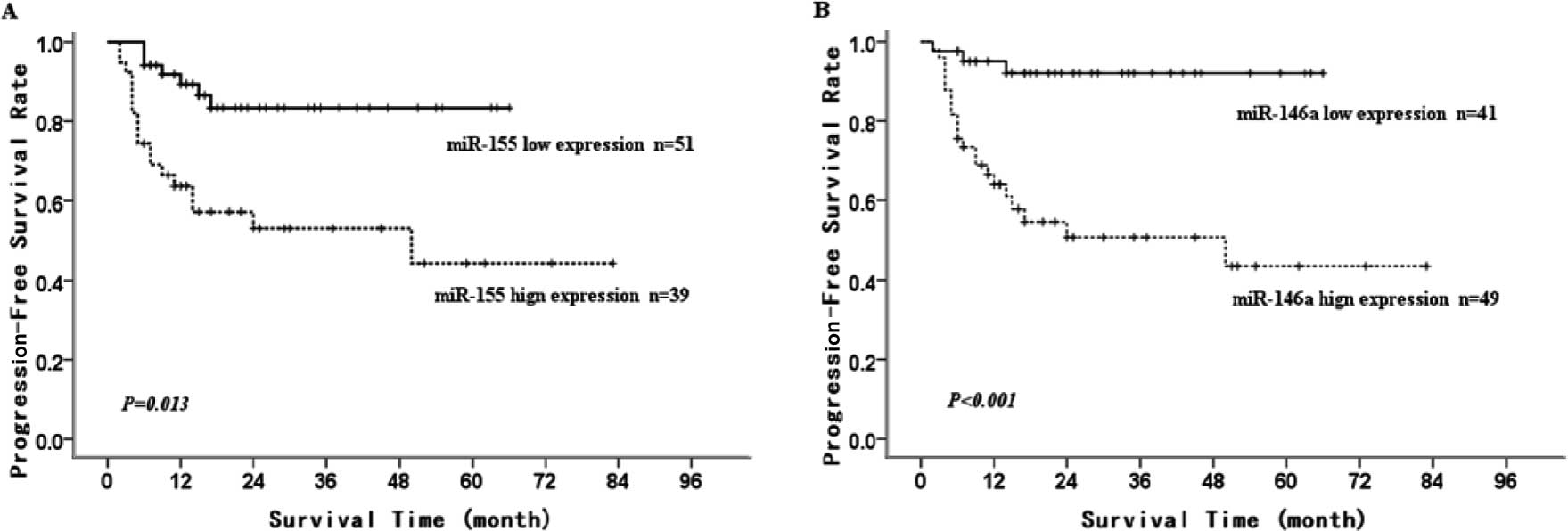
retrospective analysis. Using Kaplan-Meier survival (univariate)
analysis, we found that low miR-155 and miR-146a expression was
associated with a longer 5-year PFS in the de novo DLBCL
cases (83.3±5.9 vs. 44.3±10.8%, P=0.013; 92±4.4 vs. 43.5±9.7%,
P<0.001; Fig. 4A and B). We
further examined the effect of various factors that may affect
prognostic outcome in this cohort (i.e., gender, age, clinical
stage, B symptom and extranodal involvement IPI stage and ECOG PS),
as well as miRNA expression levels by multivariate Cox proportional
hazard regression analysis. We found that the expression levels of
miR-155 and IPI status, but not the expression levels of miR-146a,
were statistically significant independent indicators of prognosis
in this cohort (P<0.05) (Table
II).
 | Table II.Cox regression analysis of the
prognostic factors associated with progression-free survival of the
DLBCL patients. |
Table II.
Cox regression analysis of the
prognostic factors associated with progression-free survival of the
DLBCL patients.
| Cox regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| miR-155 | −1.345 | 0.260
(0.085–0.801) | 0.019 |
| miR-146a | 0.113 | 1.119
(0.977–1.282) | 0.104 |
| IPI status | −1.669 | 0.188
(0.052–0.689) | 0.012 |
Expression of miR-155 is an independent
indicator for chemotherapy protocol selection in the de novo
DLBCL
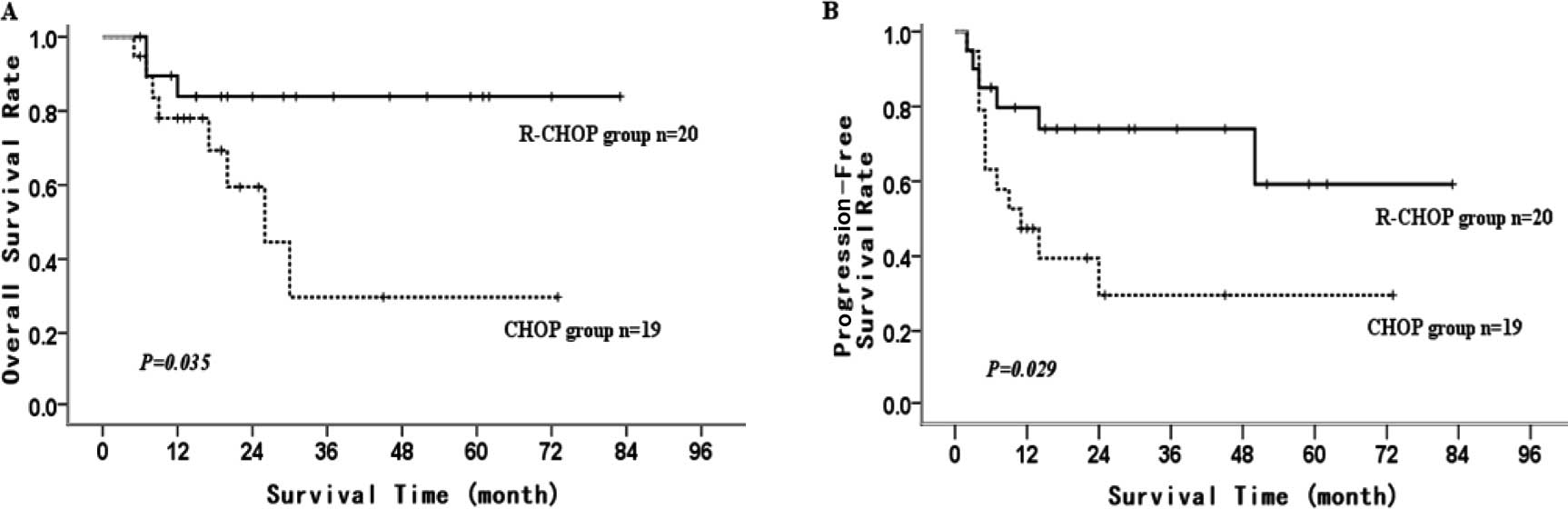
Compared to CHOP protocol, the patients in the high
miR-155 expression group who chose R-CHOP protocol for treatment
achieved better 5-year OS and 5-year PFS (CHOP vs. RCHOP: 5-year OS
29.7±16.4 vs. 83.9±8.5%, P=0.035; 5-year PFS 29.6±12.4 vs.
59.2±15.5%, P=0.029) (Fig. 5A and
B). This difference was not noted in the low miR-155 expression
group. miR-146a expression was not an indicator of chemotherapy
protocol selection in the de novo DLBCL patients (data not
shown).
Discussion
It is believed that 10–30% of all human genes are a
target for miRNA regulation (19).
Recent evidence shows that the expression of miRNA genes is
deregulated in human cancer. miRNA overexpression may result in the
down-regulation of tumor-suppressor genes, whereas their
underexpression may lead to oncogene up-regulation (20–22).
The potential importance of miRNAs in cancer is also implied by the
finding that the majority of human miRNAs are located at
cancer-associated genomic regions. Cancer-associated miRNAs have
oncogenic properties (3).
Moreover, it has been suggested that miRNA expression profiling may
distinguish cancers according to diagnostic type and developmental
stage of the tumor to a greater degree of accuracy than traditional
gene expression analysis (23).
Although some progress has been made in identifying
lymphoma-associated miRNAs by profiling a large number of lymphoma
cell lines (24), clinical samples
exhibit a discrete miRNA pattern compared to cell lines,
reinforcing the need for studies on clinical material (25).
miR-155 and miR-146a as diagnostic tools
in DLBCL patients
Seeking new tools for the diagnosis and management
of lymphoma has been an ongoing endeavor. miRNAs due to their small
size are relatively resistant to RNase degradation and, unlike
mRNA, can be successfully recovered intact from archival FFPE
material (15,16,26).
Furthermore, miRNAs can be detected in biological fluids, including
serum and plasma (6). All of these
make miRNAs appropriate candidates for lymphoma research.
Georgantas et al proposed that miR-155,
miR-146a and another four microRNAs may act to prevent early-stage
progenitor cells from differentiating to a more mature stage based
on a bioinformatics approach (27). In this study, we found that miR-155
and miR-146a expression levels in DLBCL patient tissue were higher
than levels in reactive hyperplasia lymphoid nodes. miR-155
expression has also been shown by other groups to be more highly
expressed in DLBCL patients, particularly in patients with a
non-GCB immunophenotype (16,28,29).
Yet, the research of miR-146a in lymphoma is limited.
Down-regulation of miR-146a expression by the c-Myc oncogenic
transcription factor has been previously observed in human and
mouse B-cell lymphomas (30).
Recent studies have evaluated the expression levels of miR-146a in
acute lymphocytic leukemia and chronic lymphocytic leukemia
(31–33). Their findings imply that miR-146a
expression involved in deletion or amplification may be cell
type-specific. The expression levels of miR-146a are not coincident
among hematopoietic malignancies.
Differential expression of miR-155 and miR-146a
between DLBCL patient lymphoid nodes and reactive hyperplasia
lymphoid nodes suggest that miR-155 and miR-146a may be a potential
tool for future molecular diagnostics in DLBCL.
miR-155 and miR-146a as prognostic
biomarkers in DLBCL patients
Despite the fact that up to 80% of DLBCL patients
reach a complete remission using a rituximab-based chemotherapy
protocol, the response to treatment remains variable. A substantial
proportion of DLBCL patients will still succumb to this disease
(8). We sought to identify
definite miRNAs that are associated with clinical outcome. Many
studies have shown that miRNA expression levels have potential
prognostic significance in malignant diseases, such as chronic
lymphocytic leukemia, DLBCL, lung and pancreatic cancer (5,16,23,34).
We first examined whether miR-155 and miR-146a were associated with
clinical prognostic factors, including LDH, β2-MG,
c-myc, IPI and ECOG PS. We found that the expression levels of
miR-146a, but not miR-155, were associated with these prognostic
factors. The significance of miR-155 expression is consistent with
the findings of Lawrie et al (6,16,25).
We also found that the expression of miR-146a was correlated with
miR-155. We know that the NF-κB transcription factor activity is
abnormal in DLBCL (11,13). The phenomena also provide evidence
that these two miRNAs may be regulated by the NF-κB transcription
factor (12,14). Recently Volinia et al
generated an Em/VH miR-155 transgenic mouse model. miR-146a
expression levels were up-regulated in miR-155 transgenic mice when
compared to the control wild-type mice. This suggests that miR-155
and miR-146a are involved in one complex microRNA network (35).
In order to ascertain whether miRNA expression is
able to predict outcome, we analyzed PFS and OS in the DLBCL de
novo patients. We found that low expression of miR-155 and
miR-146a was associated with a high CR rate, a high OR rate and
long PFS time. We carried out univariate analysis on the expression
levels of these miRNAs individually and found that miR-155 and IPI
were statistically significant independent prognostic indicators.
Former studies indicate that miR-155 is an independent prognostic
indicator only in non-GCB-type DLBCL patients (25,28,29).
We speculate that the high percentage of non-GCB patients in our
group may have caused this discrepancy. Recently, Korean doctors
found that NK/T cell lymphoma patients with low miR-146a expression
had a significantly poor prognosis in the clinic. miR-146a was
up-regulated in TH1 cells throughout murine hematopoietic
development (36). p53/TA-p73/p63
increased the expression of c-myc-suppressed miR-146a in B-cell
lymphogenesis (37). This seems to
contradict our results. We believe that besides the different cell
types, the effect of miRNAs on cell pathology and physiology is
likely to be complex for at least two reasons: i) their activity is
exerted in a one-to-many fashion, such that each miRNA controls the
translation of tens or even hundreds of different coding
messengers, and ii) a single messenger can be controlled by more
than one miRNA.
It is of note that a high expression level of
miR-155, but not miR-146a, was an independent indicator for
chemotherapy protocol selection in our study. Patients with high
expression of miR-155 received more survival benefits from
rituximab treatment. The reason is unclear. This may be related to
inflammatory cytokines produced by constant activity of the NF-κB
signaling pathway.
miR-155 and miR-146a as oncogenes in
DLBCL patients
miRNAs, such as miR-155 and miR-146a, are associated
with many types of cancer, including a wide range of solid and
hematological malignancies (38).
This phenomenon suggests that these miRNAs play a fundamental role
in the establishment of a general malignant phenotype.
Thai et al used both BIC/miR-155-deficient
and -knock-in mice to investigate the role of miR-155 in germinal
centre (GC) B-cell responses. They found a reduction in the total
numbers of GCB cells in the deficient mice and an increase in
numbers in the knock-in mice. In vitro activation of
wild-type B cells resulted in strong up-regulation of miR-155
expression. Immunization resulted in decreased antibody titers in
miR-155-deficient mice. It was also observed that activated
miR-155-deficient B cells expressed about a third of the normal
levels of TNF, suggesting that miR-155 controls B-cell activation
through control of cytokine production (39). Transgenic mice expressing miR-155
targeted to B cells were also shown to spontaneously develop
high-grade lymphomas (40). We
found an association between the expression level of miR-155 and
PFS in the studied patients; miR-155 was more highly expressed in
all lymphoma cases, with an average expression 18.97-fold higher
than the reactive hyperplasia lymphoid nodes.
Boldin et al suggested that miR-146a
functions as a tumor-suppressor gene. They found that aging
miR-146a-null mice developed frank tumors in lymphoid organs
(41). This conclusion is in
disagreement with our studies in DLBCL patients. We presume that
miR-146a activity can be influenced either by the reposition of
other genes close to its promoter regulatory regions (as c-myc
translocation in lymphoma), or by the relocalization of other
regulatory elements. The same gene, which behaves as an oncogene or
suppressor gene, also depends on the transcriptional and/or
post-transcriptional events.
In summary, our data suggest that miR-155 and
miR-146a may be diagnostic tools and prognostic indictors for DLBCL
patients. Genetic associations have already provided compelling
evidence that DLBCL subtypes represent discrete diseases that may
arise by distinct pathogenetic pathways (42). Additional studies, including the
characterization of miR-155 and miR-146a expression in a large
number of DLBCL patients, are required to clarify the underlying
basis for this association. Since miR-155 and miR-146a silence or
modulate gene expression in humans through the regulation of
transcription factors, an approach of targeted inhibition of
miR-155 and miR-146a may be explored as a potential therapeutic
target for controlling malignant lymphogenesis.
Acknowledgements
This study was supported by grants
from the Shanghai Municipal Commission of Sciences and Technology
fund (09ZR1418400), the Social Development fund of the Shanghai
Pudong District (PW2009D-5) and Key Discipline Project of Renji
Hospital, Shanghai Jiaotong University School of Medicine (RJ
4101306). The authors thank Dr Qing-Wen Xie for the assistance in
the statistical and survival analysis.
References
|
1.
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Kim VN: MicroRNA biogenesis: coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Lawrie CH: MicroRNA expression in lymphoid
malignancies: new hope for diagnosis and therapy? Cell Mol Med.
12:1432–1444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Calin GA, Ferracin M, Cimmino A, et al: A
microRNA signature associated with prognosis and progression in
chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumour associated microRNAs in
serum of patients with diffuse large B-cell lymphoma. Br J
Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kota SK and Balasubramanian S: Cancer
therapy via modulation of microRNA levels: a promising future. Drug
Discov Today. 15:733–740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Coiffier B: Diffuse large cell lymphoma.
Curr Opin Oncol. 13:325–334. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Alizadeh AA, Eisen MB, Davis RE, et al:
Distinct types of diffuse large B-cell lymphoma identified by gene
expression profiling. Nature. 403:503–511. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Hans CP, Weisenburger DD, Greiner TC, et
al: Confirmation of the molecular classification of diffuse large
B-cell lymphoma by immunohistochemistry using a tissue microarray.
Blood. 103:275–282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Davis RE, Brown KD, Siebenlist U, et al:
Constitutive nuclear factor kappaB activity is required for
survival of activated B cell-like diffuse large B cell lymphoma
cells. J Exp Med. 194:1861–1874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Thompson RC, Herscovitch M, Zhao I, et al:
NF-kappaB down-regulates expression of the B-lymphoma marker CD10
through a miR-155/PU.1 pathway. J Biol Chem. 286:1675–1682. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Braun T, Carvalho G, Fabre C, et al:
Targeting NF-kappaB in hematologic malignancies. Cell Death Differ.
13:748–758. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Taganov KD, Boldin MP, Chang KJ, et al:
NF-kappaB dependent induction of microRNA miR-146, an inhibitor
targeted to signaling proteins of innate immune responses. Proc
Natl Acad Sci USA. 103:12481–12486. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Li J, Smyth P, Flavin R, et al: Comparison
of miRNA expression patterns using total RNA extracted from matched
samples of formalin-fixed paraffin-embedded (FFPE) cells and snap
frozen cells. BMC Biotechnol. 7:362007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Lawrie CH, Soneji S, Marafioti T, et al:
MicroRNA expression distinguishes between germinal center B
cell-like and activated B cell-like subtypes of diffuse large B
cell lymphoma. Int J Cancer. 121:1156–1161. 2007. View Article : Google Scholar
|
|
17.
|
Nogai H, Dörken B and Lenz GJ:
Pathogenesis of non-Hodgkin’s lymphoma. Clin Oncol. 29:1803–1811.
2011.
|
|
18.
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantificiation of microRNAs by stem-loop RT-PCR.
Nucletic Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar
|
|
23.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Lawrie CH, Saunders NJ, Soneji S, et al:
MicroRNA expression in lymphocyte development and malignancy.
Leukemia. 22:1440–1446. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Lawrie CH, Chi JX, Taylor S, et al:
Expression of microRNAs in diffuse large B cell lymphoma is
associated with immunophenotype, survival and transformation from
follicular lymphoma. J Cell Mol Med. 13:1248–1260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Xi Y, Nakajima G, Gavin E, et al:
Systematic analysis of microRNA expression of RNA extracted from
fresh frozen and formalin-fixed paraffin-embedded samples. RNA.
13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Georgantas RW III, Hildreth R, Morisot S,
et al: CD34+ hematopoietic stem-progenitor cell microRNA
expression and function: a circuit diagram of differentiation
control. Proc Natl Acad Sci USA. 104:2750–2755. 2007.PubMed/NCBI
|
|
28.
|
Eis PS, Tam W, Sun L, et al: Accumulation
of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad
Sci USA. 102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Kluiver J, Poppema S, de Jong D, et al:
BIC and miR-155 are highly expressed in Hodgkin, primary
mediastinal and diffuse large B cell lymphomas. J Pathol.
207:243–249. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Chang TC, Yu D, Lee YS, et al: Widespread
microRNA repression by Myc contributes to tumorigenesis. Nat Genet.
40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Zhang H, Luo XQ, Zhang P, et al: MicroRNA
patterns associated with clinical prognostic parameters and CNS
relapse prediction in pediatric acute leukemia. PLoS One.
4:e78262009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Wang Y, Li Z, He C, et al: MicroRNA
expression signatures are associated with lineage and survival in
acute leukemias. Blood Cells Mol Dis. 44:191–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Visone R, Rassenti LZ, Veronese A, et al:
Karyotype-specific microRNA signature in chronic lymphocytic
leukemia. Blood. 114:3872–3879. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Roldo C, Missiaglia E, Hagan JP, et al:
MicroRNA expression abnormalities in pancreatic endocrine and
acinar tumors are associated with distinctive pathologic features
and clinical behavior. J Clin Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Volinia S, Galasso M, Costinean S, et al:
Reprogramming of miRNA networks in cancer and leukemia. Genome Res.
20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Monticelli S, Ansel KM, Xiao C, et al:
MicroRNA profiling of the murine hematopoietic system. Genome Biol.
6:R712005. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Boominathan L: The guardians of the genome
(p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNA
network. Cancer Metastasis Rev. 29:613–639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Thai TH, Calado DP, Casola S, et al:
Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Costinean S, Zanesi N, Pekarsky Y, et al:
Pre-B cell proliferation and lymphoblastic leukemia/highgrade
lymphoma in E (mu)-miR155 transgenic mice. Proc Natl Acad Sci USA.
103:7024–7029. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Boldin MP, Taganov KD, Rao DS, et al:
miR-146a is a significant brake on autoimmunity,
myeloproliferation, and cancer in mice. J Exp Med. 208:1189–1201.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Lenz G, Wright GW, Emre NCT, et al:
Molecular subtypes of diffuse large B-cell lymphoma arise by
distinct genetic pathways. Proc Natl Acad Sci USA. 105:13520–13525.
2008. View Article : Google Scholar : PubMed/NCBI
|