Introduction
Survivin, a new member of the inhibitor of apoptosis
protein (IAP) family, both inhibits apoptosis and regulates the
cell cycle. It is overexpressed in most tumor tissues, but hardly
expressed in most normal tissues. Therefore, it may be an
attractive target for antitumor gene therapy (1–3). As
an important technique for the research on gene function, RNA
interference (RNAi) technique is an economical, fast and highly
efficient technique for knocking down gene expression (4,5). In
the present study, we constructed a survivin small interfering RNA
(siRNA) lentiviral vector, assessed its effect on malignant
biological behaviors (proliferation, invasion and metastasis) of
lung cancer and verified the function of survivin in the
carcinogenesis and development of lung cancer, in order to carry
out further research on the mechanisms of this gene.
Materials and methods
Materials
A549 and 293T lung cancer cell strains were
purchased from the Shanghai Cell Resource Center of the Chinese
Academy of Sciences. pGC-LV vector, pHelper 1.0 vector and pHelper
2.0 vector were purchased from Shanghai GeneChem Co., Ltd. Qiagen
plasmid Midi kit was purchased from USA Qiagen Co. tryptase was
purchased from Shanghai Chemical Reagent Co., Ltd. E. coli
DH5α, SYBR Master Mixture, T4 DNA ligase and Taq DNA
polymerase were purchased from Takara Co. (Japan). AgeI and
EcoRI restriction enzymes were purchased from New England
Biolabs (NEB) Co. Liposome Lipofectamine 2000 was purchased from
Invitrogen Co (USA). DMSO was purchased from Shanghai Biological
Reagent Co., Ltd. DMEM culture medium was purchased from Gibco Co.
(USA). FBS was purchased from Shanghai Weike Biochemical Reagent
Co., Ltd. Oligo(dT) was purchased from Sangon Biotech (Shanghai)
Co., Ltd. M-MLV reverse transcriptase and dNTP were purchased from
Promega Co. (USA). Rabbit survivin monoclonal antibody against the
human was purchased from CTS (USA). The S-P IHC kit was purchased
from Fuzhou Maixin Bio Co., Ltd.
Thirty lung cancer specimen and corresponding
surgical margin specimens were obtained from the Fujian Medical
University Union Hospital.
Detection of the expression of survivin
in tissue specimens by immunohistochemistry (IHC)
The paraffin specimens were routinely dewaxed.
Antibodies were fixated by microwave. Endogenous hydrogen
peroxidase was blocked by incubation in 3%
H2O2 for 10 min. The specimens were then
incubated with rabbit survivin monoclonal antibody against human
for 2 h at 37°C. After rinsing with PBS, the specimens were
incubated with goat antibody against rabbit labeled with biotin for
15 min at room temperature. The specimens were rinsed with PBS
again, and were further incubated with streptavidin-hydrogen
peroxidase for 15 min at room temperature. After rinsing with PBS,
the specimens were processed with DAB. Then, the specimens were
redyed with hematoxylin, transparented, sealed and observed under a
light microscope. Tissue positive for survivin was used as the
positive control, and specimens for which PBS replaced the primary
antibody were used as the negative control.
Construction of survivin-siRNA lentiviral
vector and screening
We designed the target sequence according to the
survivin mRNA sequence in GenBank and the principles of siRNA
design. Four pairs of siRNA targeted with survivin and one pair of
siRNA with negative control were designed (Table I). The synthesis of siRNA was
carried out by the Shanghai GeneChem Co., Ltd. (6,7). The
siRNA was subsequently transfected into 293T cells according to the
guidelines for Lipofectamine 2000 from Invitrogen. The transfection
results were observed under a fluorescence microscope 24 h later.
The cells were collected 36 h later. At the same time, the protein
was extracted. The most efficient siRNA was chosen by western
blotting. The result showed that the first pair of siRNA was the
most efficient. Double-stranded DNA fragment, with cohesive termini
of the AgeI and EcoRI restriction enzymes, and the
hairpin sequence of 5′-GGCTGGCTT
CATCCACTGCTTCAAGAGAGCAGTGGATGAAGCCAG CC-3′ inside, was synthesized
in vitro. The fragment was ligated into pGC-LV, and then
transfected into E. coli DH5α. After amplifying and
screening, the construction was confirmed successful by sequencing.
The plasmid was extracted, and the survivin-siRNA lentiviral vector
was recombined. The A549 lung cancer cells transfected with the
survivin-siRNA lentiviral vector were considered the knockdown
group (KD). The cells with the negative control sequence were
considered to be the negative control (NC) and the cells with no
sequence were considered as the control group (CON).
 | Table I.siRNA sequence-specific to
survivin. |
Table I.
siRNA sequence-specific to
survivin.
| siRNA no. | Sequence |
|---|
| Survivin 1 |
GGCTGGCTTCATCCACTGC |
| Survivin 2 |
GGACCACCGCATCTCTACA |
| Survivin 3 |
GAAAGTGCGCCGTGCCATC |
| Negative control |
TTCTCCGAACGTGTCACGT |
Detection of the expression of survivin
mRNA by real-time quantitative PCR (RT-qPCR) test
Total RNA was extracted by TRIzol and
reverse-transcripted into cDNA. Then, the RNA was detected by
RT-qPCR. Survivin primer and actin primer (as internal control)
were synthesized by the Shanghai GeneChem Co., Ltd. The sequences
are shown in Table II. The
reaction conditions of PCR were as follows: pre-denaturation at
95°C for 15 sec; denaturation at 95°C for 5 sec; annealing at 60°C
for 30 sec; 45 cycles were completed. The mixture was denatured for
1 min at the end of the PCR, and then cooled to 55°C, at which the
double strands of DNA are able to combine sufficiently. From 55 to
95°C the light absorption value was recorded for 4 sec at every
0.5°C. From this step, the melting curve was depicted. Quantitative
analysis was performed using the ratio of the target gene to
actin.
 | Table II.Primer sequences of survivin and
actin. |
Table II.
Primer sequences of survivin and
actin.
| Gene | Primer (5′-3′) | Product size
(bp) |
|---|
| Survivin | | 113 |
| Forward |
ACCGCATCTCTACATTCAAG | |
| Reverse |
CAAGTCTGGCTCGTTCTC | |
| Actin | | 302 |
| Forward |
GTGGACATCCGCAAAGAC | |
| Reverse |
AAAGGGTGTAACGCAACTA | |
Detection of the protein expression of
survivin by western blotting
Total protein of the A549 lung cancer cells was
isolated 72 h after transfection. Protein quantification was
performed by the BCA assay. The protein sample was normalized at
the same time. The sample load was 30 μg total protein per lane.
Protein from 10% SDS-PAGE gel was transferred to a PVDF membrane
after electrophoresis. The protein was blocked with 5% nonfat dry
milk at 4°C. Then, the primary antibodies, rabbit monoclonal
anti-survivin (1:1,000) and anti-GAPDH (1:1,000) antibodies were
added, respectively, and the mixture was incubated overnight at 4°C
on a rocking platform. After washing, the membrane was added
together with the HRP-conjugated secondary antibody (1:5,000) and
incubated for 2 h. The membrane was then developed with ECL
enhanced chemiluminescence system and exposed to X-ray film. Its
gray scales were scanned by an image analytical system.
Detection of lung cancer cell
proliferation by colony formation assay
Cells at a log phase of growth for each group were
digested with 0.25% tryptase into a single-cell suspension, diluted
and inoculated into 24-well plates at 200 cells/well, 3
wells/group. The cells were then incubated for 2 weeks. The
incubation was terminated when visible clones formed on the plates.
The clones were then washed with PBS. Paraformaldehyde (1 ml) was
added and cells were fixated for 30–60 min. The clones were washed
with PBS again and then dyed with Giemsa for 20 min. The number of
colonies in which there were >50 cells was counted under a
microscope. The colony formation rate = number of colonies/number
of incubating cells × 100%.
Determination of the lung cancer cell
proliferation curve by MTT assay
The cells at a log phase of growth for each group
were inoculated into 96-well plates at 100 μl/well. Ten microliters
of MTT (5 mg/ml) was added to each well before termination. The
plates were incubated at 37°C in 5% CO2 for 4 h. The
supernatant was discarded. DMSO (100 μl/well) was added. The
mixtures were shaken gently in order to dissolve the hyacinth in
sediment. The absorbance (A) value at a wavelength of 570 nm was
detected by a microplate spectrophotometer, and the suppression
rate of cell proliferation was then calculated. Five wells were
performed for each group. The suppression rate of proliferation of
the lung cancer A549 cells = (1 - A value of KD)/A value of CON ×
100%. Detection was carried out continuously for 5 days. The cell
proliferation curve was sketched in order to compare the cell
proliferation rate for each group.
Detection of the invasive capacity of
lung cancer cells by cell invasion assay
The invasion chamber was put into an incubator. Warm
serum-free medium (300 μl) was added into the insert. ECM was
rehydrated for 1–2 h at room temperature. Medium was carefully
removed from the insert. Medium (500 μl) rich in FBS was added to
the lower chamber. A cell suspension (300 μl) was added into each
insert. The insert was incubated for 72 h. Medium and non-invasive
cells were removed. Dye (500 μl) was added into the empty wells of
the plates. The insert was immersed in the dye for 20 min. The
lower membrane surface of invasive cells was dyed. The insert was
then immersed in a large cup, washed several times and dried in
air. Images were captured under a microscope. The membrane was
dissolved with 10% acetate and detected at OD570.
Detection of the cell migratory capacity
by wound-healing repair assay
We scratched horizontal lines cross the wells at the
back of 96-well plate with a marker pen. There were at least 2
lines for each well. Approximately 5×104 cells were
added into each well. The next day, lines perpendicular to the
horizontal lines were scratched with the head of a pipette. Cells
were washed twice with PBS, and the dead skin cells were removed.
Serum-free medium was added into the wells. The cells were
incubated for 24 h. Images were captured under a fluorescent
microscope. We replaced the medium with complete medium and further
incubation in an incubator was carried out for 20 and 26 h. Images
were captured to observe the cell distribution at the scratch zone
at different times. The vertical distance in the inner face of the
scratch zone was measured. The wound-healing repair rate =
(vertical distance of the inner face of the scratch zone before
repair - vertical distance of the inner face of the scratch zone
after repair)/vertical distance of the inner face of the scratch
zone before repair × 100%.
Statistical analysis
Data were processed using SPSS15.0 statistical
software. Quantitative data were expressed as the means ± standard
deviation. One-way ANOVA was performed for comparison between
different groups. Dunnett's t (when homogeneity of variances
existed) or Dunnett T3 (when heterogeneity of variances existed)
was calculated. The difference was significant at P-values
<0.05.
Results
Expression of survivin in lung
cancer
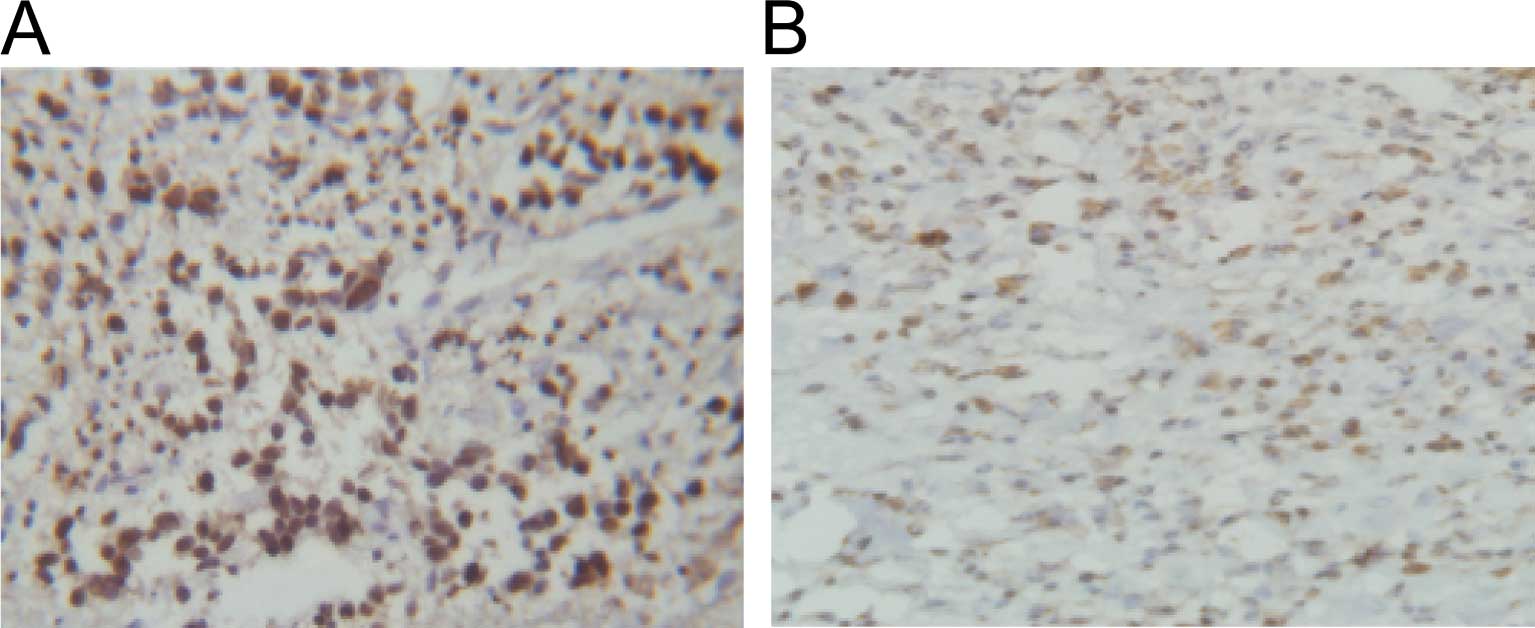
IHC results showed that the total positive rate of
survivin expression in non-small cell lung carcinoma (NSCLC) was
significantly higher than that in the paraneoplastic tissue (66.7
vs. 3.3%, P<0.05) (Fig. 1A and
B).
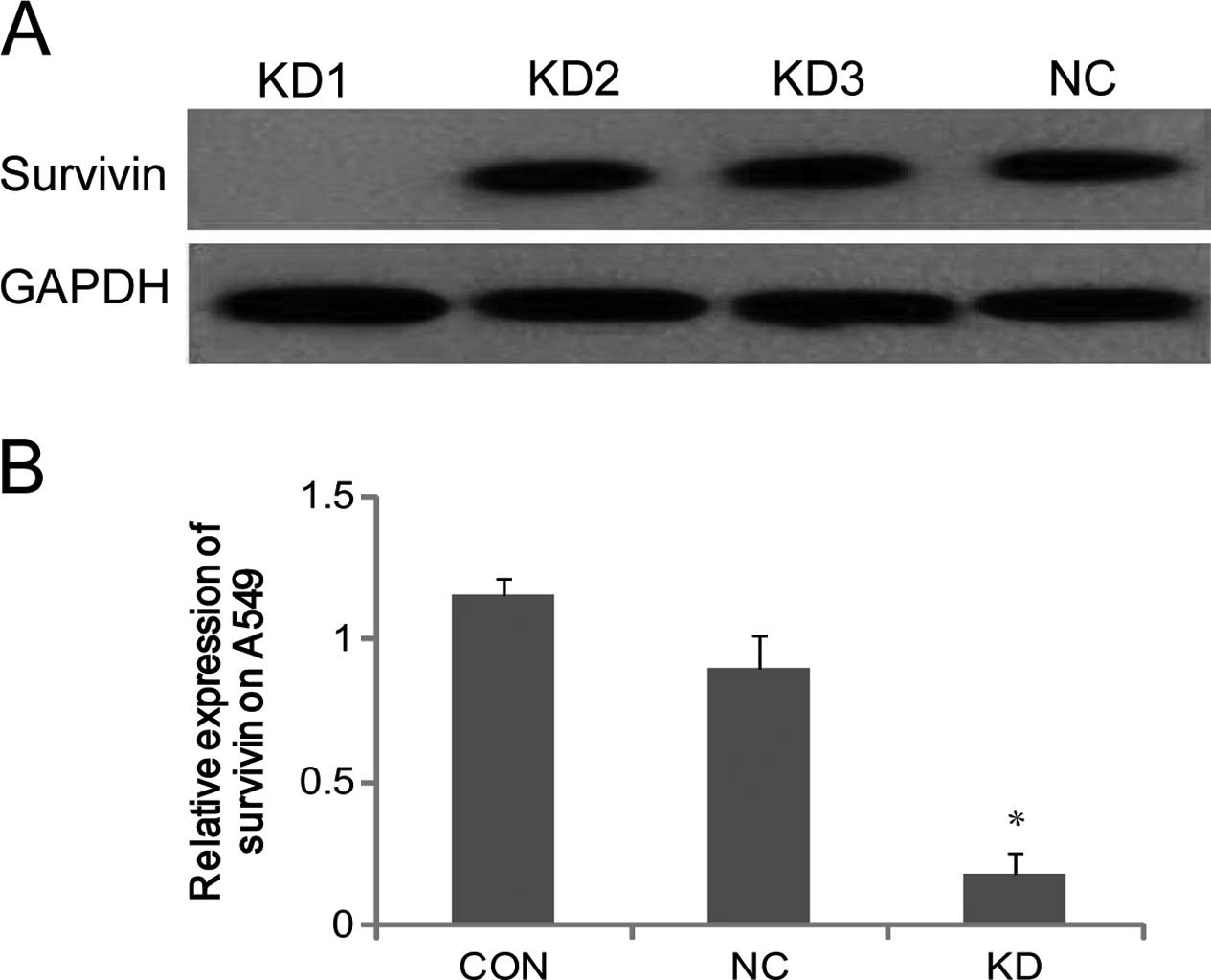
Effect of siRNA on the expression of
survivin in lung cancer cells
Western blotting confirmed that the first pair of
siRNA was the most efficient, and this was ascertained to be the
most competitive candidate to be recombined into the survivinsiRNA
lentiviral vector (Fig. 2A). As
shown by RT-qPCR, the expression of survivin mRNA in KD was
significantly lower compared to that in NC and CON (P<0.05), and
the difference between NC and CON was not significant. The result
proved that survivin-siRNA was successfully transfected into A549
lung cancer cells, and it specifically downregulated the expression
of survivin mRNA (Fig. 2B).
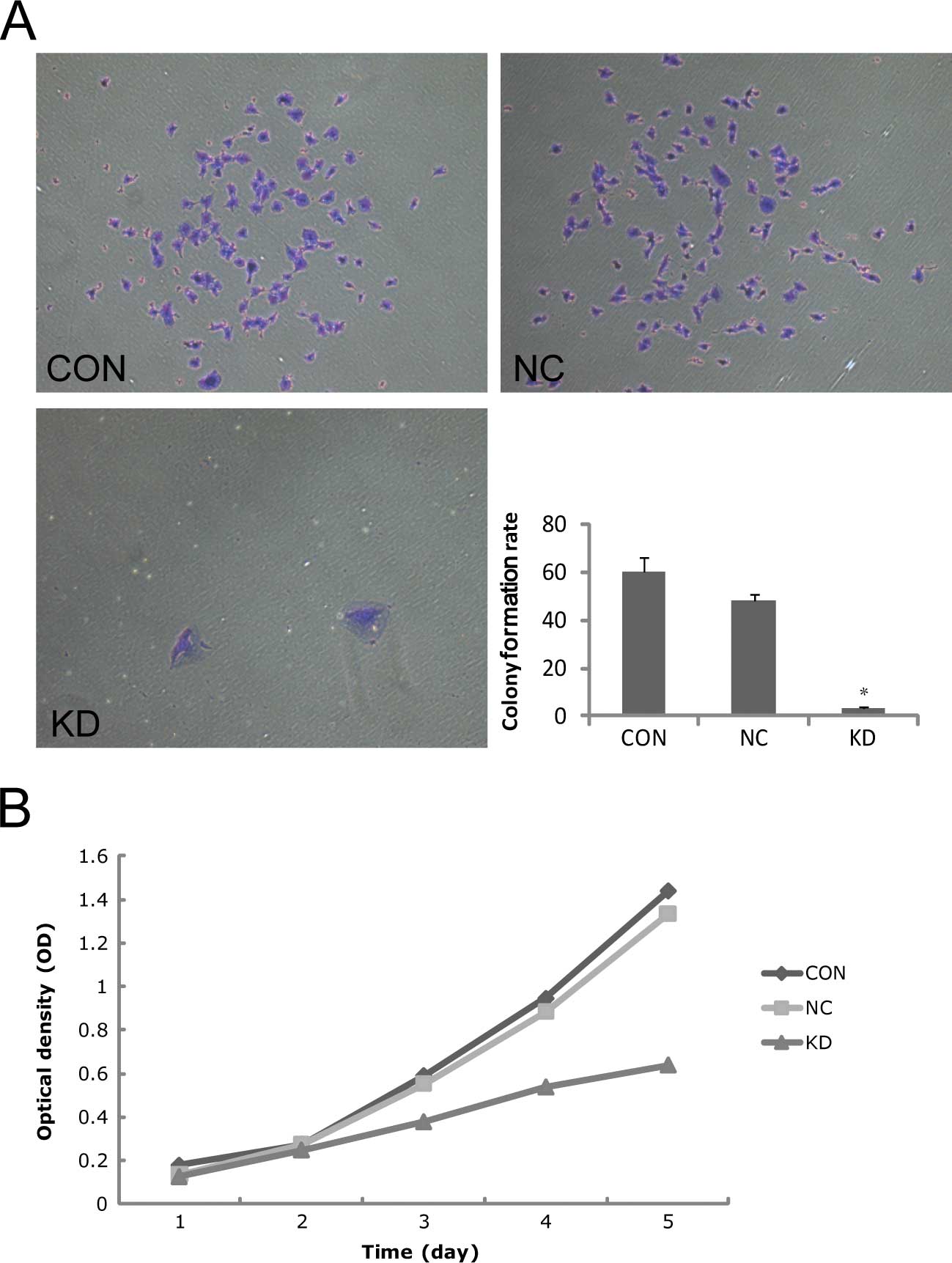
Effect of survivin-siRNA on lung cancer
cell proliferation
Two hundred cells were inoculated and then incubated
for 2 weeks. The result showed that the colony formation rates in
CON, NC and KD were 60±6, 48±3 and 3±1%, respectively. The colony
formation rate in KD was significantly lower than the rates in CON
and NC (P<0.05), which indicated that the cells in KD had
extremely low proliferating capacity (Fig. 3A). The cell proliferation curves
were sketched based on the OD values in each group from Day 1 to 5.
The initial OD values were CON: 0.18±0.02, NC: 0.17±0.01, KD:
0.16±0.01. The differences were not significant (P>0.05).
However, the OD values on Day 5 proved that the difference between
CON and NC was not significant (1.44±0.01 vs. 1.13±0.44,
P>0.05), but the proliferative activity of KD (0.80±0.03) was
significantly decreased (P<0.05). The OD values in the KD group
on Days 3, 4 and 5 were 36.0, 43.1 and 44.6%. The result proved
that the cell proliferation was markedly suppressed after
transfection with survivin-siRNA (Fig.
3B).
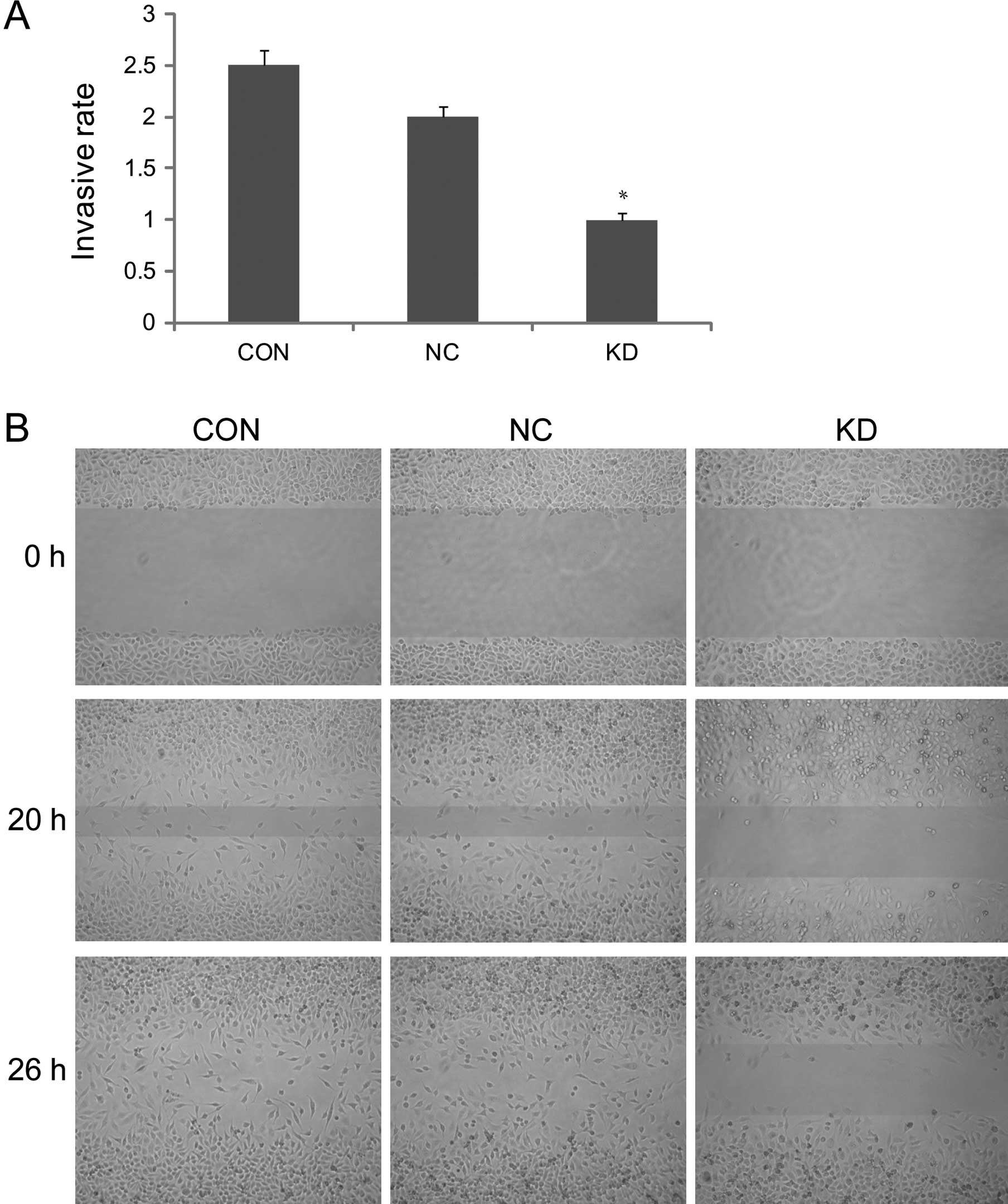
Change in the invasive capacity and cell
migratory capacity of the lung cancer cells after survivin
interference
Reconstituted basement membrane penetrating capacity
of A549 lung cancer cells reflected its invasive capacity in the
Transwell test. The OD value 72 h later was 1.07±0.01 in KD, which
was significantly lower compared to CON (2.53±0.2) and NC
(1.96±0.1, P<0.05). The result proved that the invasive capacity
of A549 cells significantly decreased after transfection with
survivin-siRNA (Fig. 4A). The
wound-healing repair rate at 20 h in the KD group was significantly
lower than that in CON (61.3±3.4%) and NC (59.3±4.1%, P<0.05).
The differences were more significant 26 h later, after scratching,
whereas the difference between NC and CON was not significant
(P>0.05). The result proved that the cell migration capacity
decreased markedly after transfection with survivin-siRNA (Fig. 4B).
Discussion
Lung cancer, one of the most malignant,
health-threatening tumors, responds poorly to existing treatments
and has an unfavorable prognosis. The identification of targets for
the gene therapy of lung cancer has been an intense focus of
research (8,9). Previous studies have shown that
survivin is highly expressed in many types of tumors, and it
directly or indirectly participates in carcinogenesis and
development of tumors (10,11).
In the present study, we detected a high expression
of survivin in tissue specimens of lung cancer by IHC, which was
consistent with the research of Hofmann et al and Akyürek
et al (12,13). The high expression of survivin in
lung cancer offers a new method for its diagnosis. In our study, we
used RNAi technique to suppress the expression of survivin to
explore its importance, role and functions. Under this condition,
we observed how survivin participates in the proliferation, growth,
invasion and migration of lung cancer cells. The RNAi technique,
the most effective antisense technique to date originated from a
hereditary phenomenon widely existing in flora and fauna, and
refers to a protective mechanism against gene instability caused by
viral infection and insertion mutations. The technique specifically
induces the degradation of target mRNA by double-strand siRNA.
Compared to other gene knockout techniques, this technique shows
high efficiency, specificity, stability, transmissibility and
hereditability, therefore it plays an important role in gene
function research and gene therapy of tumors.
In our study, RT-qPCR and western blotting confimed
that survivin siRNA effectively suppressed the expression of
survivin in A549 lung cancer cells. In addition, the colony
formation assay and mononuclear cell direct cytotoxicity assay
proved that the survivin-siRNA sequence altered the proliferation
and growth of the cells. In particular, the suppression rate of
lung cancer A549 cell proliferation was 32.2%. In brief, our study
proved the importance of survivin in maintaining and promoting the
proliferation and growth of lung cancer cells at the mRNA and
protein levels. After the transfection of survivin siRNA into lung
cancer cells, the invasion and migration capacities were
significantly altered, as shown by the markedly decreased cell
membrane-penetrating capacity and wound-healing repair rate. All of
the data proved the importance of survivin in invasion and
migration of lung cancer. Dohi et al (14) proved that survivin could enhance
the invasive capacity of tumor cells, independent of its
anti-apoptosis capacity. Mehrotra et al (15) knocked out the expression of
survivin and XIAP, two members of the IAPs, from invasive breast
cancer MDA-MB-231 cells and prostate cancer PC3 cells using RNAi
technique. They found that the invasive capacities of the two types
of cancer cells markedly decreased. The same phenomenon was
observed in intestinal cancer HCT116 cells. However, the invasive
capacity of non-invasive MCF-7 cells was enhanced markedly after
the cells were transfected with survivin. The animal models (mice)
showed that the intermolecular cooperation between survivin and
XIAP activated NF-κB, independent of IAP inhibition of cell death,
which in turn led to increased fibronectin gene expression,
signaling by β1 integrins, and activation of cell motility kinases
FAK and Src. In this way, the invasion and migration of tumor cells
were promoted. The researchers considered that antagonists against
IAPs may provide anti-metastatic activity in patients with
cancer.
In brief, downregulation of the expression of
survivin may effectively suppress the malignant biological behavior
of lung cancer, but more details regarding the mechanisms require
further research (16–19).
Acknowledgements
This study was supported by grants
from the Natural Science Foundation of Fujian Province (no.
2008J0284), and the Scholastic Development Foundation of Fujian
Medical University (no. JA09121).
References
|
1.
|
Altieri DC: New wirings in the survivin
networks. Oncogene. 27:6276–6284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Li C, Wu Z, Liu M, Pazgier M and Lu W:
Chemically synthesized human survivin does not inhibit caspase-3.
Protein Sci. 17:1624–1629. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Srinivasula SM and Ashwell JD: IAPs:
what's in a name? Mol Cell. 30:123–135. 2008.
|
|
4.
|
Ashihara E: RNA interference for cancer
therapies. Gan To Kagaku Ryoho. 37:2033–2041. 2010.(In
Japanese).
|
|
5.
|
Sioud M: Promises and challenges in
developing RNAi as a research tool and therapy. Methods Mol Biol.
703:173–187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Zheng W, Ma X, Wei D, Wang T, Ma Y and
Yang S: Molecular cloning and bioinformatics analysis of a novel
spliced variant of survivin from human breast cancer cells. DNA
Seq. 16:321–328. 2005.PubMed/NCBI
|
|
7.
|
Horn ME and Waterhouse PM: Rapid
match-searching for gene silencing assessment. Bioinformatics.
26:1932–1937. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Alberg AJ and Nonemaker J: Who is at high
risk for lung cancer? Population-level and individual-level
perspectives. Semin Respir Crit Care Med. 29:223–232. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Lee MW, Kim DS, Min NY and Kim HT: Akt1
inhibition by RNA interference sensitizes human non-small cell lung
cancer cells to cisplatin. Int J Cancer. 122:2380–2384. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kanwar JR, Kamalapuram SK and Kanwar RK:
Targeting survivin in cancer: patent review. Expert Opin Ther Pat.
20:1723–1737. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Small S, Keerthivasan G, Huang Z,
Gurbuxani S and Crispino JD: Overexpression of survivin initiates
hematologic malignancies in vivo. Leukemia. 24:1920–1926. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Hofmann HS, Simm A, Hammer A, Silber RE
and Bartling B: Expression of inhibitors of apoptosis (IAP)
proteins in non-small cell human lung cancer. J Cancer Res Clin
Oncol. 128:554–560. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Akyurek N, Memis L, Ekinci O, Kokturk N
and Ozturk C: Survivin expression in pre-invasive lesions and
non-small cell lung carcinoma. Virchows Arch. 449:164–170. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Dohi T, Xia F and Altieri DC:
Compartmentalized phosphorylation of IAP by protein kinase A
regulates cytoprotection. Mol Cell. 27:17–28. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Mehrotra S, Languino LR, Raskett CM,
Mercurio AM, Dohi T and Altieri DC: IAP regulation of metastasis.
Cancer Cell. 17:53–64. 2010. View Article : Google Scholar
|
|
16.
|
Jang JS, Kim KM, Kang KH, et al:
Polymorphisms in the survivin gene and the risk of lung cancer.
Lung Cancer. 60:31–39. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Nabilsi NH, Broaddus RR and Loose DS: DNA
methylation inhibits p53-mediated survivin repression. Oncogene.
28:2046–2050. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Varfolomeev E, Blankenship JW, Wayson SM,
et al: IAP antagonists induce autoubiquitination of c-IAPs,
NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell.
131:669–681. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar : PubMed/NCBI
|