Introduction
Esophageal cancer is the sixth most leading cause of
cancer-related mortality worldwide (1). Esophageal adeno-carcinoma (EAC) and
esophageal squamous cell carcinoma are the two main histological
types of esophageal cancers (1).
Of all esophageal cancer types in Western countries, 30-50% of
cases are esophageal adenocarcinoma (1). The incidence of EAC has increased
faster than that of any malignancy in Western countries, with an
increase of 400% over the past 40 years (2). However, there has been no increase in
the prevalence of proximal gastric cancers and distal esophageal
adenocarcinomas in the Turkish population (3). The prognosis for EAC is poor and the
overall 5-year survival rate is less than 10% (4). Risk factors for EAC include dietary
factors, alcohol and tobacco use, obesity, gastroesophageal reflux
disease and Barrett’s esophagus (BE) (5). Gastroesophageal reflux disease (GERD)
is very common worldwide (6). The
prevalence of GERD is high, especially in developed Western
countries (7). Bor et al
found the prevalence of GERD to be 20% in Izmir (8) and 22.8% in Turkey, similar to the
rates in the US (7). Chronic GERD
is one of the main risk factors for the development of Barrett’s
esophagus (BE) and BE is one of the strongest risk factors for EAC
(5,9). Mechanisms that control chromatin
stucture and gene expression in normal mammalian cells are DNA
methylation, covalent histone modifications, nucleosome position,
histone variants and miRNAs (10–12).
Recent epigenetic studies have shown the effect of epigenetic
alterations in carcinogenesis as well as genetic alterations. A
number of studies suggest that epigenetic alterations may even be
initiating factors for certain types of cancer (13). Genetic alterations are irreversible
but epigenetic alterations are reversible and this fact supports
future hope for epigenetic therapy (14).
Studies that have investigated the relationship
between erosive esophagitis (EE) and EAC usually focus on
symptom-related evidence or on polymorphisms. There are no
epigenetic gene expression studies on this topic. We aimed to
evaluate the relationship between EE and EAC to ascertain whether
there is a genetic tendency for EAC.
Materials and methods
Location of study
The study was conducted at the Department of
Gastroenterology, Department of Medical Biology, Celal Bayar
University, Manisa between March 2010 and September 2011. Patients
were also referred from the Department of Gastroenterology, Celal
Bayar University, the Department of Gastroenterology, Ege
University and the Department of Gastroenterology, Ataturk Research
and Training Hospital.
Ethics
This study was performed in accordance with the
Declaration of Helsinki, good clinical practice and applicable
regulatory requirements. Celal Bayar University Institutional
Review Board approved this clinical trial on June 2, 2009. Each
patient signed a consent form prior to any study-related
procedure.
Study design and subjects
Between March 2010 and September 2011 fresh paired
tissue samples from 60 patients [group 1 (20 patients) categorized
as the macroscopic and histopathologically confirmed esophageal
carcinoma group; group 2 (20 patients) categorized as the erosive
esophagitis (without histopathologically esophageal adenocarcinoma
and Barrett’s esophagus) group; and group 3 (20 patients)
categorized as the control group (who had normal esophageal mucosa
with no endoscopic or histopathological lesions)] were collected.
Typical GERD symptoms were defined as at least five years of
regurgitation and/or heartburn per week in erosive esophagitis.
Patients were excluded from the study if they had a history of
upper gastrointestinal surgery such as gastrectomy, fundoplication
or distal esophagectomy, severe gastroparesis and esophageal
varices.
Endoscopy
Erosive esophagitis and control
group
Esophagogastroduodenoscopies were undertaken for the
EE and control group at the Department of Gastroenterology, Celal
Bayar University by the same two endoscopists (E.K., H.Y.) who
performed the study. During upper gastrointestinal endoscopy, the
distal 5 cm of the esophagus mucosal morphology at the
squamo-columnar junction was visualized using conventional
endoscopy followed by the Narrow Band Imaging (NBI) system using
video endoscopes. During standard white-light endoscopy and NBI
examination, erosions, mucosal breaks and other complications were
graded according to the Los Angeles classification (15). Two biopsies were taken 2 cm above
the esophagogastric junction from patients in the control group,
and two biopsies were taken from mucosal breaks in patients with
EE.
Esophageal adenocarcinoma
Esophagogastroduodenoscopies were undertaken for EAC
cases at the Department of Gastroenterology, Celal Bayar University
(11 patients), Department of Gastroenterology, Ege University and
Department of Gastroenterology (5 patients), Ataturk Research and
Training Hospital (4 patients). Two biopsies were taken from
patients pathologically diagnosed as having EACs using Olympus
biopsy forceps.
EAC was evaluated according to thoracic and
abdominal computed tomography (CT) in three stages: stage 1,
esophageal adenocarcinoma located in the esophagus; stage 2,
esophageal adenocarcinoma located in the esophagus and with
pathological lymphadenopathy; stage 3, esophageal adenocarcinoma
located in the esophagus with pathological lymphadenopathy and
distant metastasis.
Samples were immediately frozen using dry ice (a
block of dry ice has a surface temperature of −78.5°C) and stored
at −80°C until RNA extraction.
Isolation of total RNA
Total RNA was extracted using the TriPure solution
as described in the manufacturer’s protocol. The fresh tissue was
resuspended in a vial of MagNA Lyser Green Beads containing 350 μl
lysis buffer and 50 μl of proteinase (Roche). Next, the suspension
was subjected to mechanical lysis in a MagNA lyser instrument
(Roche) for 45 sec at 4500 rpm. Afterwards, RNA was further
extracted and purified using a MagNA Pure LC instrument (Roche) in
combination with the MagNA Pure NA isolation kit III. All steps
were taken according to the manufacturer’s protocol.
Quantity and purity of total RNA
RNA was quantified measuring the absorbance at 260
nm (A260 nm) and RNA purity was determined by the ratio A260
nm/A280 nm using a classical spectrophotometer. RNA quality was
good, with 260/280 ratios slightly higher than 2.0 and 260/230
ratios slightly higher than 1.8.
RT2 profiler™
PCR protocol first strand cDNA synthesis
The protocol took 2 h to perform (per sample) from
start to finish. We initially had as little as 25 ng of total RNA
from our experimental samples. We first converted the experimental
RNA samples into PCR templates to prepare cDNAs with the
RT2 First Strand kit (SABioscience, Frederick, MD, USA)
according to the manufacturer’s instructions. Next we combined the
template with a specific instrument and used ready-to-use
RT2 SYBR Green qPCR Master Mix. Then we added equal
aliquots of this mixture (25 μl for 96-well) to each well of the
same PCR array plate containing the predispensed gene-specific
primer sets and performed PCR. Specialized software (SABiosciences)
was used to calculate the threshold cycle (Ct) values for the genes
on each PCR array.
Epigenetic chromatin modification
enzyme PCR array
The Human Epigenetic Chromatin Modification Enzyme
RT2 Profiler™ PCR Array (PAHS-085A)
(SABiosciences) was used to detect the expression levels of 84 key
genes (Table I) encoding enzymes
known or predicted to modify genomic DNA and histones to regulate
chromatin accessibility and therefore gene expression. These genes
exhibit differential expression profiles in tumor cells relative to
normal cells. The PCR array is a 96-well plate containing
RT2 Profiler™ PCR Primer Assays for a set of
84 related genes, plus five housekeeping genes and three
controls.
 | Table I.List of key genes. |
Table I.
List of key genes.
| Name | Description |
|---|
| KDM1A | Lysine (K)-specific
demethylase 1A |
| ASH1L | Ash1 (absent,
small, or homeotic)-like (Drosophila) |
| ATF2 | Activating
transcription factor 2 |
| AURKA | Aurora kinase
A |
| AURKB | Aurora kinase
B |
| AURKC | Aurora kinase
C |
| CARM1 |
Coactivator-associated arginine
methyltransferase 1 |
| CDYL | Chromodomain
protein, Y-like |
| CIITA | Class II, major
histocompatibility complex, transactivator |
| CSRP2BP | CSRP2 binding
protein |
| DNMT1 | DNA
(cytosine-5-)-methyltransferase 1 |
| DNMT3A | DNA
(cytosine-5-)-methyltransferase 3 α |
| DNMT3B | DNA
(cytosine-5-)-methyltransferase 3 β |
| DOT1L | DOT1-like, histone
H3 methyltransferase (S. cerevisiae) |
| DZIP3 | DAZ interacting
protein 3, zinc finger |
| EHMT2 | Euchromatic
histone-lysine |
| N-methyltransferase
2 |
| ESCO1 | Establishment of
cohesion 1 homolog 1 (S. cerevisiae) |
| ESCO2 | Establishment of
cohesion 1 homolog 2 (S. cerevisiae) |
| HAT1 | Histone
acetyltransferase 1 |
| HDAC1 | Histone deacetylase
1 |
| HDAC10 | Histone deacetylase
10 |
| HDAC11 | Histone deacetylase
11 |
| HDAC2 | Histone deacetylase
2 |
| HDAC3 | Histone deacetylase
3 |
| HDAC4 | Histone deacetylase
4 |
| HDAC5 | Histone deacetylase
5 |
| HDAC6 | Histone deacetylase
6 |
| HDAC7 | Histone deacetylase
7 |
| HDAC8 | Histone deacetylase
8 |
| HDAC9 | Histone deacetylase
9 |
| KDM5B | Lysine (K)-specific
demethylase 5B |
| RPS6KA3 | Ribosomal protein
S6 kinase, 90 kDa, polypeptide 3 |
| RPS6KA5 | Ribosomal protein
S6 kinase, 90 kDa, polypeptide 5 |
| SETD1A | SET domain
containing 1A |
| SETD1B | SET domain
containing 1B |
| SETD2 | SET domain
containing 2 |
| SETD3 | SET domain
containing 3 |
| SETD4 | SET domain
containing 4 |
| SETD5 | SET domain
containing 5 |
| SETD6 | SET domain
containing 6 |
| SETD7 | SET domain
containing (lysine methyltransferase) 7 |
| SETD8 | SET domain
containing (lysine methyltransferase) 8 |
| SETDB1 | SET domain,
bifurcated 1 |
| SETDB2 | SET domain,
bifurcated 2 |
| SMYD3 | SET and MYND domain
containing 3 |
| KDM5C | Lysine (K)-specific
demethylase 5C |
| KDM4A | Lysine (K)-specific
demethylase 4A |
| KDM4C | Lysine (K)-specific
demethylase 4C |
| KDM6B | Lysine (K)-specific
demethylase 6B |
| KAT2A | K(lysine)
acetyltransferase 2A |
| KAT2B | K(lysine)
acetyltransferase 2B |
| KAT5 | K(lysine)
acetyltransferase 5 |
| MBD2 | Methyl-CpG binding
domain protein 2 |
| MLL | Myeloid/lymphoid or
mixed-lineage leukemia (trithorax homolog, Drosophila) |
| MLL3 | Myeloid/lymphoid or
mixed-lineage leukemia 3 |
| MLL5 | Myeloid/lymphoid or
mixed-lineage leukemia 5 (trithorax homolog,
Drosophila) |
| MYSM1 | Myb-like, SWIRM and
MPN domains 1 |
| KAT8 | K(lysine)
acetyltransferase 8 |
| KAT7 | K(lysine)
acetyltransferase 7 |
| KAT6A | K(lysine)
acetyltransferase 6A |
| KAT6B | K(lysine)
acetyltransferase 6B |
| NCOA1 | Nuclear receptor
coactivator 1 |
| NCOA3 | Nuclear receptor
coactivator 3 |
| NCOA6 | Nuclear receptor
coactivator 6 |
| NEK6 | NIMA (never in
mitosis gene a)-related kinase 6 |
| NSD1 | Nuclear receptor
binding SET domain protein 1 |
| PAK1 | P21 protein
(Cdc42/Rac)-activated kinase 1 |
| PRMT1 | Protein arginine
methyltransferase 1 |
| PRMT2 | Protein arginine
methyltransferase 2 |
| PRMT3 | Protein arginine
methyltransferase 3 |
| PRMT5 | Protein arginine
methyltransferase 5 |
| PRMT6 | Protein arginine
methyltransferase 6 |
| PRMT7 | Protein arginine
methyltransferase 7 |
| PRMT8 | Protein arginine
methyltransferase 8 |
| RNF2 | Ring finger protein
2 |
| RNF20 | Ring finger protein
20 |
| SUV39H1 | Suppressor of
variegation 3-9 homolog 1 (Drosophila) |
| SUV420H1 | Suppressor of
variegation 4-20 homolog 1 (Drosophila) |
| UBE2A |
Ubiquitin-conjugating enzyme E2A |
| UBE2B |
Ubiquitin-conjugating enzyme E2B |
| USP16 | Ubiquitin specific
peptidase 16 |
| USP21 | Ubiquitin specific
peptidase 21 |
| USP22 | Ubiquitin specific
peptidase 22 |
| WHSC1 | Wolf-Hirschhorn
syndrome candidate 1 |
| B2M |
β-2-microglobulin |
| HPRT1 | Hypoxanthine
phosphoribosyltransferase 1 |
| RPL13A | Ribosomal protein
L13a |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase |
| ACTB | Actin, β |
| HGDC | Human genomic DNA
contamination |
Data analysis
Data were analyzed using RT2 profiler PCR
array data analysis software (http://www.sabiosciences.com/pcrarraydataanalysis.php).
The website also allowed online analysis. For each PCR reaction,
the Excel sheet calculated two normalized average cycle threshold
(Ct) values, a paired t-test p-value and a fold-change. PCR array
quantification was based on the Ct number. A gene was considered
not detectable when Ct >32. Ct was defined as 35 for the ΔCt
calculation when the signal was under detectable limits.
Fold-change and fold-regulation values >2 were
indicative of upregulated gene; fold-change values <0.5 and
fold-regulation values <-2 were indicative of downregulated
genes.
Statistics
Data were statistically analyzed with RT2
profiler PCR array data analysis software (http://www.sabiosciences.com/pcrarraydataanalysis.php).
Results were expressed as the mean values ± standard deviation and
the p-values were calculated based on a Student’s t-test of the
replicate 2−ΔCt values for each gene in the control
group, esophageal adenocarcinoma group and erosive esophagitis
group. A p-value <0.05 was accepted as statistically
significant.
Results
A total of 60 patients were divided into three
groups: 20 patients as a control group (who had normal esophageal
mucosa with no esophagogastroduodenoscopic or histopathological
lesions), 20 patients with EE without Barrett’s esophagus and
microscopic adenocarcinoma and 20 patients with macroscopic and
histopathological adenocarcinoma. The mean age ± SD of the control
group was 51.5±11.3 years. The mean age ± SD of the EE group was
56.6±10.2 years. The mean age ± SD of the EAC group was 58.6±12.4
years.
For the three groups genetic analysis was used to
investigate the expression of 84 key genes (Table I) encoding enzymes known or
predicted to modify genomic DNA and histones to regulate chromatin
accessibility and therefore gene expression.
Upregulated and downregulated genes in the EAC and
control group are summarized in Table
II. AURKA, AURKB, NEK6 were expressed at significantly higher
levels in the EAC than in the control group. MBD2 was expressed
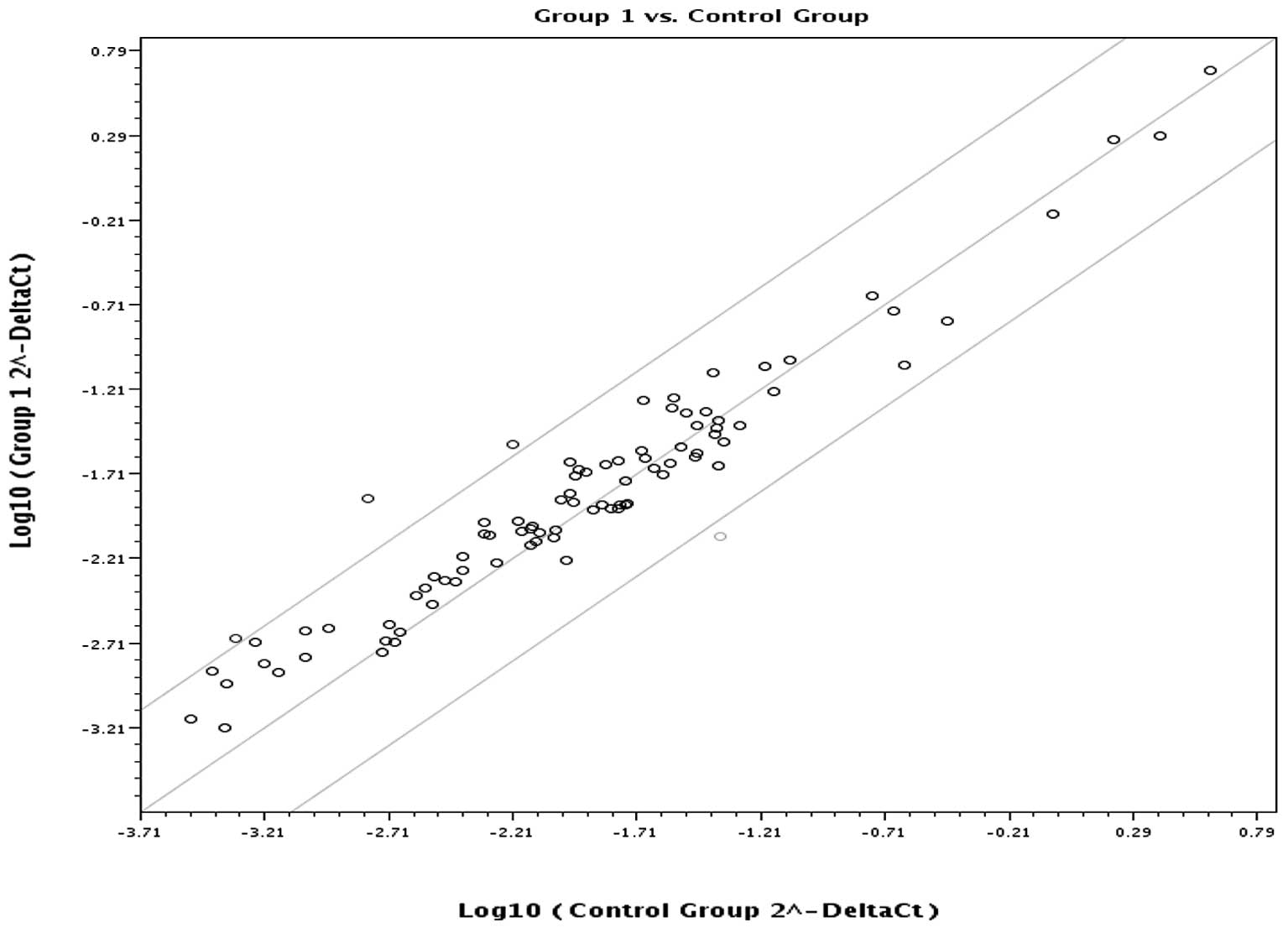
significantly lower in the EAC than in the control group. Fig. 1 is a scatter plot of the log base
10 of the hybridization intensity of each gene in the two groups
[x-axis, control group; y-axis, the EAC (group 1) group]. The
middle line indicates a fold-change (2−ΔCt) of 1. The
top and the bottom lines indicate the desired fold-change in gene
expression threshold. Expression of 80 key genes was unchanged,
showing no significant difference in expression between the two
groups. The three points above the top line indicate upregulated
(AURKA, AURKB, NEK6) genes. The one point under the bottom line
represents a down-regulated (MBD2) gene.
 | Table II.Differentially upregulated and
downregulated genes between the esophageal adenocarcinomas and
control group. |
Table II.
Differentially upregulated and
downregulated genes between the esophageal adenocarcinomas and
control group.
| Fold-change | 95% CI | p-value |
|---|
| Upregulated
genes | | | |
| AURKA | 2.1809 | (0.74–3.62) | 0.041697 |
| AURKB | 2.5729 | (1.33–3.81) | 0.004832 |
| NEK6 | 8.6782 | (1.79–15.56) | 0.002312 |
| Downregulated
genes | | | |
| MBD2 | 0.3682 | (0.20–0.54) | 0.000193 |
| Housekeeping genes
for internal control | | | |
| HPRT1 | 1.3076 | (0.87–1.74) | 0.0681 |
| RPL13A | 0.7333 | (0.49–0.97) | 0.310487 |
| GAPDH | 1.1842 | (0.81–1.56) | 0.1399 |
| ACTB | 0.7797 | (0.47–1.08) | 0.56202 |
| HGDC | 2.6061 | (0.00001–5.70) | 0.22952 |
Upregulated and downregulated genes in the EE and
control group are summarized in Table
III. Seven genes (AURKA, AURKC, HDAC9, NEK6, HDAC8, SETD5,
SETD7) were upregulated and AURKA, AURKC, HDAC9, NEK6 were
expressed at significantly higher levels in EE than in the control
group. There were no downregulated genes in the two groups.
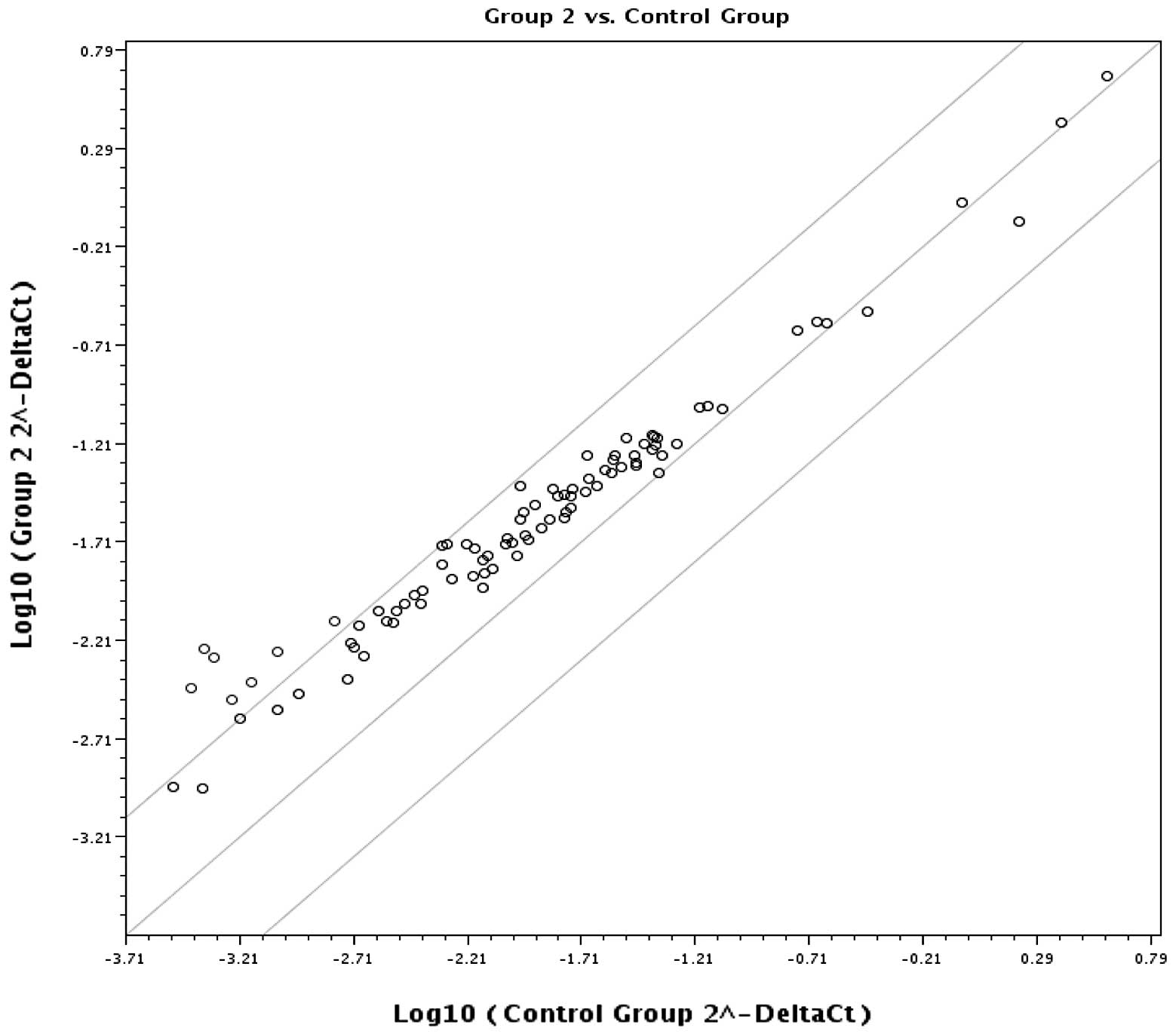
Fig. 2 is a scatter plot of the
log base 10 of the hybridization intensity of each gene in the two
groups [x-axis, control group; y-axis, EE (group 2) group]. The
middle line indicates a fold-change (2–ΔCt) of 1. The
top and the bottom lines indicate the desired fold-change in gene
expression threshold. Expression of 77 key genes was unchanged with
no significant difference in gene expression between the two
groups. The seven points above the top line represent upregulated
(AURKA, AURKC, HDAC9, NEK6, HDAC8, SETD5, SETD7) genes.
 | Table III.Differentially upregulated and
downregulateda genes
between the erosive esophagitis and control group. |
Table III.
Differentially upregulated and
downregulateda genes
between the erosive esophagitis and control group.
| Fold-change | 95% CI | p-value |
|---|
| Upregulated
genes | | | |
| AURKA | 3.5414 | (0.86–6.23) | 0.024265 |
| AURKC | 5.3826 |
(0.00001–10.99) | 0.040191 |
| HDAC9 | 10.676 |
(0.00001–24.29) | 0.036345 |
| NEK6 | 4.771 | (0.55–8.99) | 0.025135 |
| HDAC8 | 2.3888 | (0.79–3.98) | 0.052014 |
| SETD5 | 2.0724 | (0.70–3.45) | 0.100915 |
| SETD7 | 2.492 | (1.16–3.83) | 0.8001 |
| Housekeeping genes
for internal control | | | |
| HPRT1 | 1.4659 | (0.97–1.96) | 0.084585 |
| RPL13A | 1.1343 | (0.89–1.38) | 0.615362 |
| GAPDH | 1.1331 | (0.87–1.40) | 0.154067 |
| ACTB | 1.0518 | (0.69–1.41) | 0.571422 |
| HGDC | 12.8065 |
(0.00001–29.89) | 0.05754 |
Upregulated and downregulated genes in EAC and EE
are summarized in Table IV. There
was no significant difference in gene upregulation between the two
groups. Two genes (MBD2 and MYSM1) were downregulated and MBD2 was
significantly downregulated in EAC compared to the EE group.
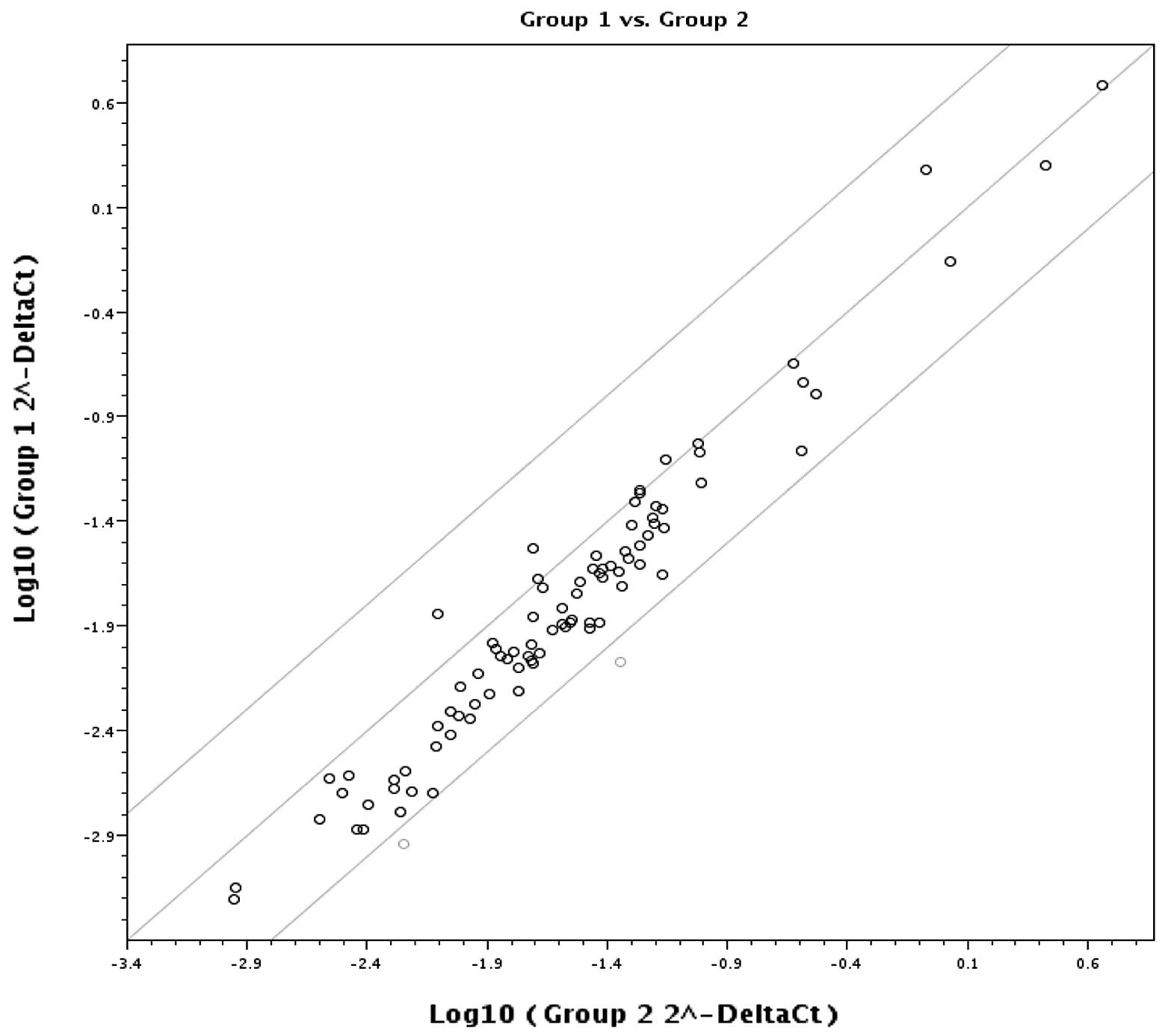
Fig. 3 is a plot of the log base
10 of the hybridization intensity of each gene in the two groups
(x-axis represents group 1, EAC, and the y-axis shows group 2, EE).
The middle line indicates a fold-change (2−ΔCt) of 1.
The top and bottom lines indicate the desired fold-change in gene
expression threshold. The expression of 82 key genes was unchanged
and no significant difference in gene upregulation was noted
between the two groups. The two points under the bottom line
represent downregulated (MBD2, MSYM1) genes.
 | Table IV.Differentially upregulateda and downregulated genes
between esophageal adenocarcinoma and erosive esophagitis. |
Table IV.
Differentially upregulateda and downregulated genes
between esophageal adenocarcinoma and erosive esophagitis.
| Fold-change | 95% CI | p-value |
|---|
| Downregulated | | | |
| MBD2 | 0.3849 | (0.17–0.60) | 0.008897 |
| MYSM1 | 0.3418 | (0.12–0.57) | 0.178543 |
| Housekeeping genes
(for internal control) | | | |
| HPRT1 | 1.0676 | (0.57–1.56) | 0.984553 |
| RPL13A | 0.6926 | (0.42–0.96) | 0.290211 |
| GAPDH | 1.0459 | (0.68–1.41) | 0.561437 |
| ACTB | 0.6882 | (0.39–0.99) | 0.185571 |
| B2M | 1.8789 | (1.18–2.58) | 0.221942 |
The NEK6 and AURKA genes were significantly
upregulated in the EAC and EE groups compared to the control
group.
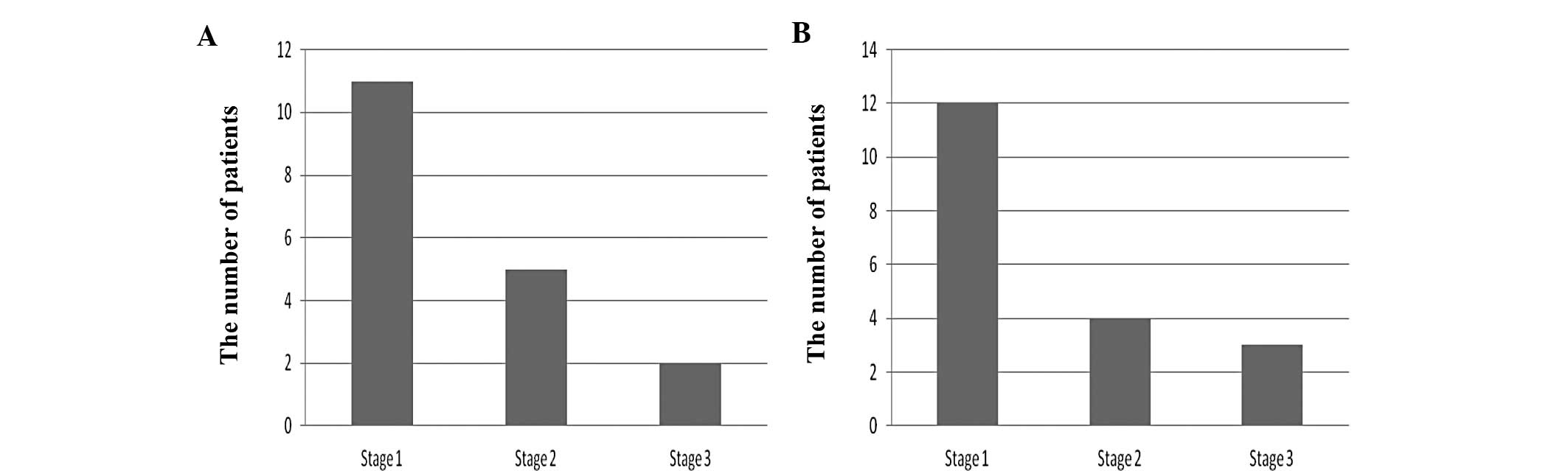
The correlation between the AURKA gene and the stage
of EAC is summarized in Fig. 4A.
There were more patients in the stage 1 group than in the stage 2
and stage 3 groups (p<0.05).
The correlation between the NEK6 gene and the stage
of EAC is summarized in Fig. 4B.
Again, there were more patients classified in the stage 1 group
than in stage 2 and stage 3 groups (p<0.05).
The correlation between expression of the AURKA gene
and the Los Angeles classification of erosive esophagitis are
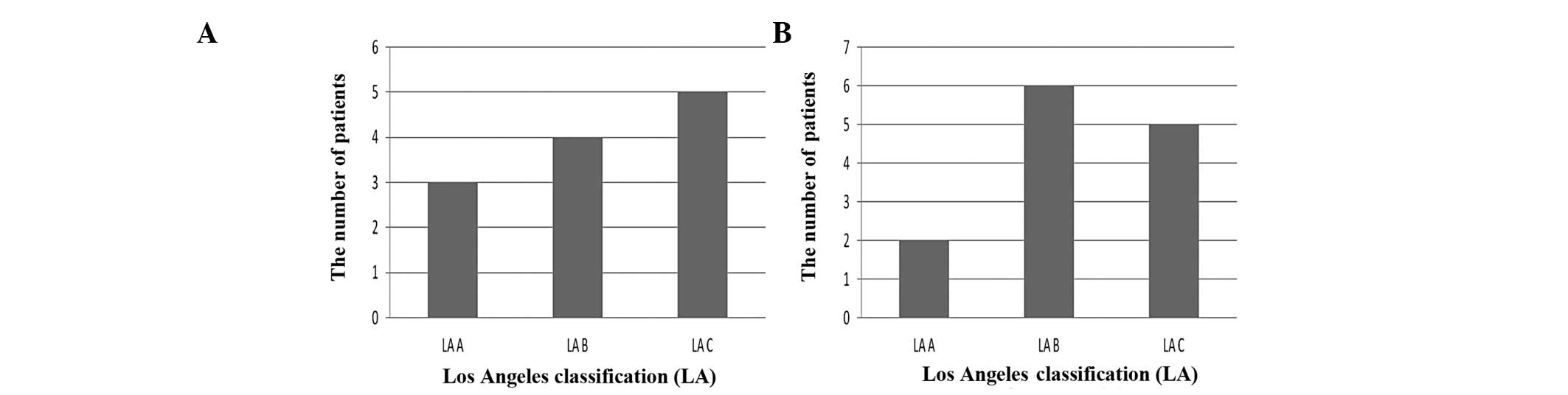
summarized in Fig. 5A. Expression
of the AURKA gene was found to be elevated dependent on the grade
of EE according to the Los Angeles Classification.
The correlation between expression of the NEK6 gene
and the Los Angeles classification of EE is summarized in Fig. 5B. The patients with overexpression
of the NEK6 gene were more prevalent in the LA Grade B than LA
Grade A group (p>0.05).
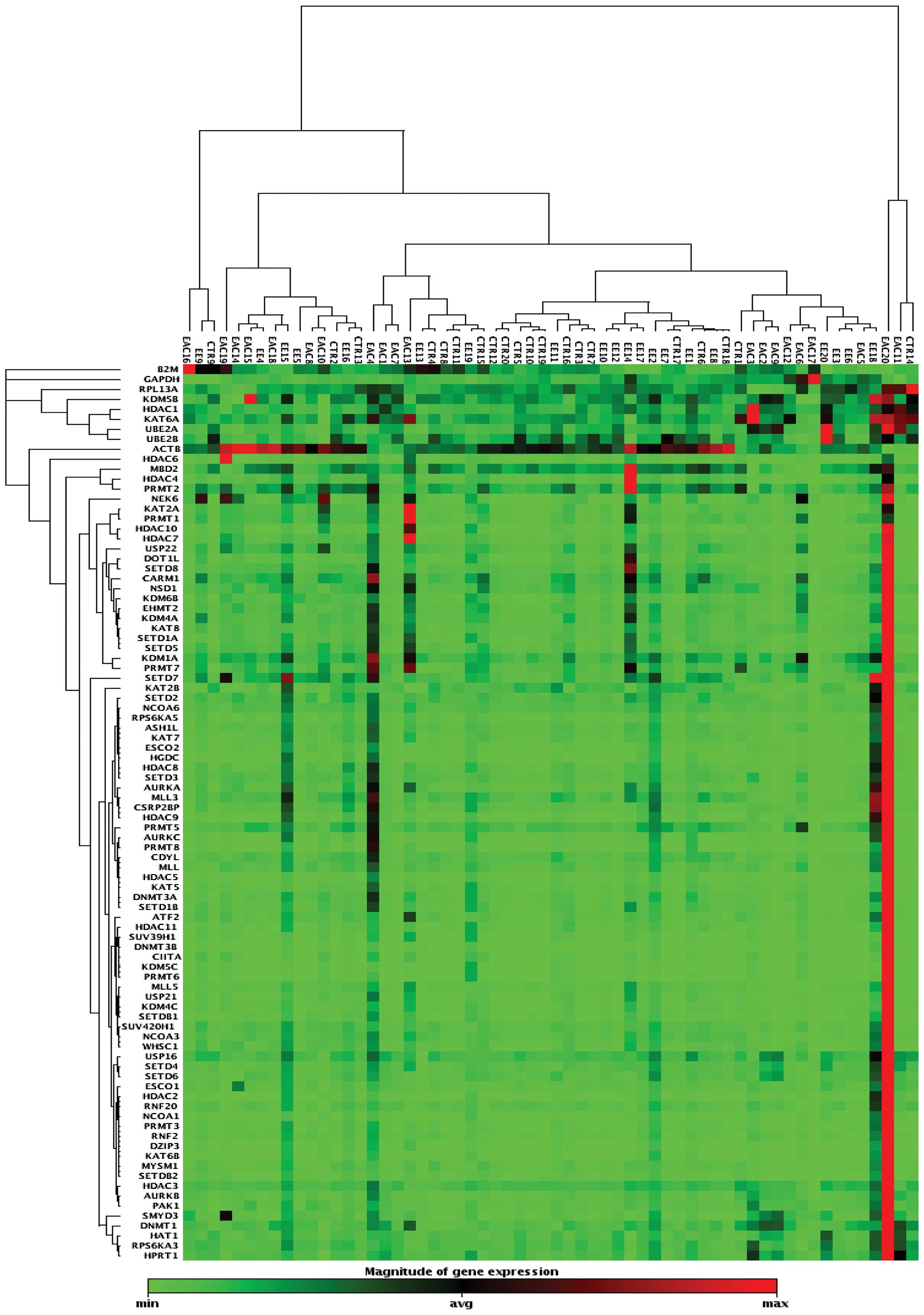
A clustergram analysis based on differentially
expressed genes between the three groups is shown in Fig. 6. The cluster-gram creates a heat
map with dendrograms to show genes that are co-regulated. The color
saturation reflects the magnitude of the change in gene expression.
Green squares represent lower gene expression in the experimental
samples (ratios less than 1); black squares represent genes equally
expressed (ratios near 1); red squares represent higher than
control levels of gene expression (ratios greater than 1); gray
squares indicate insufficient or missing data.
Discussion
Understanding the epigenetic structure of carcinomas
provides important information on carcinogenesis. Expanding the
information on the molecular biology of cancer may result in better
follow-up of precancerous and cancer lesions (13,16).
Epigenetic changes in stem cells provide important information
concerning cancer aetiology, and epigenetic alterations such as
gene expression have been used as biomarkers in recent years
(17).
In the present study, three genes were overexpressed
in the EAC group, with AURKA, AURKB and NEK6 being significantly
more highly expressed than levels in the control group. Four genes
(AURKA, AURKC, HDAC9, NEK6) were significantly more highly
expressed in the EE group than levels in the control group. AURKA
and NEK6 genes were significantly more highly expressed in the EE
group and in the EAC group than levels in the control group.
Recent studies have shown that the aurora kinase
family, polo-like kinase family and NIMA (never in mitosis gene A)
kinase family control cell cycle (18,19).
Aurora kinases are activated through autophosphorylation at the
activation loop unlike most kinases within the cell (20) and NimA promotes mitotic chromosome
condensation through phosphorylation of histone H3 at serine 10 and
may compose the nuclear membrane division during mitotic exit
(21).
Enzymes in the Aurora kinase family are encoded by
the AURKA (also called
AIK/ARK1/AURA/AURORA2/BTAK/MGC34538/STK15/STK6/STK7) gene, which is
localized on 20q13.2 (22,23). These enzymes play very important
roles in mitosis and meiosis for healthy cell proliferation. AURKA
is a serine/threonine kinase acting as a regulator of centro-some
function/duplication, mitotic entry, and bipolar spindle assembly
(24). AURKA protein levels and
kinase activity are low in the G1/S phase; accumulate during G2/M
and decrease rapidly following mitosis (25). AURKA is an important
kinase-encoding gene involved in centrosome duplication and
distribution; its overexpression leads to centrosome amplification,
chromosomal instability and aneuploidy in several cancer types
(26,27). AURKA overexpression has been found
in numerous tumor cells and tissues including gastric cancer,
breast cancer, colorectal cancer, bladder cancer, pancreatic
cancer, ovarian cancer, prostate cancer and esophageal
squamous-cell carcinoma, esophageal adenocarcinoma and Barrett’s
esophagus (27,28). Dar et al demonstrated
overexpression of mitotic kinase encoding gene in upper
gastrointestinal adenocarcinomas through the immunohistochemical
analysis of 130 tumors. This overexpression was more prevalent in
gastroesophageal junction adenocarcinomas and lower in esophageal,
Barrett-related adenocarcinomas (BAS) than in antrum and body
gastric adenocarcinomas. They also found that the expression of
AURKA caused an anti-apoptotic effect in gastrointestinal cancer
cells with drug-induced apoptosis in an in vitro model
(27). Rugge et al found
that AURKA immunostaining increased significantly along with the
Barrett’s carcinogenesis, from Barrett’s mucosa even without
metaplasia towards Barrett’s adenocarcinoma (24). AURKA appears to play an important
role in the carcinogenesis of esophageal adenocarcinoma and will be
an important target for surveillance, diagnosis, treatment and
prognosis in Barrett’s esophagus. The positive relationship between
Barrett’s esophagus and AURKA is important for our results since in
this study we demonstrated that AURKA is upregulated in erosive
esophagitis and the AURKA gene was found to be elevated dependent
on the grade of erosive esophagitis based on the Los Angeles
Classification. GERD can yield to complications such as erosive
esophagitis and strictures; furthermore, it can cause Barrett’s
esophagus, which can progress to adenocarcinoma (29). We believe that ascertaining whether
the AURKA gene may be used as an early marker in erosive
esophagitis toward the development of EAC is crucial.
NIMA is another gene found to be related to cell
cycle dysfunction when it is overexpressed or underexpressed. NIMA
(never in mitosis gene a)-related kinase 6 (NEK6; also called
SID6-1512) is localized on chromosome 9q33-34 and is a
serine/threonine kinase that belongs to the Neks (NIMA-related
kinases) family, which has been implicated in mitosis control
(30). Yin et al previously
found that human Nek6 is required for metaphase-anaphase transition
during cell cycle progression (31). It is believed that interfering with
Nek6 function causes mitotic arrest and triggers apoptosis
(32,33). The negative mutant form of Nek6 was
found to induce spindle defects, abnormal chromosome segregation,
mitotic arrest and apoptosis (34). Overexpression of Nek6 was shown in
hepatocellular carcinoma as compared with the adjacent normal
tissue as an evidence of its antiapoptotic effect (18). Overexpression of NEK6 was
associated with histological grade, level of α feto protein and
poor prognosis. NEK6 was shown to mediate human cancer cell
transformation and was proposed as a potential cancer therapeutic
marker in a previous study (34).
Takeno et al stated that NEK6 is a potential marker of
gastric cancer regardless of stage and since conventional staging
of tumors are not adequate to predict individual prognosis, genetic
analyses of tumor tissues may provide better opportunities to
predict disease outcome for each individual and may even predict
response to therapy (35). The
authors selected seven focus genes showing a 2-fold change; four
had not been previously evaluated for the association with gastric
tumors. NEK6 was one these four new genes (35). They concluded that mapping of gene
expression data on large sample numbers helped to identify two
novel candidate genes, INHBA and NEK6, that are promising potential
markers of gastric cancer. Nassirpour et al revealed that
the protein level and kinase activity of Nek6 are highly elevated
in a variety of malignant human cancers including breast, uterus,
colon, stomach, ovary, lung, kidney, rectum, thyroid, cervix,
prostate, pancreas, small intestine cancer cells, and knockdown of
Nek6 resulted in reduction of tumors in a nude mouse xenograft
model (34). They concluded that
since inhibition of NEK6 specifically induces cell death in tumor
cells and not in normal tissues, NEK6 inhibitors are a better
therapeutic option with lower side effects than cytotoxic antitumor
agents.
This is the first study investigating the impact of
NEK6 in esophageal adenocarcinoma. Our data show the significant
upregulation of NEK6 in erosive esophagitis and esophageal
adenocarcinoma. NEK6 was more prevalent in samples with Los Angeles
classification B than A in erosive esophagitis demonstrating that
NEK6 and AURKA are more evident in more severe forms of
esophagitis.
Erosive esophagitis is chronic damage of the
esophagus caused by acid, pepsin and biliary salts. Environmental
insults cause genetic and epigenetic alterations and they affect
the expression of tumor-progenitor genes. Chronic injury is a major
cause of cancer even though it is not inherently mutagenic
(16). There are studies with
large number of patients and long follow-up periods for GERD
patients investigating whether they are at risk of developing
esophageal adenocarcinoma. In a Swedish nationwide case-control
study, gastroesophageal reflux and obesity were identified as
strong and independent risk factors for esophageal adenocarcinoma.
The risk increased with duration and severity of reflux symptoms
and with increasing body mass index (36). Erichsen et al performed a
nationwide cohort study in Denmark using data from 33,849 GERD
patients and concluded that erosive but not non-erosive reflux
disease has an impact on the development of adenocarcinoma
emphasizing inflammation as an important factor in carcinogenesis
(37).
In our study, in addition to AURKA and NEK6, HDAC9
and AURKC were significantly upregulated in EE compared to the
control group. These genes were not expressed in EAC. Wu et
al stated that AURKC was overexpressed in inflamed cervical
tissue specimens and HDAC inhibitors are therapeutic for several
inflammatory conditions (38,39).
Do these genes have a role in addition to defect of the defense
mechanism of the esophagus in patients with EE in gastroesophageal
disease?
MBD2 was significantly downregulated in EAC compared
to EE and the control group. MBD2 is a member of the MBD protein
family. MBD2 binds to methylated promoter CpG islands and acts as a
methylation-dependent transcriptional repressor (40). MBD2 has a role in the activation of
methylated and unmethylated genes (42). Expression of MBD2 was particularly
low in brain tumors, immune thrombocytopenia and in colorectal and
gastric carcinomas (42–44) as found in our study. The reasons
for the loss of MBD2 expression and the functional consequences are
unknown (44). Thus, MBD2 should
be studied further in relation to its association with esophageal
adenocarcinoma.
This is the first study concerning the epigenetic
chromatin histone modification in EE and EAC patients. Compared to
the other genes investigated, AURKA and NEK6 were notably
upregulated in EAC and EE. AURKA was proven to be associated with
EAC in previous studies, and we found similar results for AURKA.
This is the first study investigating the impact of NEK6 in
esophageal adenocarcinoma, and our data revealed the significant
upregulation of NEK6 in erosive esophagitis and esophageal
adenocarcinoma. Our study is also the first study aiming to detect
the presence of genetic upregulation in EE, a lesion which is not
considered to be a precancerous lesion. These results pave the way
for future studies with larger numbers of patients and longitudinal
studies with longer follow-up periods. In a recent study (7), we found that low prevalence of
Barrett’s esophagus was demonstrated in a Western Turkish
population. Based on these data, we intend to explore expression of
AURKA and NEK6 genes in the future at the national level, using
another group with Barrett’s esophagus.
It is hoped that future studies may address the
following questions: i) Is erosive esophagitis a precancerous
lesion and once detected, is surveillance required? ii) Can AURKA
and NEK6 be used as screening tests for esophageal adeno-carcinoma
in erosive esophagitis? iii) What are the ranges of AURKA and NEK6
overexpression predictive of the prognosis and the outcome of
antitumor therapy? iv) Can AURKA and NEK6 be used as therapeutic
targets?
In conclusion, we demonstrated overexpression of
AURKA and NEK6 in erosive esophagitis and esophageal adenocarcinoma
in a Turkish population. Understanding the molecular
pathophysiology of the disease will aid in elucidating the steps
for diagnosis, therapy and prognosis. AURKA and NEK6 are two
promising genetic markers for erosive esophagitis and esophageal
adenocarcinoma.
Abbreviations:
|
AURKA
|
aurora kinase A
|
|
AURKB
|
aurora kinase B
|
|
AURKC
|
aurora kinase C
|
|
BE
|
Barrett’s esophagus
|
|
EAC
|
esophageal adenocarcinoma
|
|
EE
|
erosive esophagitis
|
|
GERD
|
gastroesophageal reflux disease
|
|
HDAC9
|
histone deacetylase 9
|
|
MBD2
|
methyl-CpG binding domain protein
2
|
|
NEK6
|
never in mitosis gene A-related kinase
6
|
Acknowledgements
This study was supported by the Celal
Bayar University Coordinator of the Scientific Research Projects
(2009-053) Manisa, Turkey.
References
|
1
|
Ekiz F, Ormeci N, Coban S, et al:
Association of methylenetetrahydrofolate reductase C677T-A1298C
polymorphisms with risk for esophageal adenocarcinoma, Barrett’s
esophagus, and reflux esophagitis. Dis Esophagus. Sep 23–2011.(Epub
ahead of print).
|
|
2
|
Yoon HH, Khan M, Shi Q, et al: The
prognostic value of clinical and pathologic factors in esophageal
adenocarcinoma: a Mayo cohort of 796 patients with extended
follow-up after surgical resection. Mayo Clin Proc. 85:1080–1089.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bor S, Vardar R, Ormeci N, et al:
Prevalence patterns of gastric cancers in Turkey: model of a
developing country with high occurrence of Helicobacter
pylori. J Gastroenterol Hepatol. 22:2242–2245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reavis KM, Morris CD, Gopal DV, Hunter JG
and Jobe BA: Laryngopharyngeal reflux symptoms better predict the
presence of esophageal adenocarcinoma than typical gastroesophageal
reflux symptoms. Ann Surg. 239:849–856. 2004. View Article : Google Scholar
|
|
5
|
Das A: Tumors of the Esophagus. In:
Sleisenger and Fordtran’s Gastrointestinal and Liver Disease.
Feldman M, Friedman L, Friedman S and Brandt JL: Saunders,
Elsevier; Philadelphia: pp. 745–773. 2010
|
|
6
|
Zendehdel N, Biramijamal F, Hossein-Nezhad
A, Zendehdel N, Sarie H, Doughaiemoghaddam M and Pourshams A: Role
of cytochrome P450 2C19 genetic polymorphisms in the therapeutic
efficacy of omeprazole in Iranian patients with erosive reflux
esophagitis. Arch Iran Med. 13:406–412. 2010.PubMed/NCBI
|
|
7
|
Bayrakçi B, Kasap E, Kitapçioğlu G and Bor
S: Low prevalence of erosive esophagitis and Barrett esophagus in a
tertiary referral center in Turkey. Turk J Gastroenterol.
19:145–151. 2008.PubMed/NCBI
|
|
8
|
Bor S, Mandıracıoglu A, Kitapcıoglu G,
Caymaz-Bor C and Gilbert RJ: Gastroesophageal reflux in a
low-income region in Turkey. Am J Gastroenterol. 100:744–754.
2005.PubMed/NCBI
|
|
9
|
Peng D, Hu TL, Jiang A, Washington MK,
Moskaluk CA, Schneider-Stock R and El-Rifai W: Location-specific
epigenetic regulation of the metallothionein 3 gene in esophageal
adenocarcinomas. PLoS One. 6:e220092011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
11
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
12
|
Jones PA and Baylin SBl: The epigenomics
of cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feinberg AP, Ohlsson R and Henikoff S: The
epigenetic progenitor origin of human cancer. Nat Rev Genet.
7:21–33. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoo CB and Jones PA: Epigenetic therapy of
cancer: past, present and future. Nat Rev Drug Discov. 5:37–50.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lundell LR, Dent J, Bennett JR, et al:
Endoscopic assessment of oesophagitis: clinical and functional
correlates and further validation of the Los Angeles
classification. Gut. 45:172–180. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tzao C, Tung HJ, Jin JS, et al: Prognostic
significance of global histone modifications in resected squamous
cell carcinoma of the esophagus. Mod Pathol. 22:252–260. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao X, Xia Y, Yang J, et al: Clinical and
biological significance of never in mitosis gene A-related kinase 6
(NEK6) expression in hepatic cell cancer. Pathol Oncol Res.
18:201–207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Malumbres M and Barbacid M: Cell cycle
kinases in cancer. Curr Opin Genet Dev. 17:60–65. 2007. View Article : Google Scholar
|
|
20
|
Fu J, Bian M, Jiang Q and Zhang C: Roles
of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res.
5:1–10. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moniz L, Dutt P, Haider N and Stambolic V:
Nek family of kinases in cell cycle, checkpoint control and cancer.
Cell Div. 6:182011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sen S, Zhou H and White RA: A putative
serine/threonine kinase encoding gene BTAK on chromosome 20q13 is
amplified and overexpressed in human breast cancer cell lines.
Oncogene. 14:2195–2200. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reiter R, Gais P, Jütting U, et al: Aurora
kinase A messenger RNA overexpression is correlated with tumor
progression and shortened survival in head and neck squamous cell
carcinoma. Clin Cancer Res. 12:5136–5141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rugge M, Fassan M, Zaninotto G, et al:
Aurora kinase A in Barrett’s carcinogenesis. Hum Pathol.
41:1380–1386. 2010.
|
|
25
|
Lo Iacono M, Monica V, Saviozzi S, Ceppi
P, Bracco E, Papotti M and Scagliotti GV: Aurora Kinase A
expression is associated with lung cancer histological-subtypes and
with tumor de-differentiation. J Transl Med. 9:1002011.PubMed/NCBI
|
|
26
|
Sillars-Hardebol AH, Carvalho B, de Wit M,
et al: Identification of key genes for carcinogenic pathways
associated with colorectal adenoma-to-carcinoma progression. Tumour
Biol. 31:89–96. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dar AA, Zaika A, Piazuelo MB, et al:
Frequent overexpression of Aurora Kinase A in upper
gastrointestinal adenocarcinomas correlates with potent
antiapoptotic functions. Cancer. 112:1688–1698. 2008. View Article : Google Scholar
|
|
28
|
El-Rifai W and Powell SM: Molecular and
biologic basis of upper gastrointestinal malignancy. Gastric
carcinoma. Surg Oncol Clin N Am. 11:273–291. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shaheen N and Ransohoff DF:
Gastroesophageal reflux, Barrett esophagus, and esophageal cancer:
scientific review. JAMA. 287:1972–1981. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jee HJ, Kim AJ, Song N, Kim HJ, Kim M, Koh
H and Yun J: Nek6 overexpression antagonizes p53-induced senescence
in human cancer cells. Cell Cycle. 9:4703–4710. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin MJ, Shao L, Voehringer D, Smeal T and
Jallal B: The serine/threonine kinase Nek6 is required for cell
cycle progression through mitosis. J Biol Chem. 278:52454–52460.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Belham C, Roig J, Caldwell JA, Aoyama Y,
Kemp BE, Comb M and Avruch J: A mitotic cascade of NIMA family
kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol
Chem. 278:34897–34909. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
O’Regan L and Fry AM: The Nek6 and Nek7
protein kinases are required for robust mitotic spindle formation
and cytokinesis. Mol Cell Biol. 29:3975–3990. 2009.PubMed/NCBI
|
|
34
|
Nassirpour R, Shao L, Flanagan P, Abrams
T, Jallal B, Smeal T and Yin MJ: Nek6 mediates human cancer cell
transformation and is a potential cancer therapeutic target. Mol
Cancer Res. 8:717–728. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takeno A, Takemasa I, Doki Y, et al:
Integrative approach for differentially overexpressed genes in
gastric cancer by combining largescale gene expression profiling
and network analysis. Br J Cancer. 99:1307–1315. 2008. View Article : Google Scholar
|
|
36
|
Lagergren J: Increased incidence of
adenocarcinoma of the esophagus and cardia. Reflux and obesity are
strong and independent risk factors according to the SECC study.
Lakartidningen J. 97:1950–1953. 2000.(In Swedish).
|
|
37
|
Erichsen R, Robertson D, Farkas DK,
Pedersen L, Pohl H, Baron J and Sorensen HT: Erosive reflux disease
increases risk for esophageal adenocarcinoma risk, compared with
non-erosive reflux. Clin Gastroenterol Hepatol. Jan 12–2012.(Epub
ahead of print).
|
|
38
|
Wu SR, Li CF, Hung LY, Huang AM, Tseng JT,
Tsou JH and Wang J: MCCAAT/enhancer-binding protein delta mediates
tumor necrosis factor alpha-induced Aurora kinase C transcription
and promotes genomic instability. J Biol Chem. 286:28662–28670.
2011. View Article : Google Scholar
|
|
39
|
Shakespear MR, Halili MA, Irvine KM,
Fairlie DP and Sweet MJ: Histone deacetylases as regulators of
inflammation and immunity. Trends Immunol. 32:335–343. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Berger J and Bird A: Role of MBD2 in gene
regulation and tumorigenesis. Biochem Soc Trans. 33:1537–1540.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fujita H, Fujii R, Aratani S, Amano T,
Fukamizu A and Nakajima T: Antithetic effects of MBD2a on gene
regulation. Mol Cell Biol. 23:2645–2657. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Müller-Tidow C, Kügler K, Diederichs S, et
al: Loss of expression of HDAC-recruiting methyl-CpG-binding domain
proteins in human cancer. Br J Cancer. 85:1168–1174.
2001.PubMed/NCBI
|
|
43
|
Kanai Y, Ushijima S, Nakanishi Y and
Hirohashi S: Reduced mRNA expression of the DNA demethylase, MBD2,
in human colorectal and stomach cancers. Biochem Biophys Res
Commun. 2:962–966. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen ZP, Gu DS, Zhou ZP, et al: Decreased
expression of MBD2 and MBD4 gene and genomic-wide hypomethylation
in patients with primary immune thrombocytopenia. Hum Immunol.
72:486–491. 2011. View Article : Google Scholar : PubMed/NCBI
|