Introduction
Hepatocellular carcinoma (HCC) is one of the most
common types of cancer in the world and a significant cause of
mortality in sub-Saharan Africa and Eastern Asia (1). The overall five-year survival rate
following resection has remained as low as 35–50% (2–4). The
extremely poor prognosis of HCC is largely the result of a high
rate of recurrence following surgery and of metastasis (5,6).
Exploring the mechanisms involved in the process of HCC metastasis
is vital as it may provide new therapeutic targets for HCC and is
likely to be useful in further improving the long-term survival of
patients with HCC (7).
It is well documented that the specific organ
microenvironment not only affects the proliferation, angiogenesis
and invasion of cancer but also influences the expression of
metastasis-regulating genes (8,9).
Malignant cells in solid tumors communicate with the
microenvironment via a complex network of extracellular signals,
which includes a large number of cytokines (10). Therefore, investigation of the role
of these cytokines in the interaction between the organ
microenvironment and the tumor may aid the revelation of the
mechanism of tumor metastasis. Transforming growth factor-β (TGF-β)
is a known regulator of epithelial cells and of autonomous tumor
initiation, progression and metastasis (11–13).
Components of the TGF-β/Smad pathway are considered to be major
tumor suppressor genes; the absence or malfunction of the genes is
believed to lead to loss of growth regulation. Previous studies
have indicated that TGF-β is significantly involved in the
interactions between cancer cells and the tumor microenvironment.
Specifically, the loss of TGF-β signaling in stromal components may
result in an ‘activated’ microenvironment that supports and
initiates the transformation of adjacent epithelial cells (14). In HCC, TGF-β is a useful
serological marker for the early detection of cancer (15) and plays a dual role in the
progression of HCC. TGF-β is able to stimulate non-invasive HCC
cells to acquire invasive phenotypes (16) and also induces in vitro
apoptosis of hepatoma cells (17).
However, little is known regarding the interaction
between the organ environment and HCC. Moreover, the role of
TGF-β/Smad in the course of HCC has yet to be elucidated. In the
current study, we demonstrate that the organ microenvironment
regulates the growth and invasion of liver cancer cells via
TGF-β/Smad.
Materials and methods
Cell lines and culture
The MHCC97-H cell line was established from human
HCC cells in the Live Cancer Institute of Fudan University
(Shanghai, China) (18,19). The cells were cultured in high
glucose Dulbecco’s modified Eagle’s medium (H-DMEM; Gibco-BRL,
Carlsbad, CA, USA) and supplemented with 10% fetal calf serum
(Gibco-BRL) at 37°C in a humidified incubator containing 5%
CO2.
Establishment of animal models of
HCC
BALB/c nude mice, average weight 25 g, were used in
this experiment. MHCC97-H models were established by inoculating
6×106 MHCC97-H cells subcutaneously into the right sides
of the backs of the nude mice (n=26). Xenograft models were
established (n=17) via orthotopic implantation of MHCC97-H
subcutaneous tumor tissues (volume ∼2×2×1 mm3) into the
livers of the mice as previously reported (20). These experiments were approved by
the Shanghai Medical Experimental Animal Care Commission.
Collection of samples and analysis of
pulmonary metastasis
After feeding for 35 days, the animals were
sacrificed. The tumor tissues were removed and weighed. The lungs
were removed, fixed in paraformalin and embedded in paraffin. Each
sample was sliced into 20 sections, each 5 μm in thickness
with 50-μm intervals between successive sections. After
staining with hematoxylin and eosin (H&E), the sections were
independently observed under a microscope by two pathologists to
evaluate pulmonary metastasis.
RNA extraction and real-time PCR
The total RNA of the tumor tissues was extracted
using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the instructions of the product. Using
SYBR-Green mix (Toyobo Co., Ltd., Osaka, Japan), real-time RT-PCR
analysis was performed to identify the expression levels of TGF-β,
Smad2 and Smad7. The primers were designed by software (Primer
Premier 5.0) as follow: TGF-β sense, 5′-GGC GATACCTCAGCAACCG-3′ and
antisense, 5′-CTAAGG CGAAAGCCCTCAAT-3′; Smad2 sense, 5′-TACTACTCT
TTCCCCTGT-3′ and antisense, 5′-TTCTTGTCATTTCTA CCG-3′; Smad7 sense,
5′-CAACCGCAGCAGTTACCC-3′ and antisense, 5′-CGAAAGCCTTGATGGAGA-3′;
and β-actin sense, 5′-TCGTGCGTGACATTAAGGAG-3′ and antisense,
5′-ATGCCAGGGTACATGGTAAT-3′. The amplification conditions were: 95°C
for 9 min, followed by 45 cycles of 95°C for 30 sec, 57°C for 30
sec and 72°C for 15 sec, followed by an extension at 72°C for 5
min. β-actin was used as a control for the presence of amplifiable
cDNA. The mRNA expression level was assessed by 2−ΔΔCt.
In brief, the Ct value for the target gene was subtracted from the
Ct value of β-actin to yield a ΔCt value. The average ΔCt was
calculated for the control group and this value was subtracted from
the ΔCt of all other samples (including the control group). This
resulted in a ΔΔCt value for all samples which was then used to
calculate the fold-induction of mRNA expression of the target gene
using the formula 2−ΔΔCt, as recommended by the
manufacturer (Bio-Rad, Hercules, CA, USA). In the current study,
MHCC97-H model samples were used as the control.
Protein extraction and western blot
analysis
Sections of frozen tumor samples (n=14) were lysed
in RIPA buffer (50 mM Tris-HCl pH 7.5; 150 mM NaCl; 0.5% NaDOC; 1%
NP-40; and 0.1% SDS) with protease inhibitors. Protein was
extracted by spinning in a microcentrifuge for 30 min. Protein
concentrations were determined using the Bradford reagent. Equal
amounts of each sample (20 μl) and 10 μl markers were
run on 10% SDS-PAGE gels and electro-transferred onto PVDF
membranes using the Mini-Genie blotting system (Bio-Rad). The
membranes were incubated with primary antibody, mouse anti-human
TGF-β1 antibody (Chemicon, Temecula, CA, USA; 1:1000 diluted),
mouse anti-human β-actin antibody (Chemicon; 1:2000 dilution) and
HRP-conjugated goat anti-mouse IgG secondary antibody (Sigma, St.
Louis, MO, USA; 1:2000 dilution), The membranes were washed,
incubated with 10 ml LumiGLO and exposed to film.
Immunohistochemistry
Paraffin-embedded tumor tissues were sliced into 5
μm-thick sections and mounted on glass. The slides were
deparaffinized and rehydrated over 10 min through a graded alcohol
series to deionized water; 1% Antigen Unmasking solution (Vector
Laboratories, Burlingame, CA, USA) and microwave treatment were
used to enhance antigen retrieval. The slides were incubated with
the specific TGF-β1 primary antibody and with HRP-conjugated
secondary antibody and then stained with DAB.
ELISA
The total protein of all tumor tissues was extracted
as described in a previous section. TGF-β1 protein levels in the
tumors were determined using the Quantikine ELISA TGF-β1
immunoassay kit (R&D Systems, Minneapolis, MN, USA). The
operational approach was according to the manufacturer’s
instructions.
Statistical analysis
Statistical analysis of the data was performed using
SPSS 11.5 software (SPSS, Inc., Chicago, IL, USA). The correlation
between TGF-β/Smad and tumor weight was estimated using linear
regression analysis and correlation coefficients were evaluated by
the Student’s t-test. Multiple covariance analysis was used for
interaction between TGF-β/Smad and metastasis. All statistical
tests were two-sided; P<0.05 was considered to indicate a
statistically significant result.
Results
Tumor weight and pulmonary metastasis in
the animal models
The mean tumor weights (g) in the MHCC9-H and
xenograft models were 1.83±0.75 and 2.89±0.84, respectively
(P<0.01). The pulmonary metastatic rate and the number of
metastatic foci in the MHCC97-H model were higher than those in the
xenograft model (69.2 vs. 64.7%; and 4.72±5.50 vs. 2.27±1.01,
respectively) but the difference was not statistically significant,
while the number of metastatic cells approximated a statistically
significant difference (119.11±185.92 vs. 85.18±79.96,
respectively, P=0.08; Table
I).
 | Table ITumor weight and pulmonary metastasis
rate in two models of hepatocellular carcinoma. |
Table I
Tumor weight and pulmonary metastasis
rate in two models of hepatocellular carcinoma.
| Model | No. of cases | Tumor weight (g)
(mean ± SD) | Metastatic rate, %
(n/total) | No. of metastatic
foci (mean ± SD) | No. of metastatic
cells (mean ± SD) |
|---|
| MHCC97-H | 26 | 1.83±0.75 | 69.2 (18/26) | 4.72±5.50 | 119.11±185.92 |
| Xenograft | 17 | 2.89±0.84a | 64.7 (11/17) | 2.27±1.01 | 85.18±79.96b |
Organ microenvironment may affect the
expression of TGF-β1 and Smad
After implanting the MHCC97-H subcutaneous tumor
into the mouse liver, the mRNA levels of TGF-β1 were significantly
decreased (1.92±1.70 vs. 0.97±0.73; P=0.035). However, the Smad2
and Smad7 mRNA levels in the MHCC97-H model were not statistically
different from those in the xenograft model (Table II). The protein levels of TGF-β1 in
the MHCC97-H model mice were revealed to be higher than those in
the xenograft model mice by ELISA with OD values of 0.04±0.01 and
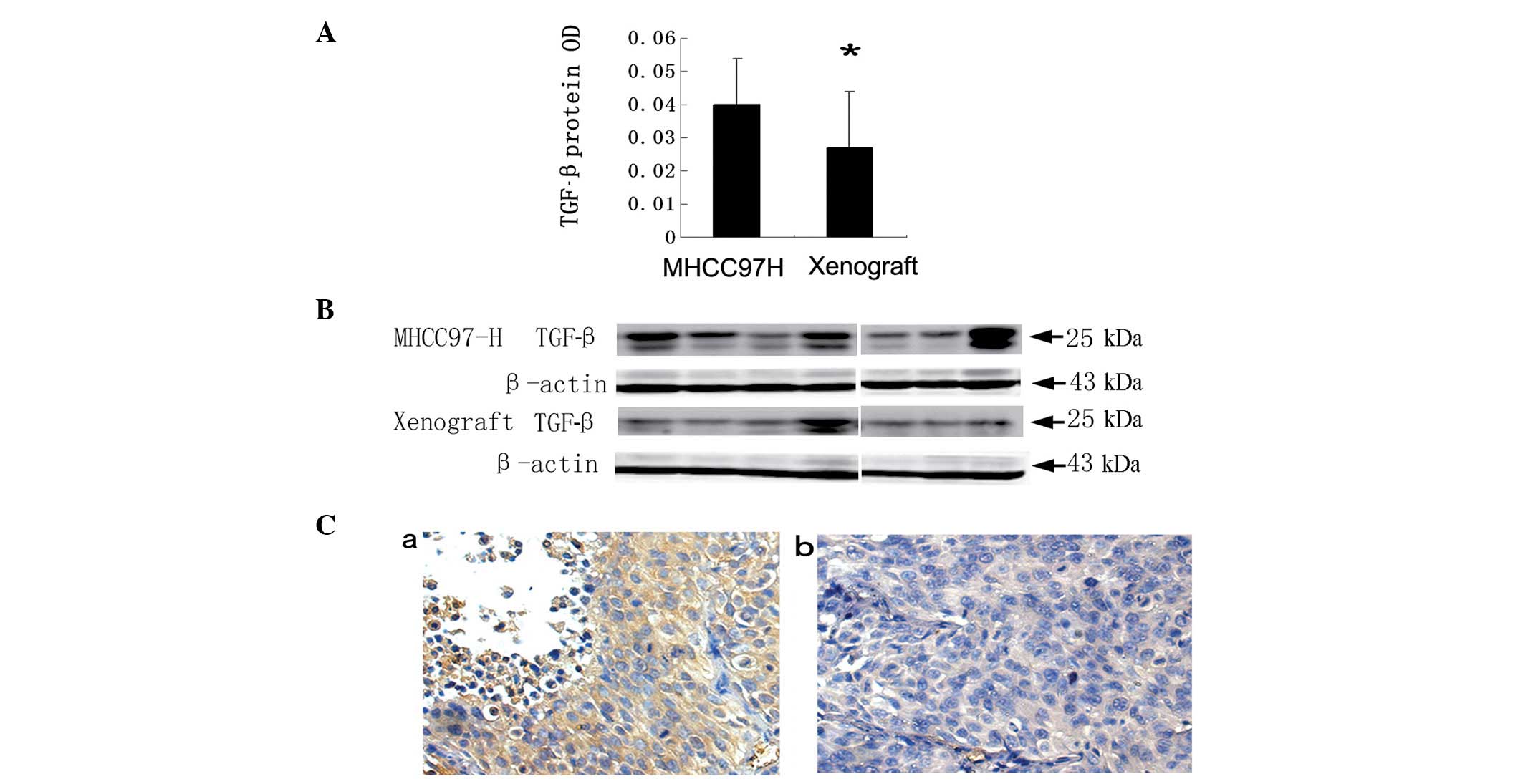
0.03±0.01, respectively, (P=0.003; Fig. 1A); similar results were obtained by
western blotting (Fig. 1B) and
immunohistochemical staining (Fig.
1C).
 | Table IIIn vivo expression of
TGF-β1/Smad mRNA in two mice models. |
Table II
In vivo expression of
TGF-β1/Smad mRNA in two mice models.
| | | 95% CI
| |
|---|
| mRNA | Models | 2−ΔΔCt
(mean ± SD) | Lower bound | Higher bound | P-value |
|---|
| TGF-β1 | MHCC97 | 1.92±1.70 | 1.23 | 2.61 | 0.035 |
| Xenograft | 0.97±0.73 | 0.59 | 1.35 | |
| Smad2 | MHCC97 | 1.17±1.09 | 0.73 | 1.61 | 0.89 |
| Xenograft | 1.13±0.31 | 0.98 | 1.29 | |
| Smad7 | MHCC97 | 1.46±0.97 | 1.07 | 1.85 | 0.17 |
| Xenograft | 1.10±0.52 | 0.83 | 1.37 | |
Expression of Smad7 correlated with tumor
size
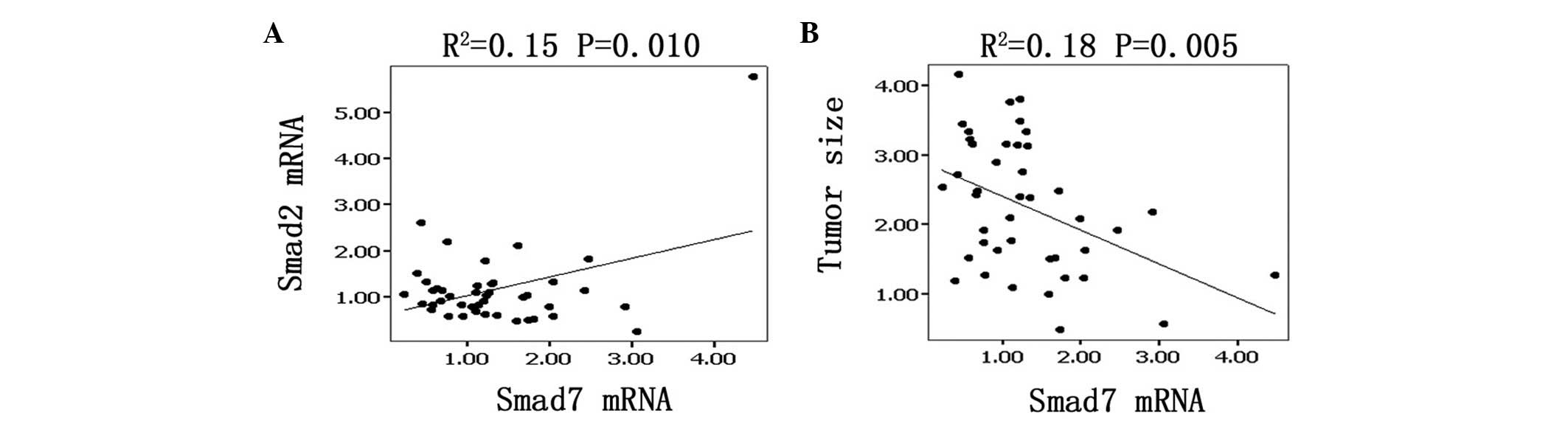
The mRNA levels of Smad7 linearly and positively
correlated with those of Smad2 by linear regression analysis. The
correlation coefficient R2=0.15, which has a statistical
significance according to the Student’s t-test (P=0.005; Fig. 2A). Moreover, Smad7 mRNA levels were
linearly and negatively correlated with tumor weight
(R2=0.18, P=0.005; Fig.
2B).
Expression of TGF-β1 mRNA correlated with
metastasis
We divided all the samples (n=43) into three groups
according to the median metastatic cell number: non-metastatic,
lower metastatic and higher metastatic groups. We identified that
the higher metastatic group had a higher mean TGF-β level than the
non-metastatic and lower metastatic groups by multiple covariance
analysis. The mean TGF-β1 mRNA levels were 2.32±2.11, 1.10±0.83 and
1.16±0.63, respectively, P= 0.024 (Table III).
 | Table III.Comparison of TGF-β1/Smad mRNA between
metastatic and non-metastatic samples. |
Table III.
Comparison of TGF-β1/Smad mRNA between
metastatic and non-metastatic samples.
| | | | 95% CI
| |
|---|
| mRNA | Metastasis | No. | 2−ΔΔCt
(mean ± SD) | Lower bound | Higher bound | P-value |
|---|
| TGF-β1 | None | 14 | 1.10±0.83 | 0.61 | 1.58 | 0.024 |
| Lower | 14 | 1.16±0.63 | 0.79 | 1.53 | |
| Higher | 15 | 2.32±2.11 | 1.16 | 3.49 | |
| Smad2 | None | 14 | 1.23±1.35 | 0.45 | 2.01 | 0.65 |
| Lower | 14 | 0.97±0.55 | 0.65 | 1.30 | |
| Higher | 15 | 1.25±0.47 | 0.99 | 1.52 | |
| Smad7 | None | 14 | 1.28±1.95 | 0.59 | 1.97 | 0.89 |
| Lower | 14 | 1.26±0.62 | 0.91 | 1.62 | |
| Higher | 15 | 1.40±0.62 | 1.06 | 1.75 | |
Discussion
Paget’s ‘seed and soil’ hypothesis suggests that the
interaction between tumor cells and target organ determines whether
metastasis will occur (21).
Metastasis depends on multiple interactions of cancer cells with
host homeostatic mechanisms. In this present study, when the
subcutaneous tumor tissues were transplanted into the liver,
pulmonary metastasis was reduced. These results suggest that the
organ microenvironment may alter the invasive potential of HCC.
We also found that the expression levels of TGF-β1
in the MHCC97-H and xenograft models were statistically different.
In addition, TGF-β1 was highly expressed in the higher metastatic
group and the expression of Smad7 negatively correlated with tumor
size. These result indicate that the TGF-β/Smad pathway plays an
important role in the interaction between HCC and the organ
microenvironment and affects the progression of HCC. Similar
results have been published for renal cancer, which demonstrate
that the basic fibroblast growth factor (bFGF) levels of tumors
implanted in kidney were 10–20 times higher than in subcutaneous
tumors (22). Other studies have
revealed that tumors growing in the stomach express more vascular
endothelial cell growth factor (VEGF) than ectopically placed
tumors and only the tumors in the stomach were able to undergo
metastasis (6,23).
It has been reported TGF-β plays a dual role in the
progression of tumors. During the early stages of tumor formation,
TGF-β acts as a tumor suppressor, inhibiting proliferation and
inducing apoptosis of tumor cells. However, during the later stages
of tumorigenesis, a number of tumor cells become unresponsive to
the growth inhibitory functions of TGF-β and become more motile and
invasive (24). Our findings that
the location of the tumor in the liver correlated with bigger size
and lower metastasis are consistent with the dual role of TGF-β1
following implantation and support this view.
The molecular mechanism for the downregulation of
TGF-β1 production in HCC in the liver remains unclear. With the
exception of the delicate balance between TGF-β and the tumor
microenvironment (14), TGF-β1
expression is affected by various growth factors secreted by normal
and tumor cells. Signaling by TGF-β family members occurs mainly
through Smad proteins (25), and
TGF-β and Smad may cross-talk with other pathways (26,27).
Future studies are necessary to determine whether the expression
levels of other factors are also modulated by the organ
microenvironment.
A number of tumors have a selectivity for metastasis
to specific organs; the precise cellular and molecular mechanisms
involved are unknown. It has been reported that differences in
tumor-secreted humoral factors, the upregulation of fibronectin and
site-specific delivery of VEGFR1+ cells within target
organs may promote metastatic spread in specific distant organs
(28). Our results indicate that
TGF-β1 is significant in the preference for metastasis to the
lung.
The results of the current study suggest that the
organ environment affects the progression of HCC. For many years,
all efforts to treat cancer have concentrated on the inhibition or
destruction of tumor cells, but none of them have been able to
alter the natural history of the disease (8,29).
Strategies to modulate the TGF-β levels of the host
microenvironment may provide a better approach for HCC
treatment.
Acknowledgements
This study was supported in part by
the China National Natural Science Foundation for Distinguished
Young Scholars (30325041), the China National ‘863’ R&D
High-Tech Key Project. The authors would like to thank Dr Qiong
Xue, Dongmei Gao and Jun Chen for assistance with the animal
experiments, Dr Ruixia Sun and Jie Chen for helpful suggestions
concerning cell culture and Dr Haiying Zeng and Tengfang Zhu for
performing the pathological experiments.
References
|
1
|
Parkin DM, Pisani P and Ferlay J: Global
cancer statistics. CA Cancer J Clin. 49:33–64. 1999. View Article : Google Scholar
|
|
2
|
Kurokawa Y, Matoba R, Takemasa I, et al:
Molecular-based prediction of early recurrence in hepatocellular
carcinoma. J Hepatol. 41:284–291. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lai EC, Fan ST, Lo CM, Chu KM, Liu CL and
Wong J: Hepatic resection for hepatocellular carcinoma. An audit of
343 patients Ann Surg. 221:291–298. 1995.PubMed/NCBI
|
|
4
|
Ono T, Yamanoi A, Nazmy El Assal O, Kohno
H and Nagasue N: Adjuvant chemotherapy after resection of
hepatocellular carcinoma causes deterioration of long-term
prognosis in cirrhotic patients: metaanalysis of three randomized
controlled trials. Cancer. 91:2378–2385. 2001. View Article : Google Scholar
|
|
5
|
Genda T, Sakamoto M, Ichida T, Asakura H,
Kojiro M, Narumiya S and Hirohashi S: Cell motility mediated by rho
and Rho-associated protein kinase plays a critical role in
intrahepatic metastasis of human hepatocellular carcinoma.
Hepatology. 30:1027–1036. 1999. View Article : Google Scholar
|
|
6
|
Ye QH, Qin LX, Forgues M, et al:
Predicting hepatitis B virus-positive metastatic hepatocellular
carcinomas using gene expression profiling and supervised machine
learning. Nat Med. 9:416–423. 2003. View
Article : Google Scholar
|
|
7
|
Qin LX and Tang ZY: Recent progress in
predictive biomarkers for metastatic recurrence of human
hepatocellular carcinoma: a review of the literature. J Cancer Res
Clin Oncol. 130:497–513. 2004.PubMed/NCBI
|
|
8
|
Fidler IJ: The organ microenvironment and
cancer metastasis. Differentiation. 70:498–505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fidler IJ, Kim SJ and Langley RR: The role
of the organ microenvironment in the biology and therapy of cancer
metastasis. J Cell Biochem. 101:927–936. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Leek RD, Harris AL and Lewis CE: Cytokine
networks in solid human tumors: regulation of angiogenesis. J
Leukoc Biol. 56:423–435. 1994.PubMed/NCBI
|
|
11
|
Giannelli G, Fransvea E, Marinosci F, et
al: Transforming growth factor-beta1 triggers hepatocellular
carcinoma invasiveness via alpha3beta1 integrin. Am J Pathol.
161:183–193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iyer S, Wang ZG, Akhtari M, Zhao W and
Seth P: Targeting TGFbeta signaling for cancer therapy. Cancer Biol
Ther. 4:261–266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oft M, Peli J, Rudaz C, Schwarz H, Beug H
and Reichmann E: TGFbeta1 and Ha-Ras collaborate in modulating the
phenotypic plasticity and invasiveness of epithelial tumor cells.
Genes Dev. 10:2462–2477. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stover DG, Bierie B and Moses HL: A
delicate balance: TGF-beta and the tumor microenvironment. J Cell
Biochem. 101:851–861. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song BC, Chung YH, Kim JA, et al:
Transforming growth factor-beta1 as a useful serologic marker of
small hepatocellular carcinoma. Cancer. 94:175–180. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Katabami K, Mizuno H, Sano R, et al:
Transforming growth factor-beta1 upregulates transcription of
alpha3 integrin gene in hepatocellular carcinoma cells via
Ets-transcription factor-binding motif in the promoter region. Clin
Exp Metastasis. 22:539–548. 2005. View Article : Google Scholar
|
|
17
|
Lin JK and Chou CK: In vitro apoptosis in
the human hepatoma cell line induced by transforming growth factor
beta 1. Cancer Res. 52:385–388. 1992.PubMed/NCBI
|
|
18
|
Li Y, Tang Y, Ye L, et al: Establishment
of a hepatocellular carcinoma cell line with unique metastatic
characteristics through in vivo selection and screening for
metastasis-related genes through cDNA microarray. J Cancer Res Clin
Oncol. 129:43–51. 2003.
|
|
19
|
Li Y, Tang ZY, Ye SL, et al: Establishment
of cell clones with different metastatic potential from the
metastatic hepatocellular carcinoma cell line MHCC97. World J
Gastroenterol. 7:630–636. 2001.PubMed/NCBI
|
|
20
|
Zhou J, Tang ZY, Fan J, et al:
Capecitabine inhibits postoperative recurrence and metastasis after
liver cancer resection in nude mice with relation to the expression
of platelet-derived endothelial cell growth factor. Clin Cancer
Res. 9:6030–6037. 2003.
|
|
21
|
Paget S: The distribution of secondary
growths in cancer of the breast. 1889 Cancer Metastasis Rev.
8:98–101. 1989.PubMed/NCBI
|
|
22
|
Singh RK, Bucana CD, Gutman M, Fan D,
Wilson MR and Fidler IJ: Organ site-dependent expression of basic
fibroblast growth factor in human renal cell carcinoma cells. Am J
Pathol. 145:365–374. 1994.PubMed/NCBI
|
|
23
|
Takahashi Y, Mai M, Wilson MR, Kitadai Y,
Bucana C and Ellis L: Site-dependent expression of vascular
endothelial growth factor, angiogenesis and proliferation in human
gastric carcinoma. Int J Oncol. 8:701–705. 1996.PubMed/NCBI
|
|
24
|
Welm AL: TGFbeta primes breast tumor cells
for metastasis. Cell. 133:27–28. 2008. View Article : Google Scholar
|
|
25
|
Itoh S, Itoh F, Goumans MJ and Ten Dijke
P: Signaling of transforming growth factor-b family members through
Smad proteins. Eur J Biochem. 267:6954–6967. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kwak HJ, Park MJ, Cho H, et al:
Transforming growth factor-beta1 induces tissue inhibitor of
metalloproteinase-1 expression via activation of extracellular
signal-regulated kinase and Sp1 in human fibrosarcoma cells. Mol
Cancer Res. 4:209–220. 2006. View Article : Google Scholar
|
|
27
|
Watanabe H, de Caestecker MP and Yamada Y:
Transcriptional cross-talk between Smad, ERK1/2, and p38
mitogen-activated protein kinase pathways regulates transforming
growth factor-beta-induced aggrecan gene expression in chondrogenic
ATDC5 cells. J Biol Chem. 276:14466–14473. 2001.
|
|
28
|
Kaplan RN, Riba RD, Zacharoulis S, et al:
VEGFR1-positive haematopoietic bone marrow progenitors initiate the
pre-metastatic niche. Nature. 438:820–827. 2005. View Article : Google Scholar
|
|
29
|
Loberg RD, Bradley DA, Tomlins SA,
Chinnaiyan AM and Pienta KJ: The lethal phenotype of cancer: the
molecular basis of death due to malignancy. CA Cancer J Clin.
57:225–241. 2007. View Article : Google Scholar : PubMed/NCBI
|