Introduction
Killer cell immunoglobulin-like receptor (KIR),
expressed by natural killer (NK) cells and certain T cells, is a
type of CD158 glucoprotein (1).
Through the recognition of human leukocyte antigen (HLA)-I, KIR
regulates the cytoactivity of the effector cell, begins the
immunological process and regulates cytokine secretion. KIR gene
clusters have extremely complex gene structures gradually formed
from the single gene structure of KIR3DX1 (2). The human KIR gene cluster contains
fifteen genes and two silencers. The KIR gene cluster encoding the
receptor contains protein guide sequences (exons 1 and 2), the
extracellular domain (exons 3–5), stem (exon 6), transmembrane
domain (exon 7) and intracellular domain (exons 8 and 9). The KIR
gene cluster genes are named according to their extracellular
domain and intracellular domain structural features. An
extracellular domain with two similar immunoglobulin domains would
be named 2D, or 3D if it had three similar immunoglobulin domains.
KIR genes with longer intracellular domains and transduction
inhibitory signals are designated L, while those with shorter
intracellular domains and transduction activition signals are
designated S. The main ligand of KIR is the HLA-I class antigen.
Following the recognition of various HLAs, the KIR receptor
determines whether the object belongs to the ‘self’ or not, thus
making it extremely important in immune regulation (3,4). The
KIRs may be divided into two categories: stimulatory KIRs (sKIRs)
and inhibitory KIRs (iKIRs).
The specific receptors of certain inhibitory KIR
class HLA-I molecules are already known. The ligand of the
inhibitory genes KIR2DL1, -2DL2 and -2DL3 is HLA-C. Based on the
amino acid combination at the heavy chain loci 80, HLA-C has been
divided into two groups. In the HLA-C1 group, the amino acid of
site 80 is lysine, while in the HLA-C2 group it is aspartic acid.
The ligand of KIR2DL1 is HLA-C2 and the ligand of KIR2DL2/2DL3 is
HLA-C1 (5). KIR2DL1+HLA-C2
combined excitatory and inhibitory signals are stronger than those
of KIR2DL2/2DL3+HLA-C1 (6). The
ligand of KIR3DL1/S1 is the HLA-B antigen which contains a Bw4
structure. It has been reported that KIR3DL1/S1 is able to combine
with the HLA-A antigen, which also contains a Bw4 structure
(7). The ligand of KIR3DL2 is
HLA-A3 or HLA-A11. The ligand of KIR2DL4 is HLA-G. The ligand of
KIR2DS1-5, an activating gene of KIR, may be an HLA class I antigen
although there is no direct evidence to support this theory.
Individuals with variations in the KIR gene cluster number and type
may be divided into the haploid A and B subtypes, according to its
haploid configuration. Haploid A contains frame gene KIR2DL1,
KIR2DL3, KIR3DL1 suppressor gene, and KIR2DS4 activating gene.
Other configurations are collectively referred to as haploid B.
Haploid B contains a number of activating genes.
HLA-Cw is distributed widely in human tissue cells,
specifically recognized by KIR and involved in the regulation of NK
cell function, which is the basis of NK cell recognition of self
and non-self molecules. When the same HLA-Cw molecule is combined
with different KIRs, it produces different signals to activate or
inhibit the killing function of the NK cells (8). Observations have revealed that there
are differences between individuals in the combined forms of HLA-Cw
and sKIR or iKIR, which not only affect susceptibility to disease
(9) but also disease progression
(10). In order to understand the
method of the recognition of HLA-Cw and KIR in a Chinese
population, the KIR and its HLA-Cw ligands of 154 normal Han
individuals from Jilin were examined by PCR-SSP and the
identification of HLA-Cw and KIR was analyzed, to provide a the
theoretical basis for future research concerning its role in
disease progression.
Materials and methods
Objective of the study
A total of 154 normal specimens were selected from
unpaid blood donors from a Jilin Han population. This group
contained 84 males and 70 females (no blood relations) aged between
21 and 50 years old. Venous blood (5 ml) was obtained, placed in
test tubes containing EDTA anticoagulants and stored in a −80°C
refrigerator. The experimental protocol was established according
to the guidelines of the Declaration of Helsinki and was approved
by Human Ethics Committee of Jilin University. Informed consent was
obtained from each subject prior to their inclusion in the
study.
Genomic DNA extraction
Using the TIANamp Blood DNA kit (Tiangen Biotech
Beijing Co. Ltd., Beijing, China), genomic DNA was extracted
according to the manufacturer’s instructions. The DNA concentration
was ≥80 mg/l and the purity was A260/280 =
1.65–1.90.
Primer synthesis
A previously published method for synthesizing the
amplified KIR genes using PCR-SSP genotyping primers (Table I) was used (11). The internal control primers of
reactions 1 to 15 were FGH (5′-GCCTTCCCAACCATTCCCTTA-3′) and RGH
(5′-TCACGGATTTCTGTTGTGTTTC-3′). The length of the amplification
product was 429 bp; the internal contrast primers of reaction 16
were DRB1-F (5′-TGCCAAGTGGAGCACCCAA-3′) and DRB1-R
(5′-GCATCTTGCTCTGTGCAGAT-3′) and the length of the amplification
products was 796 bp. The primers of the HLA-Cw gene for PCR-SSP
genotyping were in accordance with those of Mandelboim et
al(5). The primers are shown
in Table II. All primers were
synthesized by Beijing DingGuo ChangSheng Biotechnology Co., Ltd.
(Beijing, China) and verified by BLAST.
 | Table IPrimers used for KIR genotyping by
PCR-SSP. |
Table I
Primers used for KIR genotyping by
PCR-SSP.
| Gene | Primer | Sequence | Primer | Sequence | Size (bp) |
|---|
| 2DL1 | F1 |
GTTGGTCAGATGTCATGTTTGAA | R1 |
GGTCCCTGCCAGGTCTTGCG | 146 |
| F2 |
TGGACCAAGAGTCTGCAGGA | R2 |
TGTTGTCTCCCTAGAAGACG | 330 |
| 2DL2 | F1 |
CTGGCCCACCCAGGTCG | R1 |
GGACCGATGGAGAAGTTGGCT | 173 |
| F2 |
GAGGGGGAGGCCCATGAAT | R2 |
TCGAGTTTGACCACTCGTAT | 151 |
| 2DL3 | F1 |
CTTCATCGCTGGTGCTG | R1 |
AGGCTCTTGGTCCATTACAA | 550 |
| F2 |
TCCTTCATCGCTGGTGCTG | R2 |
GGCAGGAGACAACTTTGGATCA | 800 |
| 2DL4 | F1 |
CAGGACAAGCCCTTCTGC | R1 | CTGGGTGCCGACCACT | 254 |
| F2 |
ACCTTCGCTTACAGCCCG | R2 |
CCTCACCTGTGACAGAAACAG | 288 |
| 2DS2 | F1 |
TTCTGCACAGAGAGGGGAAGTA | R1 |
GGGTCACTGGGAGCTGACAA | 175 |
| F2 | CGGGCCCCACGGTTT | R2 |
GGTCACTCGAGTTTGACCACTCA | 240 |
| 2DS3 | F1 | TGGCCCACCCAGGTCG | R1 |
TGAAAACTGATAGGGGGAGTGAGG | 242 |
| F2 |
CATTGACATGTACCATCTATCCAC | R2 |
AAGCAGTGGGTCACTTGAC | 190 |
| 2DS5 | F1 |
TGATGGGGTCTCCAAGGG | R1 |
TCCAGAGGGTCACTGGGC | 126 |
| F2 |
ACAGAGAGGGGACGTTTAACC | R2 |
ATGTCCAGAGGGTCACTGGG | 178 |
| 3DL1 | F1 | CGCTGTGGTGCCTCGA | R1 |
GGTGTGAACCCCGACATG | 191 |
| F2 |
CCCTGGTGAAATCAGGAGAGAG | R2 |
TGTAGGTCCCTGCAAGGGCAA | 186 |
| 3DL2 | F1 |
CAAACCCTTCCTGTCTGCCC | R1 |
GTGCCGACCACCCAGTGA | 211 |
| F2 |
CCCATGAACGTAGGCTCCG | R2 | CACACGCAGGGCAGGG | 130 |
| 3DS1 | F1 |
AGCCTGCAGGGAACAGAAG | R1 |
GCCTGACTGTGGTGCTCG | 300 |
| F2 |
CCTGGTGAAATCAGGAGAGAG | R2 | GTCCCTGCAAGGGCAC | 180 |
| 3DL3 | F1 |
GTCAGGACAAGCCCTTCCTC | R1 |
GAGTGTGGGTGTGAACTGCA | 232 |
| F2 |
TTCTGCACAGAGAGGGGATCA | R2 |
GAGCCGACAACTCATAGGGTA | 165 |
| 2DL5 | F1 | GCGCTGTGGTGCCTCG | R1 |
GACCACTCAATGGGGGAGC | 214 |
| F2 |
TGCAGCTCCAGGAGCTCA | R2 |
GGGTCTGACCACTCATAGGGT | 191 |
| 2DP1 | F1 |
GTCTGCCTGGCCCAGCT | R1 |
GTGTGAACCCCGACATCTGTAC | 205 |
| F2 |
CCATCGGTCCCATGATGG | R2 |
CACTGGGAGCTGACAACTGATG | 89 |
| 2DS1 | F1 |
CTTCTCCATCAGTCGCATGAA | R1 |
AGAGGGTCACTGGGAGCTGAC | 102 |
| F2 |
CTTCTCCATCAGTCGCATGAG | | | |
| 2DS4 | F1 |
CTGGCCCTCCCAGGTCA | R1 |
TCTGTAGGTTCCTGCAAGGACAG | 204 |
| F2 |
GTTCAGGCAGGAGAGAAT | R2 |
GTTTGACCACTCGTAGGGAGC | 197/219 |
| 3DP1 | F1 |
GGTGTGGTAGGAGCCTTAG | R1 |
GAAAACGGTGTTTCGGAATAC | 280 |
| F2 |
CGTCACCCTCCCATGATGTA | | | 395 |
 | Table IIPrimers used for HLA-Cw genotyping by
PCR-SSP. |
Table II
Primers used for HLA-Cw genotyping by
PCR-SSP.
| Forward primers
| Reverse primers
| |
|---|
| Gene | Primer | Sequence | Primer | Sequence | Size (bp) |
|---|
|
HLA-C1Asn80 | F |
GAGGTGCCCGCCCGGCGA | R |
CGCGCAGGTTCCGCAGGC | 332 |
|
HLA-C2Lys80 | F |
GAGGTGCCCGCCCGGCGA | R |
CGCGCAGTTTCCGCAGGT | 332 |
KIR genes and the HLA-Cw PCR-SSP
classification
Each KIR gene required two pairs of KIR gene primers
(upstream and downstream) plus an internal contrast primer system.
The HLA-Cw gene also required upstream and downstream primers and
an internal contrast primer system.
To amplify KIR genes by PCR-SSP (12), 1 μl of each primer mix (5 μM) was
dispensed into separate wells in 384-well plates. Each sample
required two vertical rows of wells but KIR2DS1 and KIR3DP1
required only one well. PCR cocktails were prepared for each sample
with a total volume of 132 μl (enough for 33 reactions) as follows:
200 ng DNA, 16.5 μl 10X PCR buffer (final concentration, 1X), 4.95
μl MgCl2 (final concentration 1.5mM), 1.32 μl dNTP
(final concentration 200 mM) and 0.825 μl Taq polymerase.
PCR cocktail (4 μl) was added to each primer mix (total PCR volume
= 5 μl). The samples were amplified using a programmable thermal
cycler with a heated lid using the following parameters: 3 min at
94°C, 5 cycles of 15 sec at 94°C, 15 sec at 65°C and 30 sec at
72°C; 21 cycles of 15 sec at 94°C, 15 sec at 60°C and 30 sec at
72°C; 4 cycles of 15 sec at 94°C, 1 min at 55°C and 2 min at 72°C
with a final 7-min extension step at 72°C.
The HLA-Cw genes were amplified using PCR-SSP
genotyping (12). The PCR system
was as follows: In each reaction, 100 ng of genomic DNA was
amplified in 15 μl of 1X PCR master mix [67 mM Tris-HCl, pH 8.8,
16.6 mM (NH4)2SO4, 0.1% Tween-20,
2 mM MgCl2, 200 mM dNTP] containing specific (0.1–0.8
μm) and control primers, as well as 0.5 units Taq
polymerase. PCR conditions were: 2 min at 94°C, 10 cycles of 10 sec
at 94°C and 60 sec at 65°C; 20 cycles of 10 sec at 94°C, 50 sec at
61°C and 30 sec at 72°C with a final 10-min extension step at
72°C.
Gel electrophoresis
The amplification products (5 μl) were
electrophoresed on 2% agarose gels with a migration distance of ∼3
cm and stained with ethidium bromide. The amplification was
evaluated on a UV transilluminator and photographed.
Analysis of the recognition of HLA-Cw and
KIR
The difference in the method of recognition between
various HLA-Cws and KIRs depended on the 80th amino acid residues
of the HLA-Cw. If the residue was lysine (Lys;
HLA-Cw2Lys80), the receptor was KIR2DL1/S1. If the
residue was asparagine (Asn; HLA-Cw1Asn80), the receptor
was KIR2DL2/L3/S2. The amino acid sequence was acquired according
to the HLA-Cw classification results. By combining this information
with the phenotype of the KIR, separate HLA-Cw and KIR recognition
statistics could be analyzed.
Statistical analysis
The statistical analysis followed that of Martin and
Carrington (11) and also included
the frequency and genotype frequency of HLA-Cw and KIR in the Han
population of Jilin. The KIR gene-frequency (F) was measured by
direct counting, the phenotype frequency (pf; %) = total number of
positive gene/total nu,ber of research group; the KIR gene
frequency was calculated as F = 1−√(1−pf).
Results
Electrophoresis of the KIR gene
Electrophoretic analysis following the conventional
amplification of the KIR gene revealed that the size of the target
fragment was consistent with the expected results (Fig. 1).
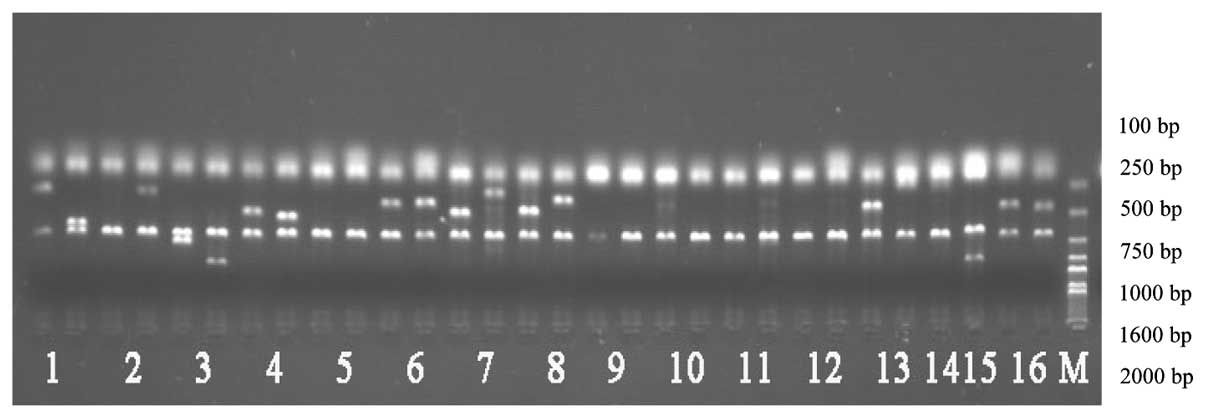 | Figure 1KIR genotyping by PCR-SSP. Lanes 1-16,
the PCR-SSP products of KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4,
KIR2DL5, KIR3DL1, KIR3DL2, KIR3DL3, KIR2DS2, KIR2DS3, KIR2DS5,
KIR3DS1, KIR2DP1, KIR2DS1 and KIR3DP1, respectively; M, Marker
2000; KIR, killer cell immunoglobulin-like receptor. |
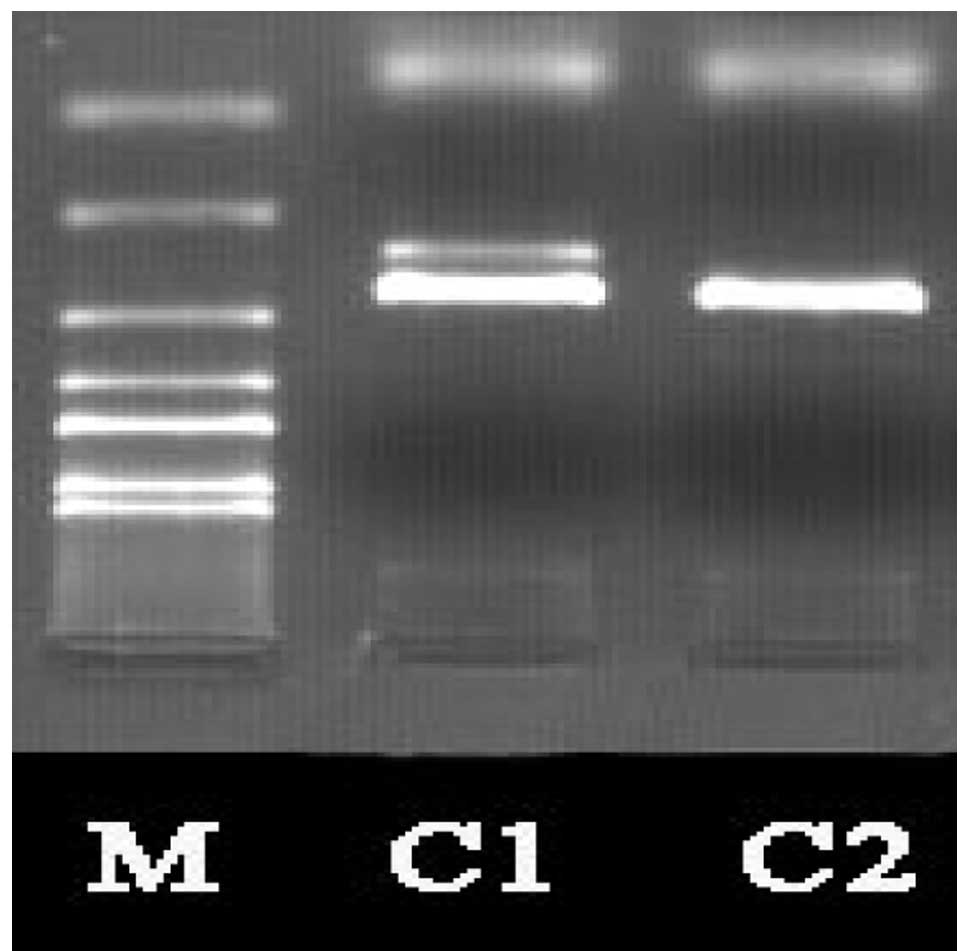
Electrophoresis of the HLA-Cw gene
Electrophoretic analysis following the conventional
amplification of the HLA-Cw gene revealed that the size of the
target fragment was consistent with the expected results (Fig. 2).
Phenotype and genotype frequencies of
HLA-Cw and KIR in the Han population in Jilin
The results are shown in Table III.
 | Table IIIPhenotype and genotype frequency of
KIR and HLA-Cw in the Jilin Han population. |
Table III
Phenotype and genotype frequency of
KIR and HLA-Cw in the Jilin Han population.
| Gene | Number | Phenotype frequency
(%) | Genotype
frequency |
|---|
| 2DL1 | 153 | 99.351 | 0.919 |
| 2DL2 | 21 | 13.636 | 0.071 |
| 2DL3 | 153 | 99.351 | 0.919 |
| 2DL4 | 154 | 100 | 1 |
| 2DL5 | 49 | 31.818 | 0.174 |
| 3DL1 | 143 | 92.857 | 0.733 |
| 3DL2 | 154 | 100 | 1 |
| 3DL3 | 154 | 100 | 1 |
| 2DS2 | 19 | 12.338 | 0.064 |
| 2DS3 | 18 | 11.688 | 0.06 |
| 2DS5 | 37 | 24.026 | 0.128 |
| 3DS1 | 43 | 27.922 | 0.151 |
| 2DP1 | 153 | 99.351 | 0.919 |
| 2DS1 | 48 | 31.169 | 0.17 |
| 3DP1*003 | 76 | 49.351 | 0.288 |
|
3DP1*001/002/004 | 153 | 99.351 | 0.919 |
| 2DS4*001/002 | 113 | 73.377 | 0.484 |
| 2DS4*003-007 | 72 | 46.753 | 0.27 |
|
HLA-C1Asn80 | 123 | 79.87 | 0.551 |
|
HLA-C2Lys80 | 43 | 27.922 | 0.151 |
Recognition of HLA-C2Lys and
KIR
The HLA-C2Lys expression frequency was
27.92%, while 72.08 and 21.43% of individuals expressed only
KIR2DL1 or KIR2DS1, respectively, without HLA-C2Lys.
Table IV shows the distribution of
the HLA-Cw KIR activating and inhibitory pairing in the Han
population of Jilin. The frequency of HLA-C2Lys80+2DL1
was 27.27% and that of 2DS1+HLA-C2Lys80 was 9.74%. Of
the studied individuals, 21.43% expressed only KIR2DS1 without
HLA-C2Lys-KIR2DL1.
 | Table IVThe combined frequencies of various
compound KIR-HLA genetypes in the Jilin Han population. |
Table IV
The combined frequencies of various
compound KIR-HLA genetypes in the Jilin Han population.
| iKIR+HLA | Frequency (%) | sKIR+HLA | Frequency (%) |
|---|
|
HLA-C1Asn80+2DL2/2DL3 | 68.83 |
2DS2+HLA-C1Asn80 | 9.74 |
|
HLA-C2Lys80+2DL1 | 27.27 |
2DS1+HLA-C2Lys80 | 9.74 |
Recognition of HLA-C1Asn
KIR
The HLA-C1Asn80 expression frequency was
79.87%, notably higher than the expression frequency of
HLA-C2Lys80 which was 27.92%, while 34.42% of
individuals expressed only HLA-C1Asn without iKIR or
sKIR. Table IV shows the
distribution of the HLA-Cw KIR activating and inhibitory pairings
in the Han population of Jilin. The frequency of
HLA-C1Asn80+2DL2/2DL3 was 68.83% and that of
2DS2+HLA-C1Asn80 was 9.74%. Of the studied individuals,
2.60% expressed only KIR2DS2 without
HLA-C1Asn-KIR2DL2/L3.
Discussion
NK cells recognize virus-infected tissue and tumor
cells, then modulate the immune response by killing the target
cells through cytotoxic effects or secreting cytokines. NK cells
constitute the first barrier of the human immune system (13,14).
Immature NK cells must be ‘educated’ by the MHC HLA-Class I
molecules, in order to become mature NK cells (15). NK cells adjust their functional
status precisely through inhibiting or activating receptors on the
membrane surface. In the long-term coevolution of the KIR and the
HLA, now highly efficient and accurate receptor-ligand system is
formed gradually. (16).
Previously, in a study of 477 cases of malaria, Hirayasu et
al(17) observed that there
was a marked correlation between the KIR2DL3 gene and its ligand
HLA-C1 in the cerebral malaria patients compared with non-cerebral
malaria patients. Further research revealed that the frequency of
the combination of the KIR2DL3-HLA-C1 in the malaria high-risk
population was maintained at a low level.
Thus, the KIR and HLA combination system is
important in regulating NK cell function. Therefore, determining
the method of recognition of the HLA-Cw and KIR is particularly
important. A total of 154 Han individuals from Jilin were selected.
The KIR and HLA-Cw genotyping results revealed that the expression
frequency of HLA-C1Asn80 was 79.87%, lower than the
97.5% of a Han population in Guangdong (18) and the 76% of a population in Iran
(12). The expression frequency of
HLA-C2Lys80 in the Han population of Jilin was 27.92%,
which was equal to that of the Guangdong Han population (18) and lower than the frequency in the
Iranian population (12). The
frequency of HLA-C1Asn80 in the Guangdong Han population
was 97.5%, which was significantly higher than that of the Jilin
Han population. Compared with other populations, the combination
frequency of the KIR receptor and its ligand in the Jilin Han
population was essentially the same. The frequency of
KIR2DS2+HLA-C1Asn80 in the Han population of Jilin was
9.74%, less than that of the Guangdong Han (14%) and Iranian
(21.5%) populations. The Jilin Han
HLA-C1Asn80+KIR2DL2/2DL3 frequency was 68.83%, higher
than in the Guangdong Han (41.8%) and Iranian (6.5%) populations.
These observations may be due to ethnic and regional differences of
the HLA and KIR.
In the 154 Han individuals from Jilin, it was also
observed that 9.74% of individuals expressed only the combination
of 2DS2+HLA-C1Asn80 and 9.74% expressed only the
combination of 2DS1+HLA-C2Lys80. For these individuals,
due to the inhibitory signal being unable to completely block the
activation of signal transduction, their NK cells may attack other
tissues, resulting in autoimmune diseases (10). However, this has yet to be
confirmed in patients through larger sample and clinical
experiments.
Martin et al(9) suggested that the ligands of sKIRs may
not be HLA antigens, but bacterial proteins. The authors observed
that the KIR2DL1 and KIR2DL2/L3 frequency was high, up to 100%.
Therefore the individual expression of HLA-Cw indicates that a
corresponding inhibitory ligand receptor is activated. NK cells
with sKIR lacking a corresponding HLA-Cw (for example, KIR2DS1
without HLA-Cw2Lys) may be activated by bacterial
proteins, resulting in autoimmune diseases. In the current study,
21.43% of individuals were observed to express KIR2DS1 without
HLA-Cw2Lys inhibitory receptor ligands. Due to the
expression frequency of HLA-Cw1Asn being up to 79.87%,
no expression of KIR2DS2 only was observed. However, 2.60% of the
population expressed KIR2DS2 but not HLA-Cw1Asn
inhibitory receptor ligands. The susceptibility to autoimmune
disease of such individuals is worthy of investigation.
Acknowledgements
This study was supported by grants
from the National Natural Science Foundation of China (No.
30972610). The authors would like to thank Medjaden Bioscience
Limited for assisting in the preparation of this manuscript.
References
|
1
|
Parham P: Immunogenetics of killer cell
immunoglobulin-like receptors. Mol Immunol. 42:459–462. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guethlein LA, Abi-Rached L, Hammond JA and
Param P: The expanded cattle KIR genes are orthologous to the
conserved single-copy KIR3DX1 gene of primates. Immunogenetics.
59:517–522. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Bergen J, Thompson A, Retière C, et
al: Cutting edge: killer Ig-like receptors mediate ‘missing self’
recognition in vivo. J Immunol. 182:2569–2572. 2009.
|
|
4
|
Lanier LL: Missing self, NK cells, and The
White Album. J Immunol. 174:65652005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mandelboim O, Reyburn HT, Valés-Goméz M,
et al: Protection from lysis by natural killer cells of group 1 and
2 specificity is mediated by residue 80 in human histocompatibility
leukocyte antigen C alleles and also occurs with empty major
histocompatibility complex molecules. J Exp Med. 184:913–922. 1996.
View Article : Google Scholar
|
|
6
|
Moesta AK, Norman PJ, Yawata M, et al:
Synergistic polymorphism at two positions distal to the
ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C
than KIR2DL3. J Immunol. 180:3969–3979. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stern M, Ruggeri L, Capanni M, et al:
Human leukocyte antigens A23, A24, and A32 but not A25 are ligands
for KIR3DL1. Blood. 112:708–710. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin Xiaolin: The evolution, expression and
function of KIR gene. Immunology. 20:S103–S105. 2004.
|
|
9
|
Martin MP, Nelson G, Lee JH, et al:
Cutting edge: susceptibility to psoriatic arthritis: influence of
activating killer Ig-like receptor genes in the absence of specific
HLA-Cw alleles. J Immunol. 169:2818–2822. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yen JH, Moore BE, Nakajima T, et al: Major
histocompatibility complex class I-recognizing receptors are
disease risk genes in rheumatoid arthritis. J Exp Med.
193:1159–1167. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martin MP and Carrington M: KIR locus
polymorphisms: genotyping and disease association analysis. Methods
Mol Biol. 415:49–64. 2008.PubMed/NCBI
|
|
12
|
Tajik N, Shahsavar F, Nasiri M and
Radjabzadeh MF: Compound KIR-HLA genotype analyses in the Iranian
population by a novel PCR-SSP assay. Int J Immunogenet. 37:159–168.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wodnar-Filipowicz A and Kalberer CP:
Function of natural killer cells in immune defence against human
leukaemia. Swiss Med Wkly. 137(Suppl 155): 25S–30S. 2007.PubMed/NCBI
|
|
14
|
Cheent K and Khakoo SI: Natural killer
cells: integrating diversity with function. Immunology.
126:449–457. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brodin P, Kärre K and Höglund P: NK cell
education: not an on-off switch but a tunable rheostat. Trends
Immunol. 30:143–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Single RM, Martin MP, Gao X, et al: Global
diversity and evidence for coevolution of KIR and HLA. Nat Genet.
39:1114–1119. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hirayasu K, Ohashi J, Kashiwase K, et al:
Significant association of KIR2DL3-HLA-C1 combination with cerebral
malaria and implications for co-evolution of KIR and HLA. PLoS
Pathog. 8:e10025652012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin Xiaolin, Xiao Lulu and Guo Kunyuan:
The analysis of HLA-Cw and KIR in 79 Han people of Guangdong. China
Immunology. 21:605–607. 2005.
|