Introduction
Breast cancer is a major public health concern
threatening the health of females worldwide and representing 4% of
all female mortalities due to cancer (1). It is the most common type of cancer
among females in developing and developed countries (2). The incidence and mortality rates of
breast cancer have considerable global variations, with the highest
rates observed in Europe and North America and the lowest in Asia
(3). Consistent with other forms
of cancer, breast cancer is a byproduct of multiple environment and
hereditary risks (4). Futhermore,
family history is an influential factor in the development of the
disease. In a population-based study, mutations in the two
predominant breast cancer susceptibility genes, BRCA1 and BRCA2,
accounted for approximately 20% of familial breast cancer diagnoses
(5). Studies have revealed that
certain rare and low-frequency variants also have an impact on the
risks of developing breast cancer, including TP53, PTEN, STK11,
ATM, CHEK2 and BRCA1-interacting protein C-terminal helicase 1
(BACH1) genes (6,7).
BACH1, also known as FANCJ or BRIP1, interacts with
the BRCA1 C-terminal (BRCT) repeats of BRCA1 and the formed complex
contributes to the BRCA1-interrelated double-strand break repair
function (8). The human BACH1 gene
is located on chromosome 17q22, distal to the BRCA1 gene located at
17q21, a region that is frequently altered in breast cancer. The
BACH1 gene spans 180 kbps, comprising 20 exons and encodes a
protein that is 1,249 amino acids long (9). Based on its interactions with BRCA1,
the BACH1 gene is considered a potential breast cancer
susceptibility gene (10). The
interrelation of the gene with cancer susceptibility was identified
by the direct and functional interaction between BACH1 and BRCA1,
known as a classic tumor suppressor (11). Previously, it was demonstrated that
the interaction of the BRCTs with BACH1 depends on the
phosphorylation of BACH1 at S990 (12). Numerous frequently-occurring
mutations in the BACH1 gene, particularly the most common
polymorphism, proline (Pro) 919 serine (Ser) (rs4986764 C>T),
have been identified and have provided indications of the function
of BACH1 in breast carcinogenesis (11).
Several studies have suggested that the BACH1
Pro919Ser polymorphism may be important in increasing
susceptibility to breast cancer (11,13–16).
By contrast, certain other studies have suggested that the BACH1
Pro919Ser polymorphism is not correlated with an increased risk of
breast cancer (17–22). A recent meta-analysis of eight
case-control studies by Pabalan et al evaluated the
correlations of three functional polymorphisms (Pro919Ser, C47G and
G64A) in the BACH1 gene with breast cancer risk (23). These findings indicated that a
heterozygous genotype (Pro/Ser) of the BACH1 Pro919Ser polymorphism
may be correlated with an increased susceptibility to breast cancer
risk in premenopausal females under the heterozygous model.
However, the study failed to observe increased risks of breast
cancer under other genetic models. There were three main reasons
for these negative results, including the fact that three
case-control studies were not searched and included by the previous
meta-analysis, which resulted in the analysis having a relatively
small sample size. Furthermore, in the previous meta-analysis, the
authors only performed subgroup analyses based on ethnicity and
menopausal status in the exploration of the sources of
heterogeneity. Numerous additional factors may also have resulted
in the observed heterogeneity, such as differences in genotyping
methods, countries and regions, the source of the cases and
controls and the quality score of the included studies. Moreover,
univariate and multivariate meta-regression analyses were not used
in the previous meta-analysis to explore possible sources of
heterogeneity among the studies. The aim of the present study was
to update previous meta-analyses, as well as to provide a more
comprehensive and reliable conclusion on the correlations between
the BACH1 Pro919Ser polymorphism and breast cancer risk.
Materials and methods
Literature search
Relevant papers published prior to March 1, 2013
were identified through a search of PubMed, Embase, Web of Science
and China BioMedicine (CBM) databases using the terms: (‘genetic
polymorphism’ or ‘polymorphism’ or ‘SNP’ or ‘single nucleotide
polymorphism’ or ‘gene mutation’ or ‘genetic variants’) and
(‘breast neoplasms’ or ‘breast cancer’ or ‘breast tumor’ or ‘breast
carcinoma’) and (‘BRCA1-interacting protein 1’ or ‘BRIP1 protein,
human’ or ‘BACH1’ or ‘BRIP1’ or ‘BRAH1’ or ‘BRCA1 interacting
protein C-terminal helicase 1’). The references from the eligible
articles or textbooks were also reviewed in order to determine
additional potential sources. Disagreements were resolved through
discussions between the authors.
Inclusion and exclusion criteria
Studies included in the present meta-analysis had to
meet the following criteria: i) case-control studies had to focus
on the correlation between the BACH1 Pro919Ser polymorphism and
breast cancer risk; ii) any diagnoses of patients with cancer had
to be confirmed by pathological examinations; iii) the published
data on the frequencies of alleles or genotypes had to be
sufficient. The exclusion criteria comprised case-control studies
not focusing on the correlation between the BACH1 Pro919Ser
polymorphism and breast cancer risk, duplicates of previous
publications, studies based on incomplete data, and meta-analyses,
letters, reviews and editorial articles.
Data extraction
Data from the published studies were extracted
independently by two authors into a standardized form. For each
study, the following characteristics were assessed: The first
author, year of publication, country, language, study design,
ethnicity of subjects, number of subjects, gender ratio, mean age,
type of cancer, detection sample, genotyping method, allele and
genotype frequencies of single-nucleotide polymorphisms (SNPs) and
evidence of the Hardy-Weinberg equilibrium (HWE) in controls. In
cases of conflicting evaluations, disagreements were resolved
through discussions between the authors.
Quality assessment of included
studies
Two authors independently assessed the quality of
the included studies according to the modified Strengthening the
Reporting of Observational Studies in Epidemiology (STROBE) quality
score systems (24). Forty
assessment items interrelated with the quality appraisal were used
in the meta-analysis, with scores of 0–40. On the basis of the
scores of the studies, the included studies were classified into
three levels: Low quality (0–19), moderate quality (20–29) and high
quality (30–40), respectively. Disagreements were resolved through
discussions between the authors.
Statistical analysis
Crude odds ratios (ORs) with 95% confidence
intervals (CIs) were calculated under five genetic models: The
allele (Ser versus Pro), dominant (Ser/Ser + Pro/Ser versus
Pro/Pro), recessive (Ser/Ser versus Pro/Pro + Pro/Ser), homozygous
(Ser/Ser versus Pro/Pro) and heterozygous (Ser/Ser versus Pro/Ser)
models. The statistical significance of the pooled ORs was assessed
using the Z-test. Interstudy variations and heterogeneities were
estimated using Cochran’s Q-test, with Ph<0.05
indicating a statistically significant heterogeneity (25). Furthermore, the effects of
heterogeneity were quantified using the I2 test (range,
0–100%), which represented the proportion of interstudy variability
that was able to be contributed to heterogeneity rather than to
chance (26). When a significant
Q-test with Ph<0.05 or I2>50% indicated
that heterogeneity existed among the studies, the random-effects
model (DerSimonian-Laird method) was conducted for the
meta-analysis; otherwise, the fixed-effects model (Mantel-Haenszel
method) was used. To explore the sources of heterogeneity, a
subgroup analysis was performed according to ethnicity, the source
of the cases, genotyping method, menopausal status, family history
and BRCA 1/2 mutations. In addition, univariate and multivariate
regression analyses were conducted (27). Sensitivity analysis was performed
through the omission of each study in turn to assess the quality
and consistency of the results, while Begg’s funnel plots were used
to detect publication biases. Egger’s linear regression test was
also used to evaluate the publication biases (28). A χ2 test was used to
test whether the genotype frequencies of the controls were in HWE.
P-values were two-sided, and analyses were calculated using Stata
software, version 12.0 (Stata Corp., College Station, TX, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characteristics of included studies
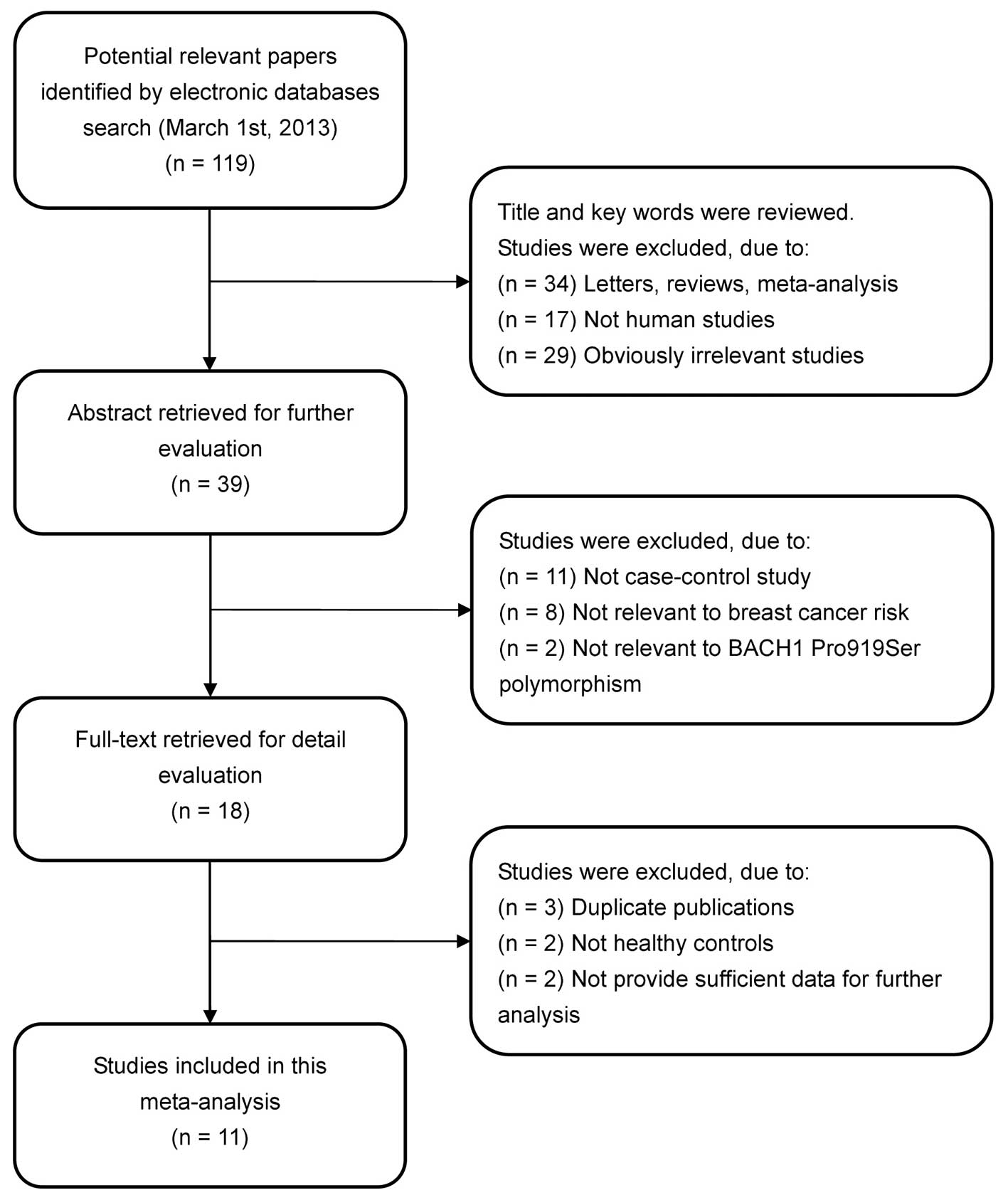
In accordance with the inclusion criteria, 11
case-control studies (11,13–22)
were included in the meta-analysis and 108 were excluded. The flow
chart of the study selection process is shown in Fig. 1. The publication years of the
included studies ranged from 2003 to 2011. A total of 15,057
subjects were involved in the meta-analysis, including 6,903 breast
cancer cases and 8,154 healthy controls. All diagnoses of breast
cancer were confirmed by pathological examinations. Six studies
used hospital-based cases, two used population-based cases and the
remaining three studies used family-based cases. The source of the
healthy controls in all the included studies was from the general
population (population-based). The DNA samples used for examination
of the BACH1 Pro919Ser polymorphism were extracted from the blood
in all the included studies. The genotyping methods included
denaturing high-performance liquid chromatography (DHPLC),
Microarray, TaqMan assay, MassArray, polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP) and
PCR-single strand conformation polymorphism (PCR-SSCP). Eight of
the studies were conducted in Caucasian populations and three in
Asian populations. The HWE test was conducted on the genotype
distribution of the controls in all 11 studies. None of the studies
deviated from the HWE (all P>0.05). The quality scores of the 11
included studies were all >20 (moderate-high quality). The
characteristics and methodological quality of the included studies
are shown in Table I.
 | Table I.Characteristics and methodological
quality of the included studies in the meta-analysis. |
Table I.
Characteristics and methodological
quality of the included studies in the meta-analysis.
| First author | Year | Country | Ethnicity | Number
| Source
| Genotyping
method | SNP ID | HWE test
(P-value) | STROBE score |
|---|
| Case | Control | Case | Control |
|---|
| Rutter et
al | 2003 | USA | Caucasian | 58 | 30 | HB | PB | DHPLC | rs4986764
(C>T) | 0.876 | 25/40 |
| García-Closas et
al | 2006 | USA | Caucasian | 1,898 | 1,514 | PB | PB | Microarray | rs4986764
(C>T) | 0.267 | 29/40 |
| Seal et
al | 2006 | UK | Caucasian | 1,212 | 2,081 | FB | PB | Microarray | rs4986764
(C>T) | 0.340 | 27/40 |
| Vahteristo et
al | 2006 | Finland | Caucasian | 888 | 736 | HB | PB | TaqMan | rs4986764
(C>T) | 0.318 | 30/40 |
| Frank et
al | 2007 | Germany | Caucasian | 571 | 712 | FB | PB | DHPLC | rs4986764
(C>T) | 0.366 | 31/40 |
| Guénard et
al | 2008 | Canada | Caucasian | 96 | 70 | FB | PB | Microarray | rs4986764
(C>T) | 0.690 | 28/40 |
| Cao et
al | 2009 | China | Asian | 357 | 864 | HB | HB | DHPLC | rs4986764
(C>T) | 0.498 | 24/40 |
| Huang et
al | 2009 | China | Asian | 50 | 150 | HB | PB | DHPLC | rs4986764
(C>T) | 0.866 | 32/40 |
| Huo et
al | 2009 | China | Asian | 568 | 624 | HB | PB | PCR-RFLP | rs4986764
(C>T) | 0.359 | 26/40 |
| Loizidou et
al | 2010 | Cyprus | Caucasian | 1,108 | 1,170 | HB | PB | MassArray | rs4986764
(C>T) | 0.242 | 32/40 |
| Silvestri et
al | 2011 | Italy | Caucasian | 97 | 203 | PB | PB | PCR-SSCP | rs4986764
(C>T) | 0.850 | 33/40 |
Quantitative data synthesis
A summary of the meta-analysis findings of the
correlation between the BACH1 Pro919Ser polymorphism and breast
cancer risk is provided in Table
II. No heterogeneity was observed with any of the genetic
models (all Ph>0.05 and I2< 50%);
therefore, the fixed effects model was used. The results of the
meta-analysis revealed that the BACH1 919Ser polymorphism was
correlated with a decreased risk of breast cancer (Ser allele
versus Pro allele: OR=0.91, 95% CI=0.87–0.96, P<0.001; Pro/Ser +
Ser/Ser versus Pro/Pro: OR=0.92, 95% CI=0.86–0.99, P=0.022; Ser/Ser
versus Pro/Pro + Pro/Ser: OR=0.83, 95% CI=0.76–0.92, P<0.001;
Ser/Ser versus Pro/Pro: OR=0.81, 95% CI=0.73–0.90, P<0.001;
Ser/Ser versus Pro/Ser: OR=0.85, 95% CI=0.77–0.94, P=0.001).
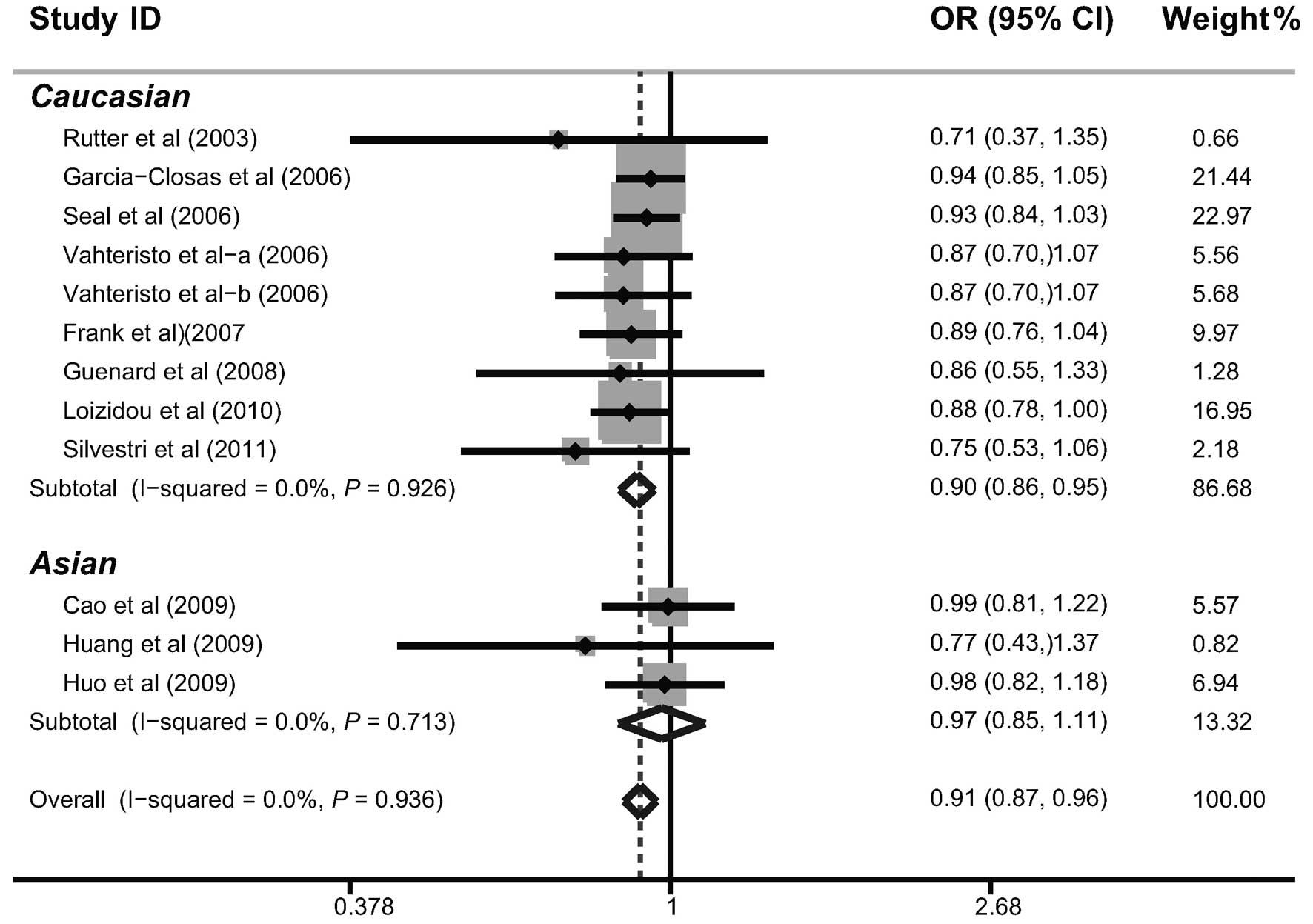
Further subgroup analysis by ethnicity indicated that the BACH1
919Ser polymorphism may decrease the risk of breast cancer among
Caucasian populations (Ser allele versus Pro allele: OR=0.90, 95%
CI=0.86–0.95, P<0.001; Pro/Ser + Ser/Ser versus Pro/Pro:
OR=0.90, 95% CI=0.84–0.98, P=0.012; Ser/Ser versus Pro/Pro +
Pro/Ser: OR=0.84, 95% CI=0.76–0.92, P<0.001; Ser/Ser versus
Pro/Pro: OR=0.81, 95% CI=0.73–0.91, P<0.001; Ser/Ser versus
Pro/Ser: OR=0.86, 95% CI=0.78–0.95, P=0.002). However, the results
did not suggest a correlation among Asian populations (Fig. 2).
 | Table II.Meta-analysis of the correlation
between the BACH1 Pro919Ser polymorphism and breast cancer
risk. |
Table II.
Meta-analysis of the correlation
between the BACH1 Pro919Ser polymorphism and breast cancer
risk.
A. Allele model:
Ser allele vs. Pro allele.
|
|---|
| Subgroup | OR | 95% CI | P-value |
Ph-value |
|---|
| Overall | 0.91 | 0.87–0.96 | <0.001 | 0.936 |
| Ethnicity | | | | |
| Caucasian
(n=8) | 0.90 | 0.86–0.95 | <0.001 | 0.926 |
| Asian (n=3) | 0.97 | 0.85–1.11 | 0.699 | 0.713 |
| Source of
cases | | | | |
| Population-based
(n=2) | 0.92 | 0.83–1.02 | 0.122 | 0.216 |
| Hospital-based
(n=6) | 0.90 | 0.84–0.98 | 0.010 | 0.817 |
| Family-based
(n=3) | 0.91 | 0.84–0.99 | 0.037 | 0.848 |
| Genotyping
method | | | | |
| DHPLC (n=4) | 0.91 | 0.81–1.03 | 0.118 | 0.635 |
| Microarray
(n=3) | 0.93 | 0.87–1.00 | 0.061 | 0.916 |
| Others (n=4) | 0.89 | 0.82–0.96 | 0.004 | 0.694 |
| Menopausal
status | | | | |
| Premenopausal
(n=4) | 0.91 | 0.79–1.04 | 0.174 | 0.613 |
| Postmenopausal
(n=4) | 0.90 | 0.82–0.99 | 0.021 | 0.760 |
| Family history of
breast cancer | | | | |
| Yes (n=6) | 0.91 | 0.85–0.97 | 0.007 | 0.938 |
| No (n=2) | 0.93 | 0.84–1.04 | 0.202 | 0.393 |
| BRCA1/2
mutations | | | | |
| Positive
(n=3) | 0.94 | 0.88–1.01 | 0.084 | 0.872 |
| Negative
(n=6) | 0.89 | 0.81–0.98 | 0.013 | 0.921 |
B. Dominant model:
Pro/Ser + Ser/Ser vs. Pro/Pro.
|
|---|
| Subgroup | OR | 95% CI | P-value |
Ph-value |
|---|
| Overall | 0.92 | 0.86–0.99 | 0.022 | 0.987 |
| Ethnicity | | | | |
| Caucasian
(n=8) | 0.90 | 0.84–0.98 | 0.012 | 0.999 |
| Asian (n=3) | 1.00 | 0.85–1.17 | 0.961 | 0.535 |
| Source of
cases | | | | |
| Population-based
(n=2) | 0.93 | 0.80–1.08 | 0.327 | 0.871 |
| Hospital-based
(n=6) | 0.93 | 0.83–1.03 | 0.157 | 0.806 |
| Family-based
(n=3) | 0.91 | 0.80–1.03 | 0.126 | 0.945 |
| Genotyping
method | | | | |
| DHPLC (n=4) | 0.90 | 0.77–1.07 | 0.228 | 0.658 |
| Microarray
(n=3) | 0.93 | 0.83–1.03 | 0.150 | 0.992 |
| Others (n=4) | 0.92 | 0.82–1.04 | 0.184 | 0.817 |
| Menopausal
status | | | | |
| Premenopausal
(n=4) | 0.91 | 0.75–1.11 | 0.347 | 0.662 |
| Postmenopausal
(n=4) | 0.93 | 0.82–1.06 | 0.270 | 0.703 |
| Family history of
breast cancer | | | | |
| Yes (n=6) | 0.91 | 0.83–1.01 | 0.079 | 0.975 |
| No (n=2) | 0.92 | 0.79–1.08 | 0.305 | 0.470 |
| BRCA1/2
mutations | | | | |
| Positive
(n=3) | 0.95 | 0.86–1.04 | 0.216 | 0.642 |
| Negative
(n=6) | 0.90 | 0.79–1.03 | 0.138 | 0.947 |
C. Recessive model:
Ser/Ser vs. Pro/Pro + Pro/Ser.
|
|---|
| Subgroup | OR | 95% CI | P-value |
Ph-value |
|---|
| Overall | 0.83 | 0.76–0.92 | <0.001 | 0.779 |
| Ethnicity | | | | |
| Caucasian
(n=8) | 0.84 | 0.76–0.92 | <0.001 | 0.712 |
| Asian (n=3) | 0.76 | 0.48–1.20 | 0.237 | 0.405 |
| Source of
cases | | | | |
| Population-based
(n=2) | 0.87 | 0.73–1.04 | 0.116 | 0.074 |
| Hospital-based
(n=6) | 0.78 | 0.67–0.91 | 0.002 | 0.998 |
| Family-based
(n=3) | 0.86 | 0.74–1.01 | 0.059 | 0.790 |
| Genotyping
method | | | | |
| DHPLC (n=4) | 0.80 | 0.61–1.06 | 0.122 | 0.802 |
| Microarray
(n=3) | 0.90 | 0.79–1.02 | 0.110 | 0.878 |
| Others (n=4) | 0.76 | 0.66–1.09 | 0.322 | 0.628 |
| Menopausal
status | | | | |
| Premenopausal
(n=4) | 0.79 | 0.58–1.08 | 0.141 | 0.817 |
| Postmenopausal
(n=4) | 0.78 | 0.66–0.92 | 0.004 | 0.996 |
| Family history of
breast cancer | | | | |
| Yes (n=6) | 0.84 | 0.74–0.95 | 0.006 | 0.909 |
| No (n=2) | 0.91 | 0.76–1.09 | 0.314 | 0.653 |
| BRCA1/2
mutations | | | | |
| Positive
(n=3) | 0.89 | 0.79–1.01 | 0.081 | 0.754 |
| Negative
(n=6) | 0.79 | 0.67–0.93 | 0.006 | 0.998 |
D. Homozygous
model: Ser/Ser vs. Pro/Pro.
|
|---|
| Subgroup | OR | 95% CI | P-value |
Ph-value |
|---|
| Overall | 0.81 | 0.73–0.90 | <0.001 | 0.920 |
| Ethnicity | | | | |
| Caucasian
(n=8) | 0.81 | 0.73–0.91 | <0.001 | 0.871 |
| Asian (n=3) | 0.79 | 0.49–1.26 | 0.317 | 0.292 |
| Source of
cases | | | | |
| Population-based
(n=2) | 0.85 | 0.69–1.04 | 0.104 | 0.112 |
| Hospital-based
(n=6) | 0.77 | 0.64–0.92 | 0.003 | 0.994 |
| Family-based
(n=3) | 0.84 | 0.70–0.99 | 0.039 | 0.812 |
| Genotyping
method | | | | |
| DHPLC (n=4) | 0.75 | 0.55–1.04 | 0.083 | 0.660 |
| Microarray
(n=3) | 0.87 | 0.76–1.01 | 0.070 | 0.949 |
| Others (n=4) | 0.75 | 0.63–0.89 | 0.001 | 0.810 |
| Menopausal
status | | | | |
| Premenopausal
(n=4) | 0.74 | 0.49–1.12 | 0.155 | 0.671 |
| Postmenopausal
(n=4) | 0.77 | 0.64–0.94 | 0.008 | 0.999 |
| Family history of
breast cancer | | | | |
| Yes (n=6) | 0.82 | 0.71–0.95 | 0.009 | 0.962 |
| No (n=2) | 0.88 | 0.71–1.08 | 0.223 | 0.512 |
| BRCA1/2
mutations | | | | |
| Positive
(n=3) | 0.87 | 0.75–1.00 | 0.051 | 0.899 |
| Negative
(n=6) | 0.77 | 0.62–0.85 | 0.013 | 0.994 |
E. Heterozygous
model: Ser/Ser vs. Pro/Ser.
|
|---|
| Subgroup | OR | 95% CI | P-value |
Ph-value |
|---|
| Overall | 0.85 | 0.77–0.94 | 0.001 | 0.722 |
| Ethnicity | | | | |
| Caucasian
(n=8) | 0.86 | 0.78–0.95 | 0.002 | 0.684 |
| Asian (n=3) | 0.72 | 0.44–1.16 | 0.177 | 0.618 |
| Source of
cases | | | | |
| Population-based
(n=2) | 0.88 | 0.73–1.07 | 0.191 | 0.093 |
| Hospital-based
(n=6) | 0.80 | 0.68–0.93 | 0.005 | 0.985 |
| Family-based
(n=3) | 0.86 | 0.75–1.04 | 0.142 | 0.797 |
| Genotyping
method | | | | |
| DHPLC (n=4) | 0.83 | 0.62–1.12 | 0.222 | 0.963 |
| Microarray
(n=3) | 0.92 | 0.80–1.06 | 0.241 | 0.867 |
| Others (n=4) | 0.77 | 0.66–0.90 | 0.001 | 0.532 |
| Menopausal
status | | | | |
| Premenopausal
(n=4) | 0.81 | 0.58–1.13 | 0.217 | 0.993 |
| Postmenopausal
(n=4) | 0.79 | 0.66–0.94 | 0.009 | 0.950 |
| Family history of
breast cancer | | | | |
| Yes (n=6) | 0.85 | 0.75–0.98 | 0.020 | 0.877 |
| No (n=2) | 0.93 | 0.77–1.13 | 0.480 | 0.829 |
| BRCA1/2
mutations | | | | |
| Positive
(n=3) | 0.91 | 0.79–1.04 | 0.166 | 0.600 |
| Negative
(n=6) | 0.80 | 0.67–0.96 | 0.015 | 0.996 |
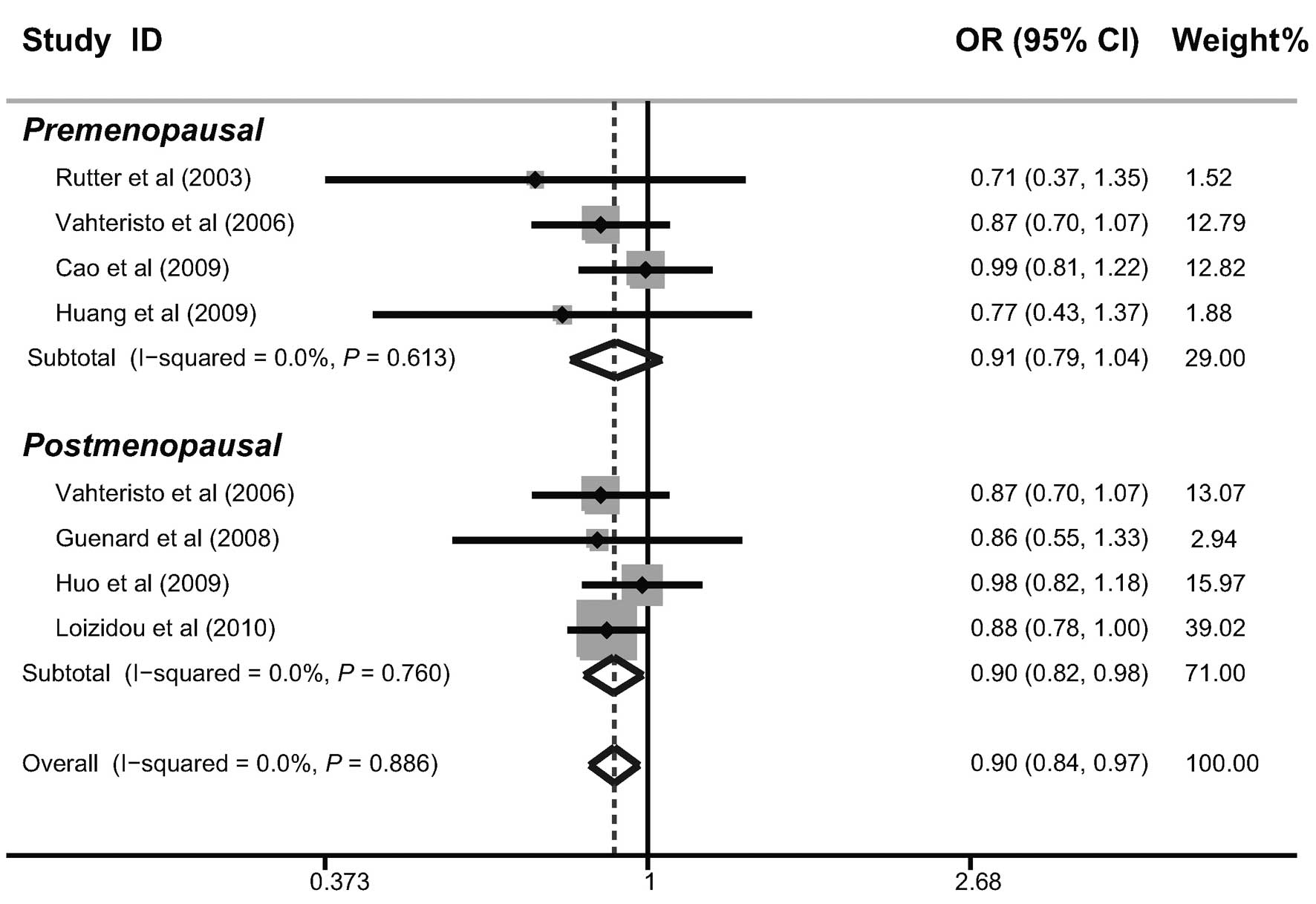
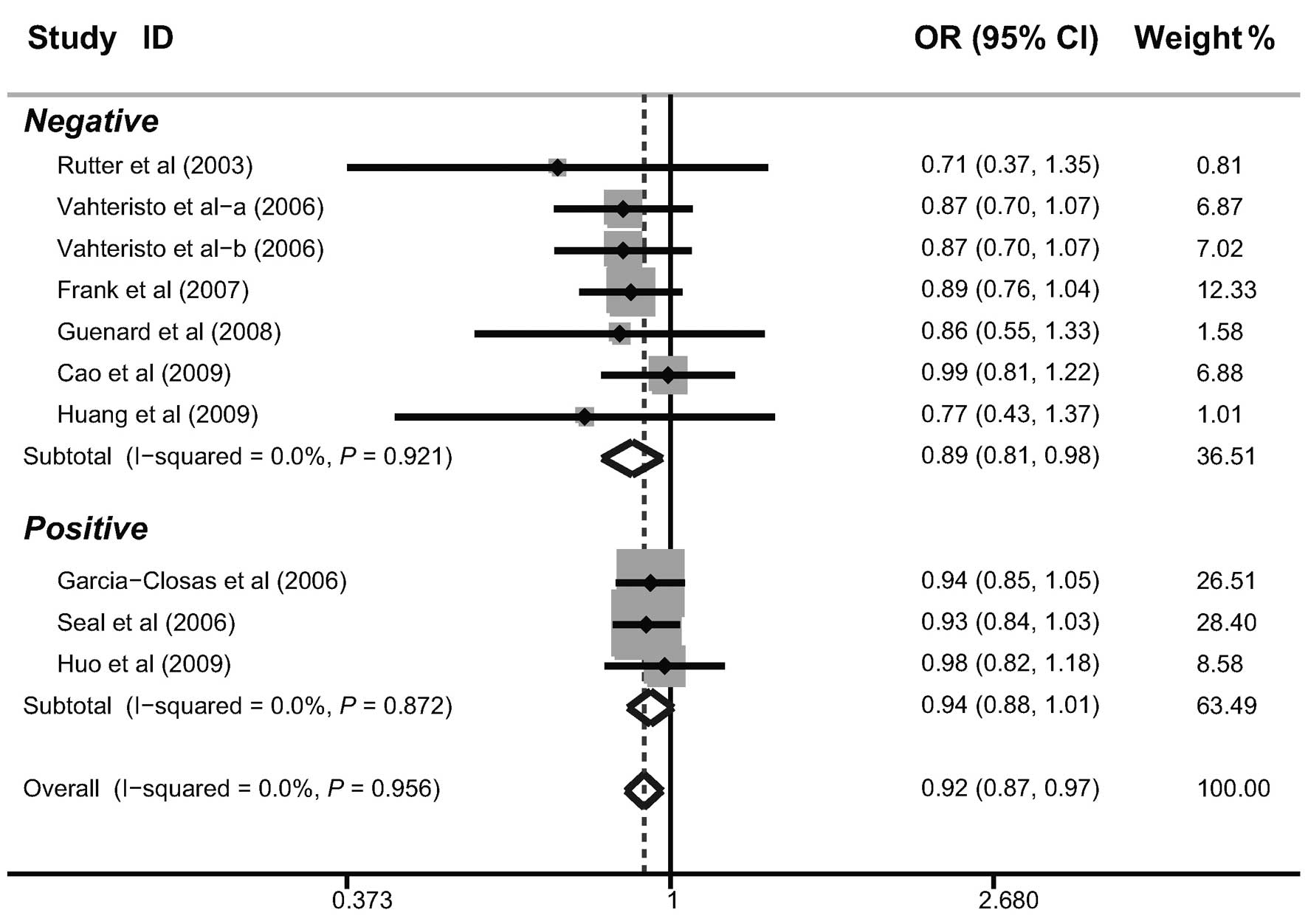
In the investigation into factors that may have had
a potential impact on the results, further subgroup analyses were
performed according to the source of the cases, genotyping method,
menopausal status, family history and BRCA1/2 mutations. The
subgroup analysis by the source of the cases indicated that there
were significant correlations between the BACH1 919Ser polymorphism
and a decreased risk of breast cancer in hospital-based and
family-based studies (as shown in Table II). Similar correlations were also
observed in post-menopausal females, females with a family history
of breast cancer and females without BRCA1/2 mutations (Figs. 3–5).
Meta-regression and sensitivity
analyses
Univariate and multivariate meta-regression analyses
were used to explore the possible sources of heterogeneity among
the studies (Table III). The
results revealed that none of the factors explained the
heterogeneity (all P>0.05). Sensitivity analysis was performed
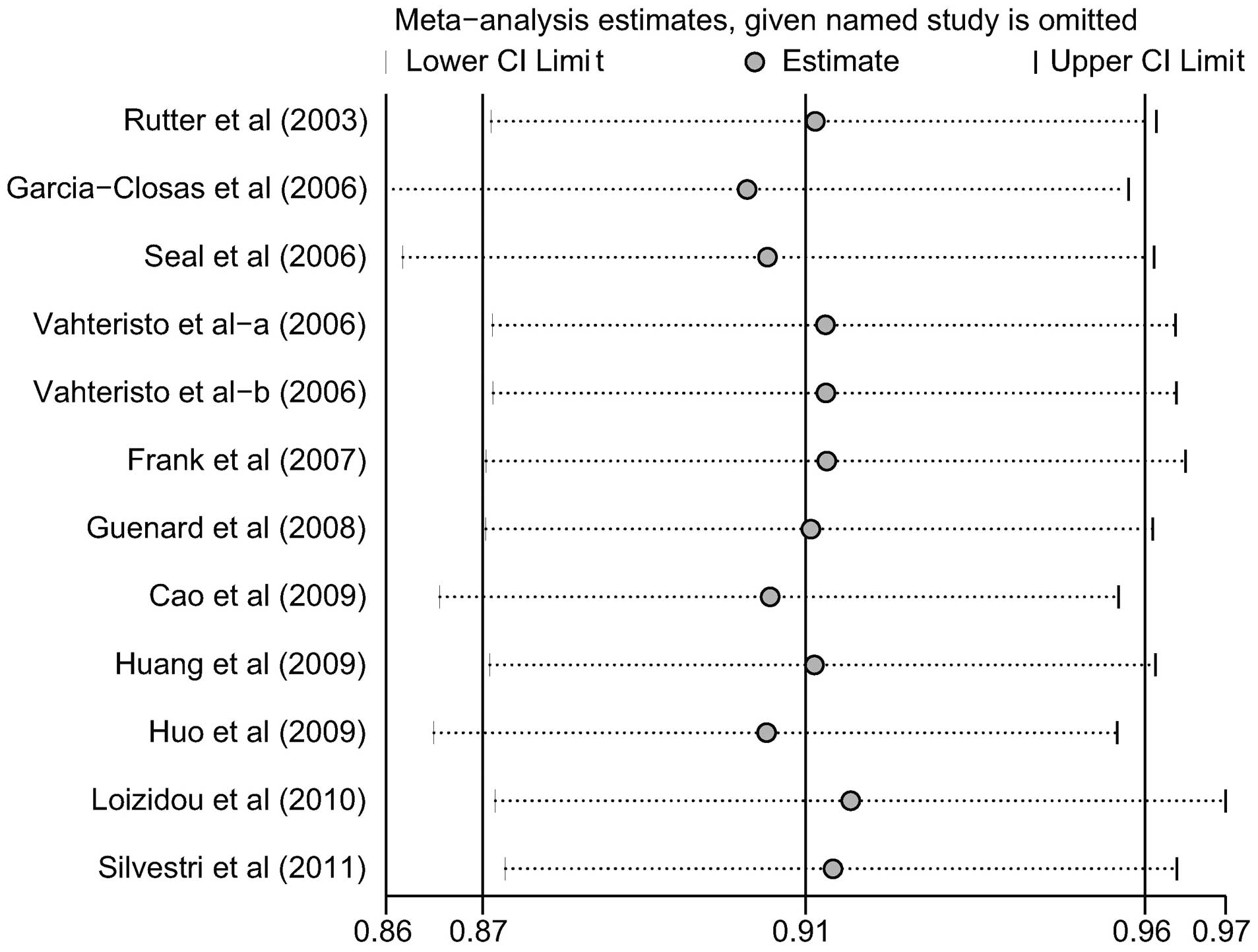
to assess the effect of each individual study on the pooled ORs by
the omission of individual studies. The analysis results suggested
that no individual studies significantly affected the pooled OR of
the correlation between the BACH1 919Ser polymorphism and breast
cancer risk under the allele model (Fig. 6), indicating that the results of
the analysis were statistically reliable.
 | Table III.Univariate and multivariate
meta-regression analyses of potential sources of heterogeneity. |
Table III.
Univariate and multivariate
meta-regression analyses of potential sources of heterogeneity.
| Heterogeneity
factor | Analysis type | Coefficient | SE | z-value | P-value | 95% CI
|
|---|
| UL | LL |
|---|
| Publication
year | Univariate | −0.005 | 0.015 | −0.34 | 0.736 | −0.034 | 0.024 |
| Multivariate | 0.032 | 0.063 | 0.51 | 0.613 | −0.092 | 0.155 |
| Ethnicity | Univariate | 0.077 | 0.073 | 1.05 | 0.293 | −0.066 | 0.220 |
| Multivariate | −0.059 | 0.256 | −0.23 | 0.817 | −0.562 | 0.443 |
| Source of
cases | Univariate | −0.004 | 0.033 | −0.12 | 0.906 | −0.069 | 0.061 |
| Multivariate | 0.001 | 0.049 | 0.02 | 0.984 | −0.095 | 0.097 |
| Genotyping
method | Univariate | −0.019 | 0.036 | −0.52 | 0.603 | −0.088 | 0.051 |
| Multivariate | −0.024 | 0.065 | −0.37 | 0.713 | −0.150 | 0.103 |
| Menopausal
status | Univariate | 0.010 | 0.036 | 0.28 | 0.777 | −0.059 | 0.080 |
| Multivariate | −0.040 | 0.105 | −0.38 | 0.704 | −0.246 | 0.166 |
| Family history of
breast cancer | Univariate | −0.004 | 0.030 | −0.13 | 0.893 | −0.063 | 0.054 |
| Multivariate | 0.004 | 0.078 | 0.05 | 0.957 | −0.148 | 0.156 |
| BRCA1/2
mutations | Univariate | −0.044 | 0.033 | −1.34 | 0.179 | −0.108 | 0.020 |
| Multivariate | −0.112 | 0.139 | −0.81 | 0.419 | −0.384 | 0.160 |
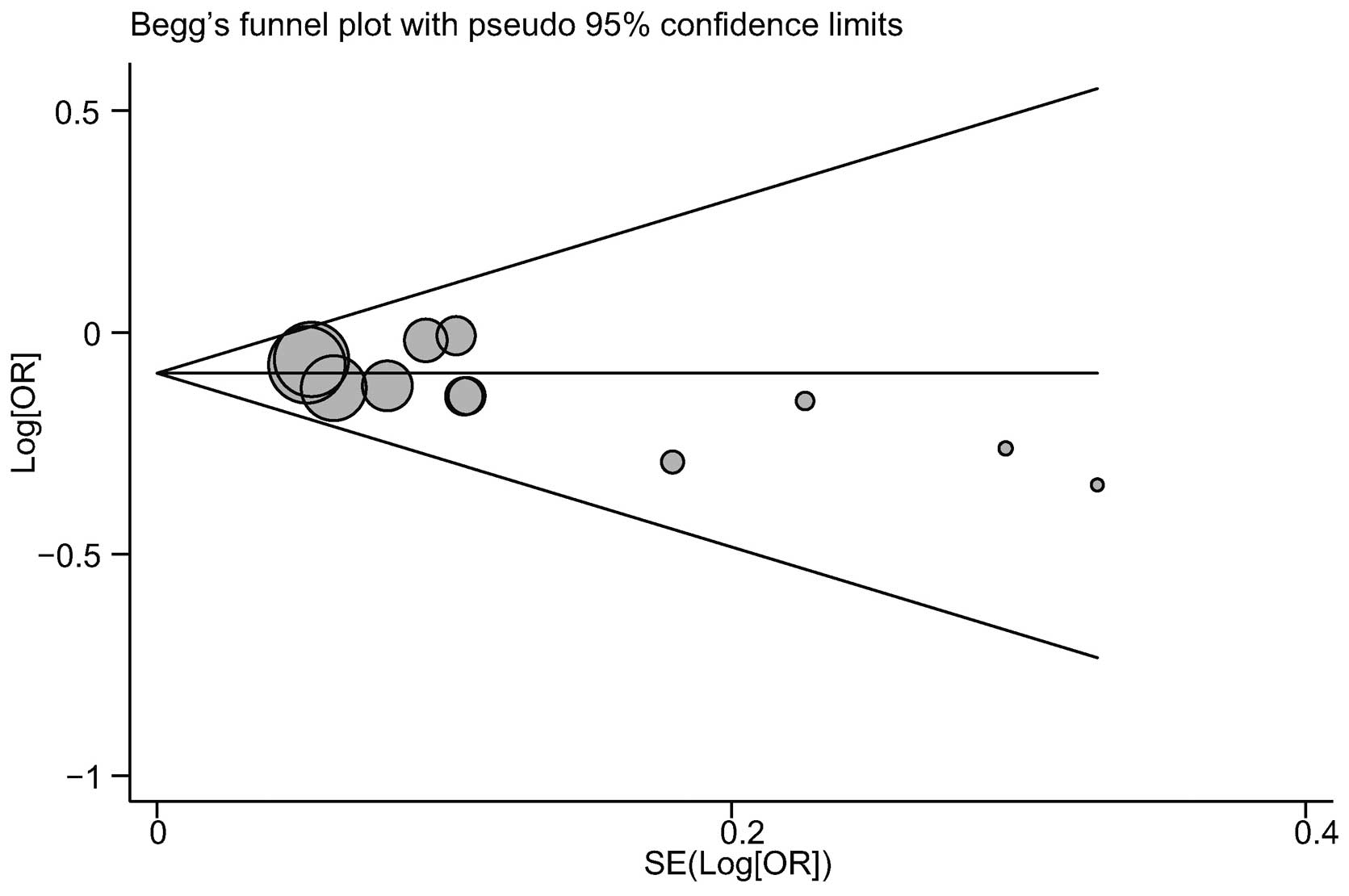
Publication bias evaluation
The publication biases within the available study
results may not have been representative of all of the results from
the study. Begg’s funnel plots and Egger’s linear regression tests
were performed to assess the publication biases in the included
studies. The shape of the funnel plot for the correlation between
the BACH1 919Ser polymorphism and breast cancer risk did not
indicate any marked asymmetry (Fig.
7). In addition, no notable suggestions of publication bias
under the allele model were observed with Egger’s test (t=−1.03,
P=0.327).
Discussion
The protein encoded by the BACH1 gene has been
demonstrated to be important in the double-strand break (DSB)
repair pathway (29). It is also
involved in the maintenance of DNA stability during transition
through interactions with BRCA1 via the BRCT repeats domain
(8). This process is required for
the establishment of the G2 cell-cycle checkpoint response to DNA
damage in the progression of the cell cycle (12). The abnormal expression of BACH1 has
been identified to be correlated with the risk of breast cancer due
to its inability to mediate DNA recombination repair (30). Furthermore, monoallelic mutations
in the BACH1 gene have been demonstrated to be the predominant
factor leading to the overexpression of BACH1, and these mutations
may increase the hereditary breast cancer susceptibility (10). Therefore, it was suggested that the
BACH1 gene polymorphisms were functional and were correlated with
breast cancer risk. At present, a total of eight BACH1 truncating
mutations have been identified worldwide, and the Pro919Ser
polymorphism, which codes for amino acid 919 of the BACH1 protein,
has been demonstrated to be closely correlated with breast cancer
susceptibility (7,17). Certain previous case-control
studies and a recent meta-analysis have suggested that the BACH1
Pro919Ser polymorphism may be important in the development of
breast cancer. However, the results from other investigations
indicated that this polymorphism did not affect the susceptibility
of an individual to breast cancer. There may be several reasons for
this controversy, such as the differences in the study designs,
sample sizes, the ethnicity of the subjects, the source of the
cases and controls, genotyping methods and menopausal status
(31). Therefore, the present
meta-analysis was performed to provide a more comprehensive and
reliable conclusion with regard to the correlation between the
BACH1 Pro919Ser polymorphism and susceptibility to breast
cancer.
In this meta-analysis, 11 case-control studies were
included with a total of 6,903 breast cancer cases and 8,154
healthy controls. When all the eligible studies were pooled into
the meta-analysis, the results indicated that the BACH1 919Ser
polymorphism decreased the risk of breast cancer among Caucasian
populations, although a similar correlation was not observed among
Asian populations. While the precise functions and effects of the
BACH1 genetic polymorphisms on an individual’s susceptibility to
breast cancer among different populations have not yet been
elucidated, a potential explanation is that inherited mutations in
BACH1 may be interrelated with the changes in expression and
function of DNA repair, thereby accounting for the interindividual
differences in susceptibility to breast cancer (11). Further subgroup analyses revealed
that there were significant correlations between the BACH1 919Ser
polymorphism and a decreased risk of breast cancer in
hospital-based and family-based studies. Similar correlations were
also observed in postmenopausal females, females with a family
history of breast cancer and females without BRCA1/2 mutations. By
contrast with the previous meta-analysis, which indicated that the
Pro/Ser genotype increased the risk of breast cancer in
premenopausal females, the present analysis revealed a significant
correlation between the BACH1 919Ser polymorphism and a decreased
risk of breast cancer in postmenopausal females (23). Furthermore, the results of the
present meta-analysis suggested that the BACH1 919Ser polymorphism
may be correlated with a decreased risk of breast cancer in females
with a family history of breast cancer and without BRCA1/2
mutations.
Consistent with previous meta-analyses (23), the present study demonstrated
certain limitations, such as the fact that only 14 investigations
were included. Therefore, the sample size was relatively small and
may not have provided sufficient statistical power. Thus,
additional studies with larger sample sizes are required to provide
an accurate and more representative statistical analysis.
Furthermore, as a type of a retrospective study, a meta-analysis
may encounter recall or selection bias, and this may have
potentially influenced the reliability of the results in the
present study (32,33). Moreover, the lack of access to the
original data from the studies limited the present meta-analysis
with regard to evaluation of potential interactions between
additional factors and breast cancer risks, such as
gene-environment and gene-gene interactions (34).
In conclusion, the present meta-analysis indicated
that the BACH1 919Ser polymorphism may decrease the risk of breast
cancer among Caucasian populations, particularly in postmenopausal
females with a family history of breast cancer and without BRCA1/2
mutations. These correlations have the potential to suggest a
functional profiling of the involvement of the BACH1 gene in the
development of breast cancer. In addition, the results may provide
a foundation for additional studies in the diagnosis and clinical
therapy of breast cancer. In consideration of the previously
mentioned limitations of this analysis, detailed studies are
required to confirm the results described. Studies investigating
the effect of gene-environment interactions on breast cancer should
also be conducted.
Acknowledgements
The authors would like to acknowledge
the comments received from the reviewers concerning this study. In
addition, the authors would like to thank their colleagues working
in the Department of Medical Oncology at the First Hospital of
China Medical University (Shenyang, China). This study was funded
by the National Science and Technology Major Project of the
Ministry of Science and Technology of China (No.
2013JX09303002).
References
|
1.
|
Benson JR and Jatoi I: The global breast
cancer burden. Future Oncol. 8:697–702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4.
|
Wernberg JA, Yap J, Murekeyisoni C,
Mashtare T, Wilding GE and Kulkarni SA: Multiple primary tumors in
men with breast cancer diagnoses: a SEER database review. J Surg
Oncol. 99:16–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Prevalence and penetrance of BRCA1 and
BRCA2 mutations in a population-based series of breast cancer
cases. Anglian Breast Cancer Study Group. Br J Cancer.
83:1301–1308. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Pharoah PD, Tyrer J, Dunning AM, Easton DF
and Ponder BA; SEARCH Investigators: Association between common
variation in 120 candidate genes and breast cancer risk. PLoS
Genet. 3:e422007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kuusisto KM, Bebel A, Vihinen M,
Schleutker J and Sallinen SL: Screening for BRCA1, BRCA2, CHEK2,
PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish
BRCA1/2-founder mutation-negative breast and/or ovarian cancer
individuals. Breast Cancer Res. 13:R202011.PubMed/NCBI
|
|
8.
|
Cantor SB, Bell DW, Ganesan S, et al:
BACH1, a novel helicase-like protein, interacts directly with BRCA1
and contributes to its DNA repair function. Cell. 105:149–160.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Song H, Ramus SJ, Kjaer SK, et al: Tagging
single nucleotide polymorphisms in the BRIP1 gene and
susceptibility to breast and ovarian cancer. PLoS One. 2:e2682007.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Cantor SB and Guillemette S: Hereditary
breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future
Oncol. 7:253–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Vahteristo P, Yliannala K, Tamminen A,
Eerola H, Blomqvist C and Nevanlinna H: BACH1 Ser919Pro variant and
breast cancer risk. BMC Cancer. 6:192006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yu X, Chini CC, He M, Mer G and Chen J:
The BRCT domain is a phospho-protein binding domain. Science.
302:639–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Huo X, Lu C, Huang X, et al: Polymorphisms
in BRCA1, BRCA1-interacting genes and susceptibility of breast
cancer in Chinese women. J Cancer Res Clin Oncol. 135:1569–1575.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Huang J, Tang LL, Hu Z, et al: BRCA1 and
BRCA2 gene mutations of familial breast cancer and early-onset
breast cancer from Hunan Province in China. China Oncology.
18:566–572. 2008.
|
|
15.
|
Cao AY, Huang J, Hu Z, et al: Mutation
analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with
early onset breast cancer or affected relatives. Breast Cancer Res
Treat. 115:51–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Seal S, Thompson D, Renwick A, et al
Breast Cancer Susceptibility Collaboration (UK): Truncating
mutations in the Fanconi anemia J gene BRIP1 are low-penetrance
breast cancer susceptibility alleles. Nat Genet. 38:1239–1241.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Silvestri V, Rizzolo P, Falchetti M, et
al: Mutation analysis of BRIP1 in male breast cancer cases: a
population-based study in Central Italy. Breast Cancer Res Treat.
126:539–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Loizidou MA, Cariolou MA, Neuhausen SL, et
al: Genetic variation in genes interacting with BRCA1/2 and risk of
breast cancer in the Cypriot population. Breast Cancer Res Treat.
121:147–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Guénard F, Labrie Y, Ouellette G, Joly
Beauparlant C, Simard J and Durocher F; INHERIT BRCAs: Mutational
analysis of the breast cancer susceptibility gene BRIP1
/BACH1/FANCJ in high-risk non-BRCA1/BRCA2 breast cancer families. J
Hum Genet. 53:579–591. 2008.PubMed/NCBI
|
|
20.
|
Frank B, Hemminki K, Meindl A, et al:
BRIP1 (BACH1) variants and familial breast cancer risk: a
case-control study. BMC Cancer. 7:832007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
García-Closas M, Egan KM, Newcomb PA, et
al: Polymorphisms in DNA double-strand break repair genes and risk
of breast cancer: two population-based studies in USA and Poland,
and meta-analyses. Hum Genet. 119:376–388. 2006.PubMed/NCBI
|
|
22.
|
Rutter JL, Smith AM, Dávila MR, et al:
Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and
BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from
breast-ovarian cancer families and among early-onset breast cancer
cases and reference individuals. Hum Mutat. 22:121–128. 2003.
View Article : Google Scholar
|
|
23.
|
Pabalan N, Jarjanazi H and Ozcelik H:
Association between BRIP1 (BACH1) polymorphisms and breast cancer
risk: a meta-analysis. Breast Cancer Res Treat. 137:553–558. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Gallo V, Egger M, McCormack V, et al:
Strengthening the reporting of observational studies in
epidemiology - molecular epidemiology (STROBE-ME): an extension of
the STROBE Statement. PLoS Med. 8:e10011172011. View Article : Google Scholar
|
|
25.
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zintzaras E and Ioannidis JP:
Heterogeneity testing in meta-analysis of genome searches. Genet
Epidemiol. 28:123–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ioannidis JP, Patsopoulos NA and Rothstein
HR: Reasons or excuses for avoiding meta-analysis in forest plots.
BMJ. 336:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Peters JL, Sutton AJ, Jones DR, Abrams KR
and Rushton L: Comparison of two methods to detect publication bias
in meta-analysis. JAMA. 295:676–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Wong MW, Nordfors C, Mossman D, et al:
BRIP1, PALB2, and RAD51C mutation analysis reveals their relative
importance as genetic susceptibility factors for breast cancer.
Breast Cancer Res Treat. 127:853–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Cantor S, Drapkin R, Zhang F, et al: The
BRCA1-associated protein BACH1 is a DNA helicase targeted by
clinically relevant inactivating mutations. Proc Natl Acad Sci USA.
101:2357–2362. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Rosenthal R and DiMatteo MR:
Meta-analysis: recent developments in quantitative methods for
literature reviews. Annu Rev Psychol. 52:59–82. 2001. View Article : Google Scholar
|
|
32.
|
Jüni P and Egger M: PRISMAtic reporting of
systematic reviews and meta-analyses. Lancet. 374:1221–1223.
2009.
|
|
33.
|
Ioannidis JP and Lau J: Pooling research
results: benefits and limitations of meta-analysis. Jt Comm J Qual
Improv. 25:462–469. 1999.PubMed/NCBI
|
|
34.
|
Dennis J, Hawken S, Krewski D, et al: Bias
in the case-only design applied to studies of gene-environment and
gene-gene interaction: a systematic review and meta-analysis. Int J
Epidemiol. 40:1329–1341. 2011. View Article : Google Scholar : PubMed/NCBI
|