Introduction
Lysophosphatidic acid (LPA) is a bioactive lipid
mediator that is released by activated platelets and is
constitutively present in serum (1,2). It
has been identified to be a potent phospholipid messenger with a
variety of biological actions, which include cell proliferation,
survival and migration, wound healing, platelet aggregation,
vascular remodeling, neurite retraction, differentiation
inhibition/reversal, membrane depolarization, formation of focal
adhesion and stress fibers (3–8),
blood pressure regulation (9) and
smooth muscle contraction (10–13).
LPA exhibits its functions mainly through binding to
its specific G protein-coupled receptors (GPCRs). Currently, there
are six GPCRs identified as specific receptors for LPA that are
referred to as LPA1-6 (14–20).
LPA1-3 were identified as members of the endothelial
differentiation gene (Edg) subfamily of GPCRS, as they share a high
homology with each other (21). By
contrast, LPA4-6 belong to the non-Edg family (22–24).
It has been demonstrated through studies in guinea
pigs, rats and cats that LPA receptors are identifiable throughout
the length of the mammalian gastrointestinal tract (11,12,25).
However, there is less information available concerning the
distribution of LPA receptors in the lower esophageal sphincter
(LES) and the studies have been mainly confined to animals.
Esophageal motility disorders, including achalasia
and diffuse esophageal spasm, are characterized by the incomplete
passage of swallowed contents into the stomach. Such conditions are
often accompanied by incomplete or failure of LES relaxation during
swallowing. Specific treatments for such conditions are not known
at present.
The aim of the present study was to examine the
expression of the LPA receptors in the human LES, in particular
within the clasp and sling fibers of the LES complex. In addition,
the potential implications of the results obtained for the
respective roles of the LPA receptors in the physiological
regulation of human LES function were considered.
Materials and methods
Patients and tissue retrieval
The experimental protocol was approved by the
Research Ethics Committee of the Fourth Hospital (Hebei Medical
University, Shijiazhuang, China). The muscle strips were collected
from 15 patients who underwent an esophagectomy for mid-third
esophageal carcinoma in the Department of Thoracic Surgery (Fourth
Hospital) between January 2012 and July 2012. There were 9 males
and 6 females, with an average age of 64 years (range, 55–68
years). Patients with a history of gastroesophageal reflux disease
or esophageal motor disorders were excluded from the study. Each
specimen was resected en bloc in the operating room and
placed immediately in ice-cold Krebs solution, the composition of
which has been described previously (26). Specimens were not included in this
study if the segment contained a macroscopically visible tumor.
In the laboratory, fresh esophagogastric junction
specimens collected in the operating room were immediately placed
in 4°C Tris-buffered saline (TBS). Following washing with 37°C
Krebs solution, specimens were pinned on a wax plate containing
TBS, with a continuous mixed gas of 95% O2 and 5%
CO2. The mucosa and submucosa were then gently removed
by sharp dissection. The sling fibers, clasp fibers and circular
muscle strips of the esophagus and stomach were separated and
prepared into 2–4 × 8–12-mm muscle strips. The LES was recognized
as a thickened band of circular muscle at the gastroesophageal
junction.
The gastric sling and clasp fibers were identified
as thickened bands of circular oriented smooth muscle in the
gastric cardia, adjacent to the greater and lesser curvature of the
stomach, respectively. The sling and clasp muscle strips were
prepared using a method described previously (27,28).
Circular muscle strips from the esophagus and stomach were prepared
as controls. These circular muscle strips were obtained 3 cm
proximal and distal to the gastroesophageal junction. The circular
muscle was not excised to the full depth of this layer to avoid
including the myenteric plexus and the longitudinal muscle in the
wall of the esophagus and stomach. The dissected muscle strips were
frozen in liquid nitrogen and stored at −80°C for subsequent RNA
extraction. Written informed consent was obtained from the
patients.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR) for LPA
receptors
Tissue was homogenized in TRIzol reagent at a ratio
of 100 mg tissue to 1 ml TRIzol (Invitrogen Life Technologies,
Carlsbad, CA, USA) and then centrifuged at 12,000 × g for 5 min.
Total RNA was extracted by acid guanidinium
thiocyanate-phenol-chloroform extraction. The quality of the RNA
was verified by agarose gel electrophoresis using ethidium bromide
staining. First-strand cDNA synthesis (reaction volume, 20 μl;
RevertAid First Strand cDNA Synthesis kit; Thermo Scientific,
Waltham, MA, USA), using 2 mg RNA, was performed in the presence of
RevertAid Moloney Murine Leukemia Virus (M-MuLV; Thermo
Scientific). Reverse transcription (Fermentas, Glen Burnie, MD,
USA) was performed using 0.5 mg oligo(dT)18 and
diethylpyrocarbonate (DEPC)-treated water to reach a volume of 11
μl. Samples were then incubated at 70°C for 5 min, prior to being
chilled on ice. Next, 4 μl reaction buffer (5X), 2 μl 4 dNTP mix
(10 mM) and 20 units RNasin were added, using DEPC-treated water to
reach a 19 μl volume and then incubated at 37°C for 5 min. Finally,
200 units (1 μl) of RevertAid M-MuLV reverse transcriptase was
added and the reaction mixture was incubated at 42°C for 60 min.
Following this, the reaction was terminated and held at 70°C for 10
min.
PCR amplification of the cDNA was performed using
primers designed specifically to match the mRNA of the LPA
receptors (primers listed in Table
I). A volume of 2 μl cDNA reaction mixture was used in each
PCR, which was performed in a 20 μl reaction volume. The
amplification conditions for each LPA receptor were different. PCR
conditions: LPA1, 36 cycles of 94°C for 5 min, 94°C for 30 sec,
58°C for 40 sec and 72°C for 1 min, followed by 72°C for 5 min;
LPA2, 38 cycles of 94°C for 5 min, 94°C for 30 sec, 56°C for 40 sec
and 72°C for 1 min, followed by 72°C for 5 min; LPA3, 40 cycles of
94°C for 5 min, 94°C for 30 sec, 57°C for 40 sec and 72°C for 1
min, followed by 72°C for 5 min; LPA4, 36 cycles of 94°C for 5 min,
94°C for 30 sec, 57°C for 40 sec and 72°C for 1 min, followed by
72°C for 5 min; LPA5, 42 cycles of 94°C for 5 min, 94°C for 30 sec,
60°C for 40 sec and 72°C for 1 min, followed by 72°C for 5 min;
LPA6, 38 cycles of 94°C for 5 min, 94°C for 30 sec, 58°C for 40 sec
and 72°C for 1 min, followed by 72°C for 5 min; β-actin, 23 cycles
of 94°C for 5 min, 94°C for 30 sec, 58°C for 40 sec and 72°C for 1
min, followed by 72°C for 5 min. A negative control in which all
the components of the reaction were added, with the exception of
the cDNA template, was tested in parallel with each sample to
identify any risk of false positive results. Amplified products
were electrophoresed on a 1.5% agarose gel and photographed under a
UV transilluminator. PCR, including positive and negative controls,
was performed in triplicate with cDNA extracted from fifteen
independent specimens.
 | Table IPrimers used for reverse transcription
polymerase chain reaction. |
Table I
Primers used for reverse transcription
polymerase chain reaction.
| Gene | Primer pair sequence
(sense/antisense) | Product size
(bp) |
|---|
| LPA1R | 5′-ATC GGG ATA CCA
TGA TGA GT C-3′
5′-TCC GTT CTA AAC CAC AGA GTG-3′ | 342 |
| LPA2R | 5′-GTC CTC ATT ACC
CAG TCA TAC CG-3′
5′-CTG ATG GAC TCC ACC CTT TAG CT-3′ | 426 |
| LPA3R | 5′-TGT CAA CCG CTG
GCT TCT-3′
5′-CAG TCA TCA CCG TCT CAT TAG-3′ | 437 |
| LPA4R |
5′-GTGGCGGTATTTCAGCCTCT-3′
5′-GAGTTGCAAGGCACAAGGTG-3′ | 402 |
| LPA5R |
5′-GATTCCGCCCTCTGAACACA-3′
5′-AACCTGGTGCTCTTCAGCTC-3′ | 409 |
| LPA6R |
5′-TGGGTTGGACTCGTTGACTG-3′
5′-TTTCGGACTTTGAGGACGCA-3′ | 458 |
| β-actin | 5′-TCC CTG GAG AAG
AGC TAC GA-3′
5′-ATC TGC TGG AAG GTG GAC AG-3′ | 362 |
Quantitative PCR (qPCR) for LPA
receptors
Primers were designed specifically to match the mRNA
of the LPA receptors (primers listed in Table II). Analysis of the LPA receptor
mRNA expression was carried out by qPCR using TransStart™ Top Green
qPCR SuperMix (Beijing TransGen Biotech Co., Ltd, Beijing, China)
and an ABI 7500 PCR system (Applied Biosystems, Inc., Foster City,
CA, USA). The final reaction mixture of 25 μl consisted of 1 μl
diluted cDNA, 12.5 μl 2X TransStart Top Green qPCR SuperMix, 0.5 μl
Passive Reference Dye, 10 μl ddH2O, 0.5 μl forward
primer and 0.5 μl reverse primer. The reactions were performed in
triplicate according to the manufacturer’s instructions. All the
reactions were performed in 96-well plates in duplicate. ABI 7500
conditions were designed as follows: Initial denaturation at 94°C
for 30 sec, followed by 42 cycles of 94°C for 5 sec, 60°C for 34
sec and melt curve stage. The relative expression levels of each
mRNA were determined using the ABI 7500 software (version 2.0.5).
The precise amount of total cDNA added to each reaction mix and its
quality are difficult to assess; therefore, the expression level of
the gene of interest in a given specimen was computed relative to
the level of β-actin mRNA, used as an invariant standard control to
normalize the variations between the sample preparations. The
threshold cycle (Ct) was used for quantification of the input
target numbers. The normalized expression levels of the target gene
were calculated by the 2−Δ(ΔCt) method, where ΔCt =
Cttarget gene − Ctβ-actin and Δ(ΔCt) =
ΔCttest sample − ΔCtcontrol.
 | Table IIPrimers used for real-time
quantitative polymerase chain reaction. |
Table II
Primers used for real-time
quantitative polymerase chain reaction.
| Gene | Primer pair sequence
(sense/antisense) | Product size
(bp) |
|---|
| LPA1R | 5′-AAT CGG GAT ACC
ATG ATG AGT CTT-3′
5′-CCA AGG AGT CCA GCA GAT GAT AAA-3′ | 77 |
| LPA2R | 5′-CAG CCT GGT CAA
GAC TGT TGT-3′
5′-TGC AGG ACT CAC AGC CTA AA-3′ | 104 |
| LPA3R | 5′-ACG GTG ATG ACT
GTC TTA GGG-3′
5′-CAC CTT TTC ACA TGC TGC AC-3′ | 113 |
| LPA4R | 5′-AAA GAT CAT GTA
CCC AAT CAC CTT-3′
5′-CTT AAA CAG GGA CTC CAT TCT GAT-3′ | 139 |
| LPA5R | 5′-CGC CAT CTT CCA
GAT GAAC-3′
5′-TAG CGG TCC ACG TTG ATG-3′ | 66 |
| LPA6R | 5′-GGT AAG CGT TAA
CAG CTC CCA CT-3′
5′-TTT GAG GAC GCA GAT GAA AAT GT-3′ | 139 |
| β-actin | 5′-ATG AAG ATC AAG
ATC ATT GCT CCTC-3′
5′-ACA TCT GCT GGA AGG TGG ACA-3′ | 94 |
Western blot analysis of LPA
receptors
Total proteins were extracted from the muscle strips
using a protein extraction kit (Solarbio, Beijing, China). Protein
concentration was determined with a colorimetric BCA protein assay
reagent (Multisciences, Hangzhou, China). Following denaturation at
100°C for 10 min, aliquots of protein samples (30 μg) were
separated by electrophoresis on SDS-polyacrylamide gel (10%
separation gel and 4% pycnotic gel) at 150 V for 1 h and then
transferred onto a polyvinylidene difluoride membrane. The membrane
was blocked for 1 h with 5% non-fat milk in Tris-buffered saline
with Tween 20 (TBST) at room temperature and incubated with a
specific primary antibody [dilutions: rabbit anti-LPA1R, 1:1,000
(GeneTex, San Antonio, CA, USA); mouse anti-LPA2R, 1:2,000 (Abcam,
Cambridge, UK); rabbit anti-LPA3R, 1:2,000 (GeneTex); rabbit
anti-LPA4R, 1:2,000 (Abcam); rabbit anti-LPA5R, 1:2,000 (Abcam);
rabbit anti-LPA6R, 1:2,000 (Abcam); and rabbit anti-β-actin,
1:10,000 (GeneTex)] at 4°C overnight. Next the membrane was washed
three times with TBST at room temperature for 30 min in total and
then incubated with a secondary antibody (1:2,000, anti-rabbit IgG;
Rockland, Philadelphia, PA, USA) for 1 h. Following three washes
with TBST, the membrane was analyzed using an infrared fluorescence
imaging instrument (Odyssey, model: 9120; LI-COR, Lincoln, NE,
USA). The relative expression level of each protein was normalized
against the value of β-actin.
Statistical Analysis
Data are expressed as mean ± SD. SPSS 19.0 (SPSS,
Inc., Chicago, IL, USA) was used for statistical analysis.
Differences in mRNA expression were analyzed with one-way ANOVA and
the Student-Newman-Keuls multiple range test was used for
comparisons within groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characterization of mRNA encoding six LPA
receptors
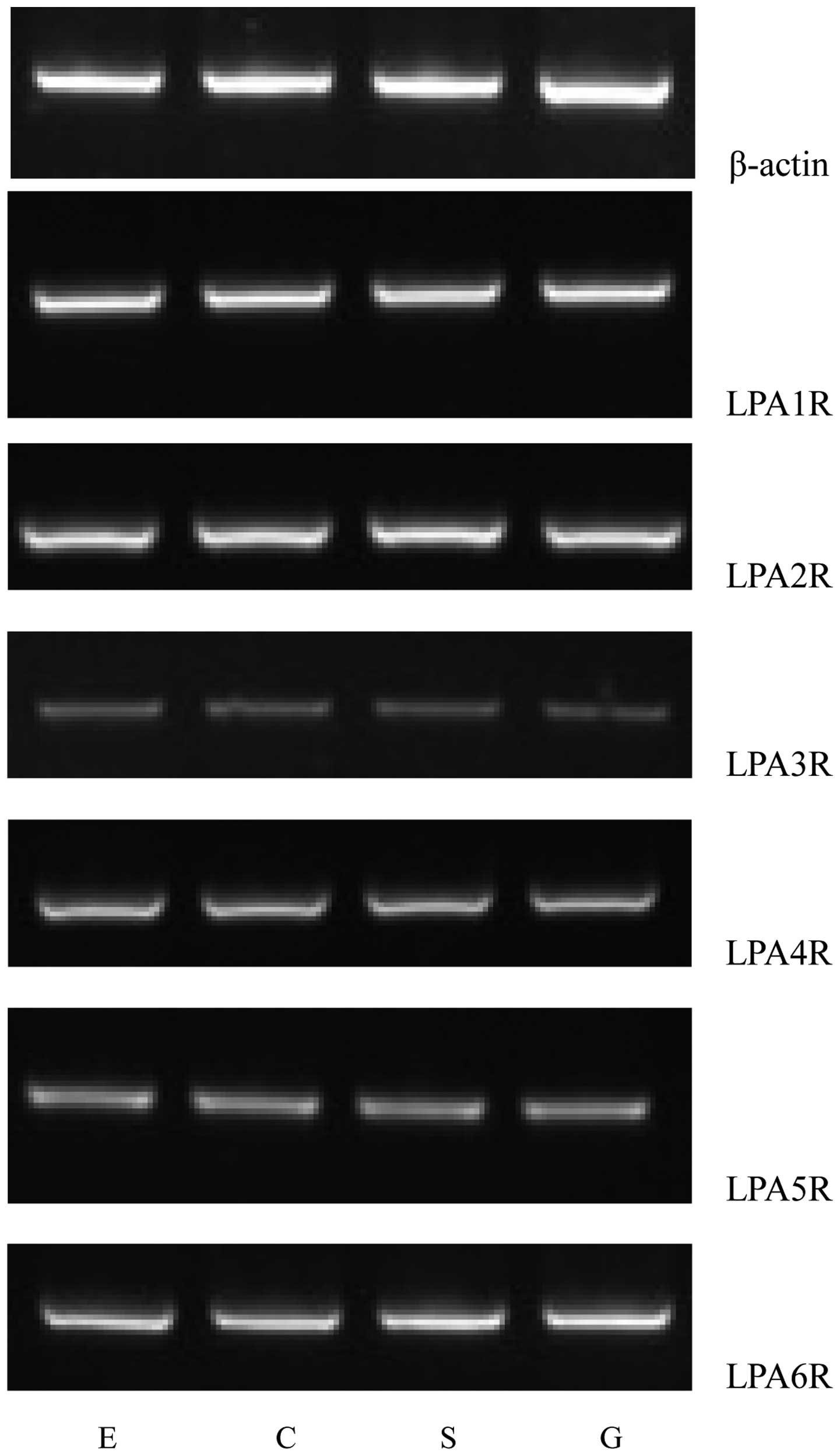
Using RT-PCR, the mRNA expression levels of six LPA
receptors in the human LES were observed, as illustrated in
Fig. 1. Distinct bands of the
expected sizes were detected for each of the six LPA receptor mRNAs
and their levels of expression were apparently different. Similar
results were obtained in all PCR assays performed on mRNA extracted
from the fifteen patients. The primer pair designed to recognize
the LPA1R mRNA generated a strong band, indicating high expression
levels of the LPA1R mRNA in the human LES. The LPA6R mRNA also
generated a comparatively strong band. However, the primer pairs
for the LPA4R, LPA5R and LPA2R mRNA produced relatively weak bands.
The lowest apparent mRNA expression level was observed for the LPA3
receptor (Fig. 1).
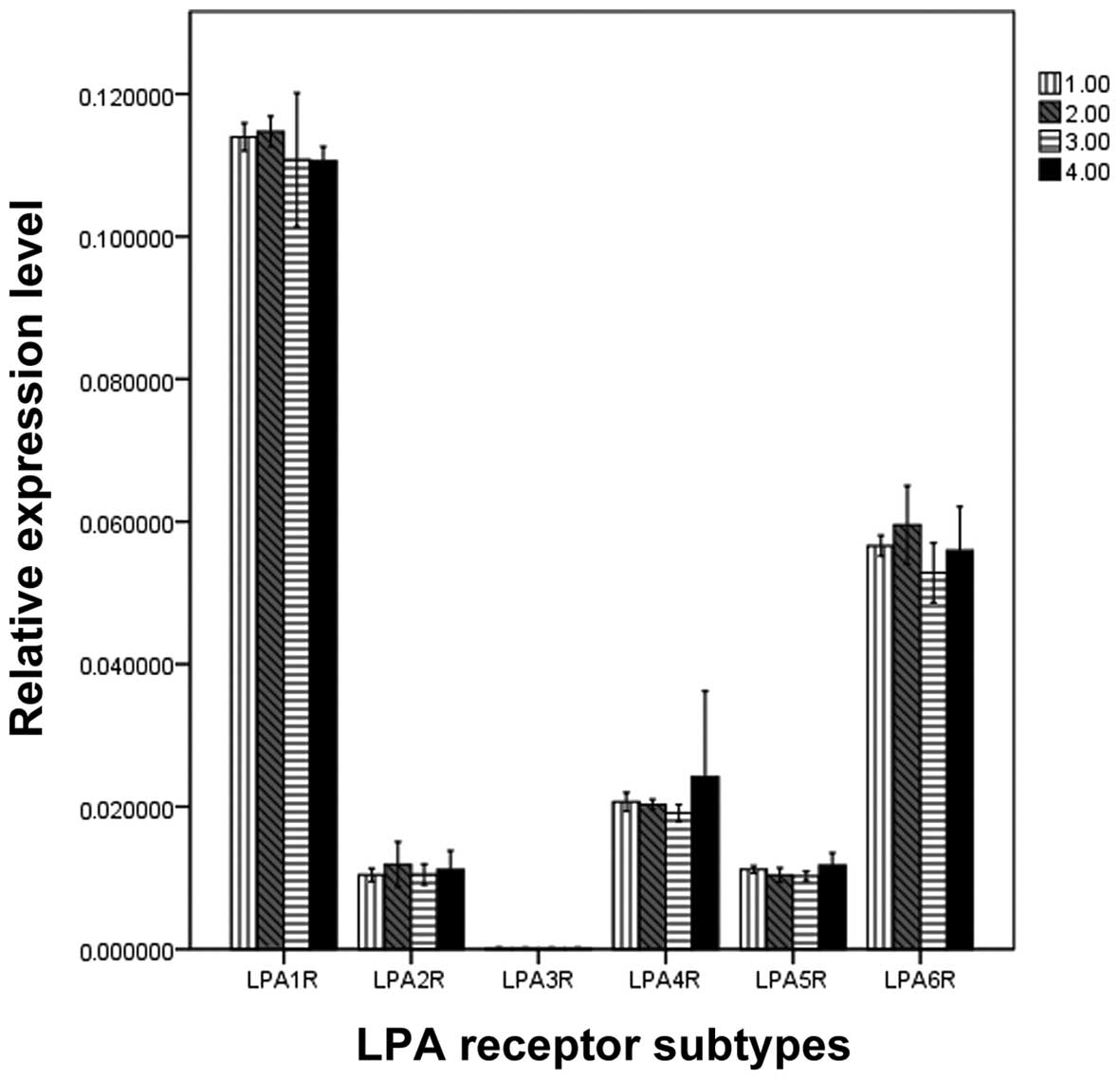
Quantification of LPA receptor mRNA
expression
To compare the levels of different LPA receptor
mRNA, qPCR was performed (Fig. 2).
Significant differences were demonstrated when the mRNA expression
levels of various LPA receptors were compared in the same muscle
strips (F, 61.034; P=0.000). The rank order of the expression was
as follows: LPA1R>LPA6R>LPA4R=LPA2R=LPA5R=LPA3R. However, no
significant difference was identified in the mRNA expression levels
of the LPA receptors between the four muscle strips. (F, 0.201;
P=0.895; Fig. 2).
Expression of LPA receptor proteins
LPA1R, LPA2R, LPA4R, LPA5R and LPA6R protein
expression was identified. Significant differences in the
integrated optical density (IOD) values for the different LPA
receptors in the same muscle strip were observed (F, 1,224.659;
P=0.000). The rank order of the IOD values was as follows:
LPA1R>LPA6R>LPA4R=LPA2R=LPA5R=LPA3R. No significant
differences in IOD values were identified among the four muscle
strips. (F, 0.039; P=0.990; Fig.
3).
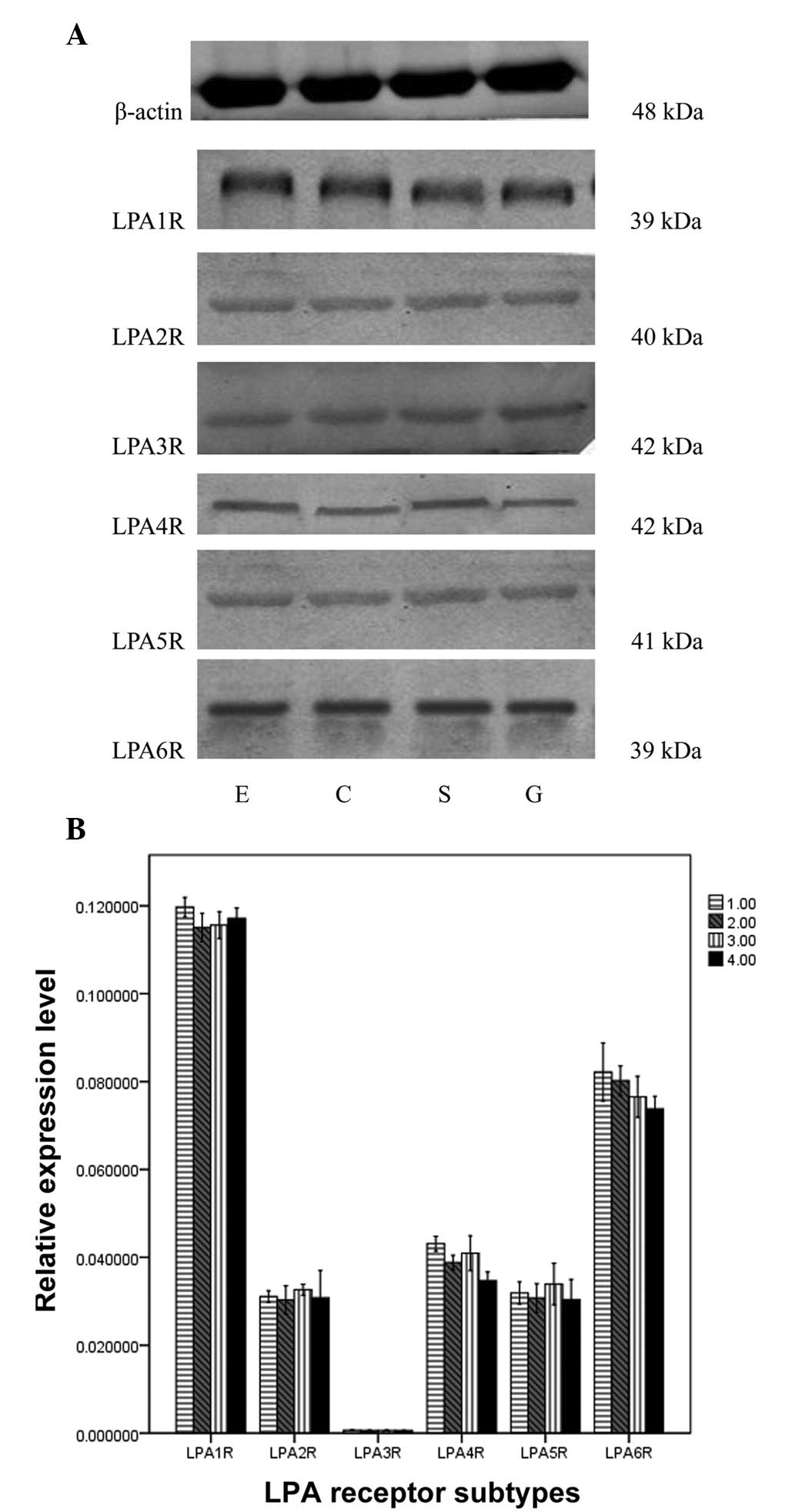 | Figure 3Expression of LPA receptor subtypes by
western blotting in the sling and clasp fibers of the LES, circular
muscle strips of esophagus and stomach. (A) Bands of the
β-adrenoceptor subtype in S, C, E and G were identified with
western blotting. (B) IOD values of the bands. There were
significant differences between the β-adrenoceptor subtype in the
same muscle strip (P<0.05), but no significant difference in a
single subtype between the four muscle strips (P>0.05). LPA,
lysophosphatidic acid; LES, lower esophageal sphincter; IOD,
integrated optical density; S, sling fibers; C, clasp fibers; E,
circular muscle strips of esophagus; G, stomach; 1.00, circular
muscle strips of esophagus; 2.00, clasp; 3.00, sling; 4.00,
circular muscle strips of stomach. |
Discussion
LPA is a bioactive phospholipid with diverse
biological functions in numerous tissues and cells. These functions
are mediated by LPA receptors, which are significant members of the
seven transmembrane domain GPCR family. LPA receptors are widely
present in the mammalian central nervous system, including the
brain, cardiovascular system and gastrointestinal tract and are
significant physiological modulators.
Various esophageal motility disorders, including
achalasia, diffuse esophageal spasm and nutcracker esophagus, are
all associated with motor disorders of the LES. Previous studies
have demonstrated that the regulatory mechanism of the LES involves
various receptors, neurotransmitters and signal transduction
pathways. CCK receptors, muscarinic receptors and dopamine
receptors have demonstrated a role in the regulation of the LES
(26–28).
The LPA receptors, widely distributed in the smooth
muscles of the body, including gastrointestinal smooth muscle, are
closely associated with the motility and secretion of the
gastrointestinal tract. Pharmacological effects of LPA on various
regions of the gut have been identified (11–13).
Previous studies have also identified the expression of LPA
receptors in the gastrointestinal tract (15,17–19,25,29).
For instance, Lee et al demonstrated that the LPA1 receptor
was significant in the regulation of the cat LES by pharmacological
study (25). An et al
identified the expression of the LPA2 receptor throughout the human
gastrointestinal tract (15). LPA3
receptor was identified in the gastric smooth muscle by Sriwai
et al(29) and Noguchi
et al(17) identified the
expression of the LPA4 receptor in the gut. Lee et
al(19) and Kotarsky et
al(18) showed that the LPA5
receptor was abundant in the gastrointestinal tract. These
observations prompted the hypothesis that LPA receptors are located
throughout the gastrointestinal tract. The present study was
designed to determine whether LPA receptors exist in the human LES.
The LES is a complex structure composed of clasp and sling fiber
muscle strips in the gastric cardia and circular muscle fibers in
the distal end of the esophagus, immediately above the
gastroesophageal junction. In the present study, the clasp and
sling fiber muscle strip component of the LES was specifically
investigated.
Several studies carried out in various human tissues
have indicated that the LPA3 receptor is not present in the
gastrointestinal tract (16,30).
However, the present study has clearly identified that the LPA3R is
present in the esophageal body, human LES and stomach. The results
obtained are in agreement with data obtained in previous studies
performed in gastric smooth muscle (29). The differences in LPA receptor
expression in the gastrointestinal tract, in the current study and
previous studies, are not fully understood and require further
study. In addition, in the present study, the LPA1 receptor was
expressed at higher levels than the other receptors and the LPA2
receptor was also identified in the gastrointestinal tract. By
contrast, a previous study has shown that the LPA1 and LPA2
receptors are not present in gastrointestinal smooth muscle
(15). Differences between the
current observations and those of other studies may be due to
species differences. In the previous studies, LPA receptors were
evaluated in animals, including rats, cats and guinea pigs. The
present study detected the distribution of LPA receptors in the
human LES and is, to the best of our knowledge, the first study to
identify LPA receptor mRNA expression in the human LES.
Among the LPA receptors, the LPA1 receptor was
identified to be the most highly expressed in the LES in the
present study. The study suggests a major involvement of LPA1
receptor in the regulation of human gastrointestinal functions. It
was also observed that the expression of the six LPA receptors
between the four muscle strips did not differ.
The use of LPA receptor agonists and antagonists has
shown that the LPA1 receptor blocks the relaxation of LES (25). Moreover, several studies show that
LPA stimulates the contraction of smooth muscles (10–13).
On the basis of previous pharmacological evidence, the expression
of LPA receptors in the human LES, as demonstrated in the present
study, implies an integral role in the modulation of the motility
balance of the LES.
The present study is, to the best of our knowledge,
the first to identify LPA receptor mRNA and protein expression in
the human LES. Although little information on the physiological and
pharmacological effects of the LPA receptors on the LES is
available, the detection of LPA receptors in this study supports
the concept that the LPA receptors are significant modulators of
esophageal motility. Information concerning the functional role of
the LPA receptors remains fragmented and requires further
investigation. Future development of more specific ligands, as well
as the use of gene deletion animal models, including knockout mice
for each LPA receptor, are likely to allow precise evaluation of
the physiological and pharmacological importance of the LPA
receptors in the LES.
Acknowledgements
This project was funded by the Hebei Provincial
Natural Science and China Natural Science Foundations.
References
|
1
|
Eichholtz T, Jalink K, Fahrenfort I and
Moolenaar WH: The bioactive phospholipid lysophosphatidic acid is
released from activated platelets. Biochem J. 291:677–680.
1993.PubMed/NCBI
|
|
2
|
Tigyi G, Hong L, Yakubu M, Parfenova H,
Shibata M and Leffler CW: Lysophosphatidic acid alters
cerebrovascular reactivity in piglets. Am J Physiol.
268:H2048–H2055. 1995.PubMed/NCBI
|
|
3
|
Jalink K, Hordijik PL and Moolenaar WH:
Growth factor-like effects of lysophosphatidic acid, a novel lipid
mediator. Biochim Biophys Acta. 1198:185–196. 1994.PubMed/NCBI
|
|
4
|
Moolenaar WH: Lysophosphatidic acid
signaling. Curr Opin Cell Biol. 7:203–210. 1995. View Article : Google Scholar
|
|
5
|
Gaits F, Fourcade O, Le Balle F, et al:
Lysophosphatidic acid as a phospholipid mediator: pathways of
synthesis. FEBS Lett. 410:54–58. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nietgen G and Durieux ME: Intercellular
signaling by lysophosphatidate. Cell Adhes Commun. 5:221–235. 1998.
View Article : Google Scholar
|
|
7
|
Ishii I, Contos JJ, Fukushima N and Chun
J: Functional comparisons of the lysophosphatidic acid receptors,
LP(A1)/VZE-1/EDG-2, LP(A2)/EDG-4, and LP(A3)/EDG-7 in neuronal cell
lines using a retrovirus expression system. Mol Pharmacol.
58:895–902. 2000.PubMed/NCBI
|
|
8
|
Moolenaar WH, Van Meeteren LA and Giepmans
BN: The ins and outs of lysophosphatidic acid signaling. Bioessays.
26:870–881. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tokumura A, Fukuzawa K and Tsukatani H:
Effects of synthetic and natural lysophosphatidic acids in the
arterial blood pressure of different animal species. Lipids.
13:572–574. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tokumura A, Fukuzawa K, Yamada S and
Tsukatani H: Stimulatory effect of lysophosphatidic acids on
uterine smooth muscles of non-pregnant rats. Arch Int Pharmacodyn
Ther. 245:74–83. 1980.PubMed/NCBI
|
|
11
|
Tokumura A, Fukuzawa K and Tsukatani H:
Contractile actions of lysophosphatidic acids with a
chemically-defined fatty acyl group on longitudinal muscle from
guineapig ileum. J Pharm Pharmacol. 34:514–516. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tokumura A, Yube N, Fujimoto H and
Tsukatani H: Lysophosphatidic acids induce contraction of rat
isolated colon by two different mechanisms. J Pharm Pharmacol.
43:774–778. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Toews ML, Ustinova EE and Schultz HD:
Lysophosphatidic acid enhances contractility of isolated airway
smooth muscle. J Appl Physiol. 83:1216–1222. 1997.PubMed/NCBI
|
|
14
|
Hecht JH, Weiner JA, Post SR and Chun J:
Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid
receptor expressed in neurogenic regions of the developing cerebral
cortex. J Cell Biol. 135:1071–1083. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
An SZ, Bleu T, Hallmark OG and Goetzl EJ:
Characterization of a novel subtype of human G protein-coupled
receptors for lysophosphatidic acid. J Biol Chem. 273:7906–7910.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bandoh K, Aoki J, Hosono H, et al:
Molecular cloning and characterization of a novel human G
protein-coupled receptor, EDG7, for lysophosphatidic acid. J Biol
Chem. 274:27776–27785. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Noguchi K, Ishii S and Shimizu T:
Identification of p2y9/GPR23 as a novel G protein-coupled receptor
for lysophosphatidic acid, structurally distant from the Edg
family. J Biol Chem. 278:25600–25606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kotarsky K, Boketoft A, Bristulf J, et al:
Lysophosphatidic acid binds to and activates GPR92, a G
protein-coupled receptor highly expressed in gastrointestinal
lymphocytes. J Pharmacol Exp Ther. 318:619–628. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee CW, Rivera R, Gardell S, Dubin AE and
Chun J: GPR92 as a new G(12/13)- and G(q)-coupled lysophosphatidic
acid receptor that increases cAMP, LPA5. J Biol Chem.
281:23589–23597. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yanagida K, Masago K, Nakanishi H, Kihara
Y, Hamano F, Tajima Y, et al: Identification and characterization
of a novel lysophosphatidic acid receptor, p2y5/LPA6. J Biol Chem.
284:17731–17741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lynch KR and Im DS: Life on the edg.
Trends Pharmacol Sci. 20:473–475. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ishii S, Noguchi K and Yanagida K: Non-Edg
family lysophosphatidic acid (LPA) receptors. Prostaglandins Other
Lipid Mediat. 89:57–65. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yanagida K and Ishii S: Non-Edg family LPA
receptors: the cutting edge of LPA research. J Biochem.
150:223–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yanagida K, Kurikawa Y, Shimizu T and
Ishii S: Current progress in non-Edg family LPA receptor research.
Biochim Biophys Acta. 1831:33–41. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JW, Kim CH, Wang YY, Yan XM and Sohn
UD: Lysophosphatidic acid presynaptically blocks NO uptake during
electric field stimulation-induced relaxation via LPA1 receptor in
cat lower esophageal sphincter. Arch Pharm Res. 34:169–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu XB and Liu JF: Expression of dopamine
receptors in human lower esophageal sphincter. J Gastroenterol
Hepatol. 27:945–950. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu JF, Gao LP, Wen SW, et al: Responses
of human sling and clasp fibers to cholecystokinin and gastrin
through CCK receptors. J Gastroenterol Hepatol. 23:1608–1612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu JF, Lu HL, Wen SW and Wu RF: Effects
of acetylcholine on sling and clasp fibers of the human lower
esophageal sphincter. J Gastroenterol Hepatol. 26:1309–1317. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sriwai W, Zhou H and Murthy KS: G(q)-
dependent signalling by the lysophosphatidic acid receptor LPA(3)
in gastric smooth muscle: reciprocal regulation of MYPT1
phosphorylation by Rho kinase and cAMP-independent PKA. Biochem J.
411:543–551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Im DS, Heise CE, Harding MA, George SR,
O’Dowd BF, Theodorescu D and Lynch KR: Molecular cloning and
characterization of a lysophosphatidic acid receptor, Edg-7,
expressed in prostate. Mol Pharmacol. 57:753–759. 2000.PubMed/NCBI
|