Introduction
Environmental factors have an important role during
embryonic development. Exposure to environmental risk factors,
including hypoxia, prenatal viral infections, drugs, smoking and
stress, results in hypoxia and hypoxic lesions prenatally in ~80%
and perinatally in 10–20% of cases (1). These early environmental risk factors
may affect the structural and/or functional development of the
fetus and neonate. In an hypoxic state, the blood flow in the fetus
is concentrated to the brain, heart and adrenals, at the expense of
the peripheral organs, particularly the lungs (2). A number of studies have demonstrated
that hypoxic exposure during development may lead to lung injury;
however, the underlying mechanisms are yet to be elucidated
(3,4).
MicroRNAs (miRNAs) have been demonstrated to
regulate a number of crucial developmental processes in a variety
of organs, with an important role during lung morphogenesis
recently established (5). However,
little is known with regard to how miRNA expression contributes to
critical events in intrauterine hypoxia. The development of the
fetal respiratory system is a complex process, and pulmonary growth
and maturation is carefully timed and regulated. The process begins
early in gestation and extends into adulthood, consisting of five
developmental phases, including embryonic, pseudoglandular,
canalicular, saccular and alveolar (6). Every phase is critical and
indispensable. A number of studies have performed miRNA profiling
at a variety of time points corresponding to various phases of rat
lung development, from which several miRNAs have been identified
that exhibited significant changes in expression (7,8).
Hypertensive disorders during pregnancy are the most common cause
of intrauterine hypoxia, with a morbidity rate of up to 9.4% in
China (9). Hypoxia usually occurs
after 20 weeks of gestation, during the canalicular phase of lung
development. In order to create an equivalent rat model, hypoxia
was initiated in the rats on embryonic day 19 (E19) in the present
study.
Numerous studies have confirmed that hypoxia
represents a serious risk factor for fetal development (4,10).
However, the majority of studies have focused on neural
development. In addition, several studies have demonstrated that
chronic hypoxia leads to fetal organ dysfunction (11,12),
although the majority have investigated miRNA expression during
normal lung development (7,13,14).
Thus, there are a limited number of studies investigating miRNA
expression during lung development following exposure to hypoxia,
particularly during pregnancy. In the present study, it was
hypothesized that intrauterine hypoxia may result in lung injury
and changes in the miRNA expression profile.
Materials and methods
Animals
All the animals were obtained from the Animal Center
of Central South University (Changsha, China). The animal
experimental protocols were approved by the Committee on Research
Animal Welfare of Central South University (SCXK-XIANG-2009-0004).
Female Sprague-Dawley rats were mated overnight at ~6 weeks of age.
The presence of a vaginal plug the following day was used to
indicate day 0 of gestation. The pregnant animals were divided
randomly into two groups (n=4 per group): Normoxic and hypoxic. All
the pregnant rats were assessed for food intake and weight gain on
a daily basis.
Maternal hypoxia
On E19, the mice in the hypoxic group were exposed
to 10.5% O2. The level of oxygen and length of exposure
during pregnancy was conducted as previously described (3). All the hypoxic rats were placed
together in a plastic exposure chamber inside an infant incubator.
The chamber was filled with room air or pure nitrogen (Changsha
Lianhu Acetylene Co. Ltd., Changsha, China). The interior of the
hypoxic chamber was continuously monitored for nitrogen
(CYX-Digital Oxygen Monitor; Shanghai Jiading Xuelian Instrument
Co., Shanghai, China), carbon dioxide concentrations (Fyrite Gas
Analyzer; Bacharach, Inc., New Kensington, PA, USA), temperature
and relative humidity. The oxygen concentration inside the hypoxic
chamber was maintained at 10.5±1.0% and the carbon dioxide
concentration was <0.5% inside the exposure chambers.
Temperature and relative humidity were maintained at 25±1°C and
50–80%, respectively. Hypoxia exposure was maintained for two days.
Rats in the normoxic group were exposed to ambient oxygen (21%)
instead. All the rats delivered vaginally at ~22 days
gestation.
Body weight and lung wet weight
The body weight of the pups was obtained on
postnatal day 1 (P1). At the end of the experiment, the animals
were sacrificed with an intraperitoneal injection of 50 mg/kg
pentobarbital (CAS 57-33-0; Sigma, St. Louis, MO, USA) and
exsanguinated by severing the femoral aorta. All offsprings were
numbered and selected randomly from each group (n=8), half of which
were males and half were females. Lung specimens were obtained from
each group randomly. Lungs were excised via a midline chest wall
incision, cleared of all nonpulmonary tissues and weighed using an
electronic scale. The lung samples were then frozen in liquid
nitrogen and stored at −70°C for further use. The lung wet
weight/body weight ratio (LW/BW) was calculated.
Lung histology
Lung samples from each group were
tracheally-perfused and fixed at 24 cm H2O pressure for
24 h, with 4% buffered paraformaldehyde (15). The lungs were sectioned into blocks
at right angles to the main bronchus for histological analysis.
Tissue blocks were embedded in paraffin and sectioned (4 μm thick).
Lung samples were stained with hematoxylin and eosin, and examined
for any histological changes. Histological analysis was performed
by a pathologist blinded to the experimental group.
Morphometric analysis
Radial alveolar counts (RACs) were analyzed as
previously described (16).
Between the center of a bronchiole lined by epithelium in one
section of the wall and the nearest connective tissue septum or
lung pleural surface, a perpendicular line was constructed using
image analysis. The number of alveoli dissected by the line was
counted. The RAC was measured for every bronchiole on a slide, from
which an average radial alveolar count for the slide was
calculated.
Mean linear intercepts (Lm) were measured using
crossed hairlines of known length (15). In total, 14 consecutive parenchymal
fields from each lung were examined at a magnification of ×200 on
4-μm sections obtained from the left lower lobe. All analyses were
performed in a blind manner, without knowledge of the experimental
group.
miRNA microarray
Following RNA isolation from the neonatal pup lungs,
a miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark)
was used for miRNA labeling, according to the manufacturer’s
instructions. Each sample (1 μg) was 3′-end-labeled with an
Hy3TM fluorescent label using T4 RNA ligase. Briefly,
RNA in 2.0 μl water was mixed with 1.0 μl calf intestinal alkaline
phosphatase (CIP) buffer and CIP (Exiqon). The mixture was
incubated for 30 min at 37°C, and the reaction was terminated by
incubation for 5 min at 95°C. Next, 3.0 μl labeling buffer, 1.5 μl
fluorescent label (Hy3TM), 2.0 μl dimethyl sulfoxide and
2.0 μl labeling enzyme were added to the mixture. The labeling
reaction was incubated for 1 h at 16°C, and terminated by
incubation for 15 min at 65°C.
Following the labeling procedure, the
Hy3TM-labeled samples were hybridized on a
miRCURYTM LNA Array (version 16.0; Exiqon), in
accordance with the manufacturer’s instructions. A total of 25 μl
hybridization buffer was added to the 25 μl mixture of
Hy3TM-labeled samples. The samples were denatured for 2
min at 95°C, incubated on ice for 2 min and hybridized to the
microarray for 16–20 h at 56°C in a 12-Bay Hybridization System
(Nimblegen Systems, Inc., Madison, WI, USA), which provided an
active mixing action and constant incubation temperature to improve
hybridization uniformity and enhance the signal. Following
hybridization, the slides were obtained, washed several times using
a wash buffer kit (Exiqon) and dried by centrifugation for 5 min at
400 rpm. The slides were scanned using an Axon GenePix 4000B
microarray scanner (Axon Instruments, Foster City, CA, USA).
Scanned images were imported into GenePix Pro 6.0
software (Axon Instruments) for grid alignment and data extraction.
Replicated miRNAs were averaged and miRNAs with intensities of ≥50
times in all samples were selected to calculate the normalization
factor. Expressed data were normalized using median normalization.
Following normalization, differentially expressed miRNAs were
identified via fold-change filtering (fold change of >2.0).
Hierarchical clustering was then performed using TIGR MeV software
(version 4.6).
Quantitative polymerase chain reaction
(qPCR)
TaqMan microRNA assays (Applied Biosystems,
Foster City, CA, USA) were performed on selected miRNAs, including
miR-465*, miR-377*, miR-327, miR-338, miR-127
and miR-210, in accordance with the manufacturer’s instructions. In
brief, total RNA was isolated from the lungs of the rats using a
mirVana miRNA Isolation kit (Ambion, Austin, TX, USA). For qPCR
with TaqMan microRNA assays, 75 ng total RNA was used as
template in each reaction with miRNA-specific reverse transcription
(RT) primers. The reactions were incubated on ice for 5 min,
followed by incubation at 16°C for 30 min, 42°C for 30 min and 85°C
for 5 min. For each PCR assay, 1.33 μl RT product was used as a
template. PCR assays were incubated at 95°C for 10 min, followed by
35 cycles of 95°C for 15 sec and 60°C for 60 sec. All the PCR
assays were run in duplicate. In addition, RT and PCR were
performed for 18S in each sample as an endogenous control using
TaqMan Ribosomal RNA Control Reagents (Applied Biosystems).
Data analysis was performed as aforementioned. RT was performed
using the poly(T) adaptor, GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTT
TTTTTTVN, while qPCR was performed using the universal reverse
primer, GCGAGCACAGAATTAATACGACTCAC, and a forward primer with the
same sequence as the mature miRNA (mir-465*,
UGAUCAGUGCCUUUCUGAGUAG; mir-377*,
AGAGGUUGCCCUUGGUGAAUUC; miR-127, UCGGAUCCGUCUGAGCUUGGCU; miR-210,
CUG UGCGUGUGACAGCGGCUGA; miR-327, CCUUGAGGG GCAUGAGGGU; miR-338,
UCCAGCAUCAGUGAUUUU GUUGA).
Statistical analysis
Results are expressed as the mean ± standard
deviation, or percentages. Differences between the groups were
analyzed using one-way analysis of variance and the
χ2-test for categorical variables, with SPSS 15.0
statistical software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
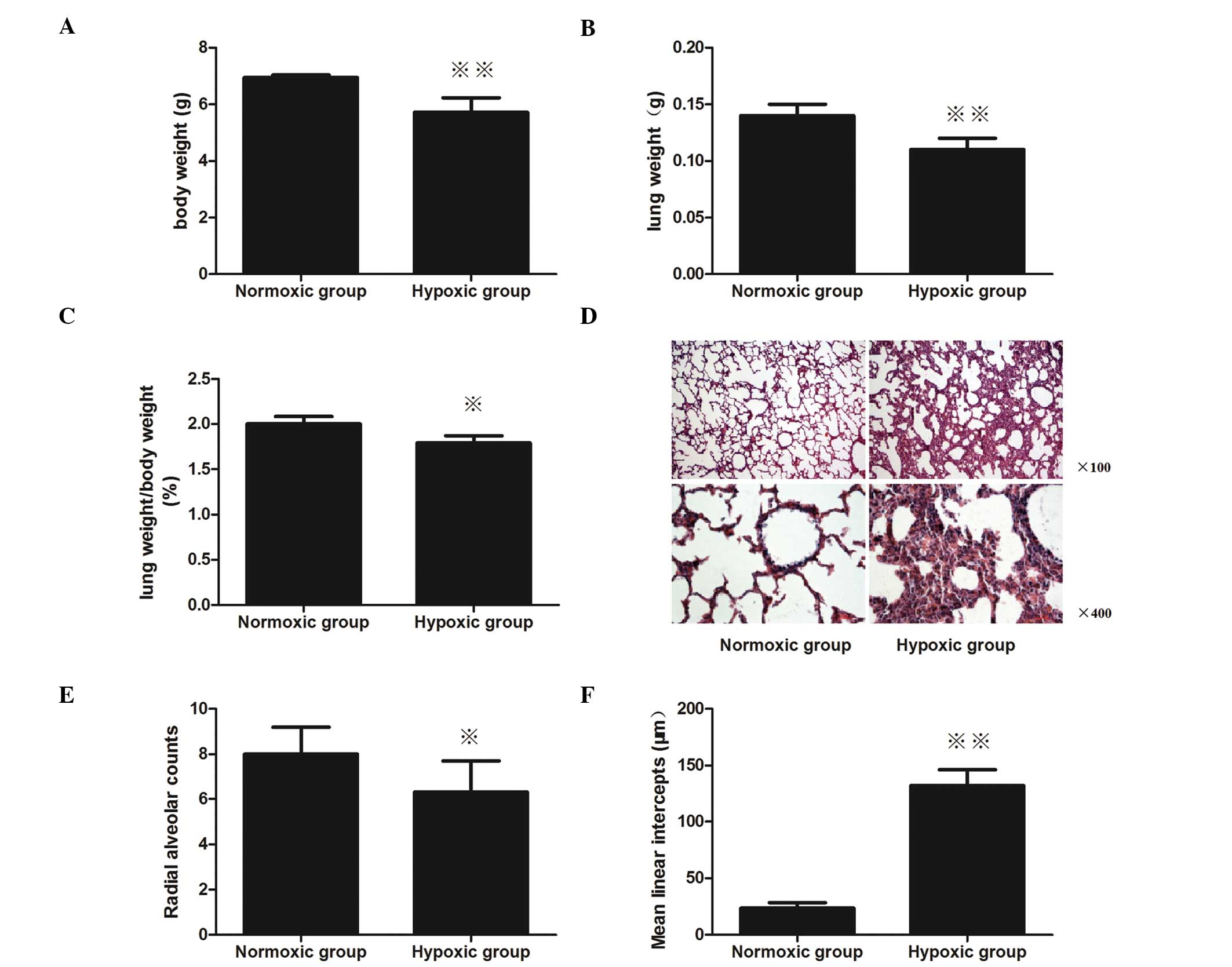
Body weight and lung wet weight
Newborn rats at P1 that had been exposed to
intrauterine hypoxia from embryonic day 19 (E19) to E20 exhibited a
significant body and lung wet weight reduction. The body weight of
the hypoxic group pups was significantly reduced compared with the
normoxic group (P<0.01; Fig.
1A), as well as the lung wet weight (P<0.01, Fig. 1B). Furthermore, the LW/BW of the
hypoxic group pups was also markedly decreased compared with the
normoxic pups (P<0.05; Fig.
1C).
Morphometric analysis
Alveolar-like structures in the newborn rat lungs in
the normoxic group on P1 were irregular, and a small number of
septa were observed (Fig. 1D).
Compared with the normoxic group, significant lung injury was
present in the animals in the hypoxic group. In the hypoxic group,
the alveolar-like structures were more irregular, the alveolar
septum was thick and a small quantity of red blood cells were
present in the alveolar septum and alveolar space.
The RAC of the hypoxic-treated lungs was markedly
decreased compared with the normoxic-treated lungs on P1
(P<0.05; Fig. 1E). In addition,
the Lm was shown to increase in the neonatal rats that had been
exposed to intrauterine hypoxia, as compared with the neonatal rats
exposed to intrauterine normoxia; the Lm significantly increased
from 23.5 to 131.7 μm (P<0.01; Fig.
1F).
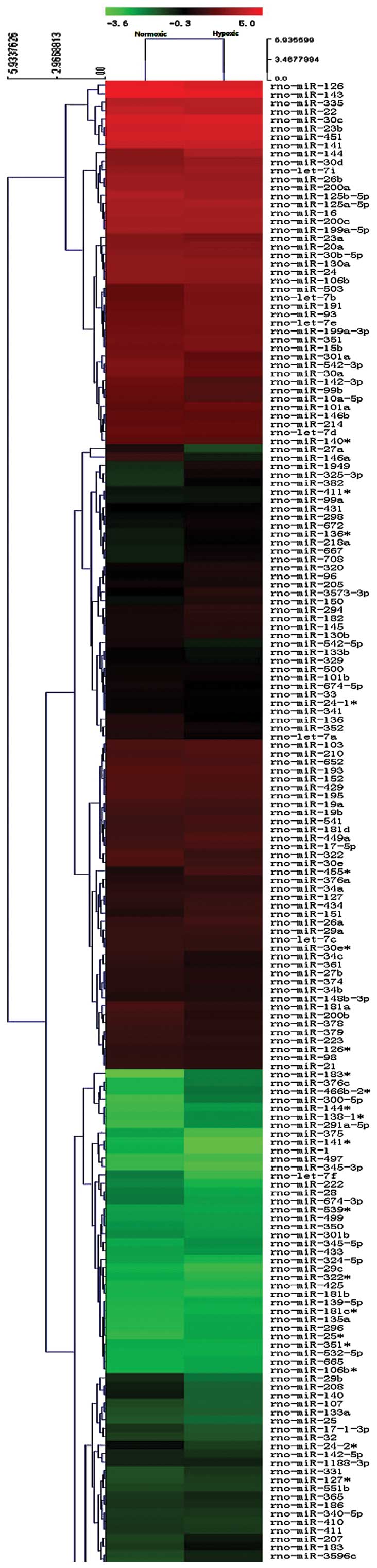
miRNA microarray
The sixth generation of the miRCURYTM LNA
Array (version 16.0) was used, which contained >1,891 capture
probes, covering all human, mouse and rat miRNAs annotated in
miRBase 16.0, as well as all viral miRNAs associated with these
species. In addition, this array contained capture probes for 66
novel miRPlusTM human miRNAs. As a result, 69
differentially expressed miRNAs between these two groups passed the
fold-change filtering (fold-change of >2.0; Fig. 2), including 55 upregulated miRNAs
and 14 downregulated miRNAs (Table
I).
 | Table IDifferentially expressed miRNAs in the
hypoxic group compared with the normoxic groupa. |
Table I
Differentially expressed miRNAs in the
hypoxic group compared with the normoxic groupa.
| miRNAs | Fold-change | Expression |
|---|
|
rno-miR-465* | 28.57707510 | Upregulated |
|
rno-miR-377* | 28.38902356 | Upregulated |
|
rno-miR-26a* | 12.85968379 | Upregulated |
| rno-miR-208b-3p | 12.24797596 | Upregulated |
| rno-miR-125b-3p | 10.71640316 | Upregulated |
|
rno-miR-375* | 10.35918972 | Upregulated |
| rno-miR-3571 | 8.463210702 | Upregulated |
| rno-miR-192 | 7.858695652 | Upregulated |
|
rno-miR-9* | 7.144268775 | Upregulated |
|
rno-miR-218a-1* | 7.057959375 | Upregulated |
| rno-miR-291b | 5.71541502 | Upregulated |
| rno-miR-124 | 5.239130435 | Upregulated |
|
rno-miR-224* | 5.080368906 | Upregulated |
|
rno-miR-3590-5p | 5.000988142 | Upregulated |
|
rno-let-7a-2* | 4.156665469 | Upregulated |
| rno-miR-3572 | 4.082439300 | Upregulated |
| rno-miR-323 | 3.969038208 | Upregulated |
|
rno-miR-3557-5p | 3.572134387 | Upregulated |
| rno-miR-344a | 3.798850313 | Upregulated |
|
rno-miR-664-2* | 3.333992095 | Upregulated |
|
rno-miR-3594-3p | 3.214920949 | Upregulated |
|
rno-miR-183* | 3.162217327 | Upregulated |
| rno-miR-31 | 2.917243083 | Upregulated |
| rno-miR-3596b | 2.857707517 | Upregulated |
| rno-miR-3592 | 2.843578545 | Upregulated |
|
rno-miR-29b-1* | 2.739856731 | Upregulated |
|
rno-miR-374* | 2.737659550 | Upregulated |
|
rno-miR-582* | 2.729787569 | Upregulated |
|
rno-miR-449c-5p | 2.698577075 | Upregulated |
|
rno-miR-448* | 2.662863816 | Upregulated |
| rno-miR-2985 | 2.619565217 | Upregulated |
|
rno-miR-296* | 2.593104963 | Upregulated |
| rno-miR-347 | 2.578906777 | Upregulated |
|
rno-miR-652* | 2.571936759 | Upregulated |
|
rno-miR-551b* | 2.500494071 | Upregulated |
|
rno-miR-331* | 2.442879000 | Upregulated |
|
rno-miR-221* | 2.429051383 | Upregulated |
|
rno-miR-493* | 2.422838976 | Upregulated |
|
rno-miR-16* | 2.381422925 | Upregulated |
| rno-miR-190b | 2.286166008 | Upregulated |
| rno-miR-463 | 2.266932395 | Upregulated |
|
rno-miR-21* | 2.262351779 | Upregulated |
| rno-miR-300-5p | 2.238537549 | Upregulated |
| rno-miR-3577 | 2.222661397 | Upregulated |
|
rno-miR-134* | 2.177300960 | Upregulated |
| rno-miR-9 | 2.143280632 | Upregulated |
|
rno-miR-202* | 2.140053487 | Upregulated |
|
rno-miR-153* | 2.139897411 | Upregulated |
| rno-miR-873 | 2.133579843 | Upregulated |
| rno-miR-1224 | 2.131559843 | Upregulated |
|
rno-miR-208* | 2.112218594 | Upregulated |
|
rno-miR-433* | 2.102979629 | Upregulated |
|
rno-miR-154* | 2.078332734 | Upregulated |
| rno-miR-1949 | 2.065437510 | Upregulated |
|
rno-miR-455* | 2.008607060 | Upregulated |
| rno-miR-327 | 0.142885375 | Downregulated |
| rno-miR-338 | 0.295624915 | Downregulated |
|
rno-miR-222* | 0.317523057 | Downregulated |
| rno-miR-27a | 0.354368736 | Downregulated |
| rno-miR-196a | 0.357213439 | Downregulated |
| rno-miR-412 | 0.357213439 | Downregulated |
|
rno-miR-299* | 0.40824393 | Downregulated |
|
rno-miR-133a* | 0.446516798 | Downregulated |
|
rno-miR-23b* | 0.476284585 | Downregulated |
|
rno-miR-27a* | 0.476284585 | Downregulated |
|
rno-miR-29c* | 0.476284585 | Downregulated |
| rno-miR-504 | 0.476284585 | Downregulated |
| rno-let-7f | 0.490939495 | Downregulated |
| rno-miR-140 | 0.495260066 | Downregulated |
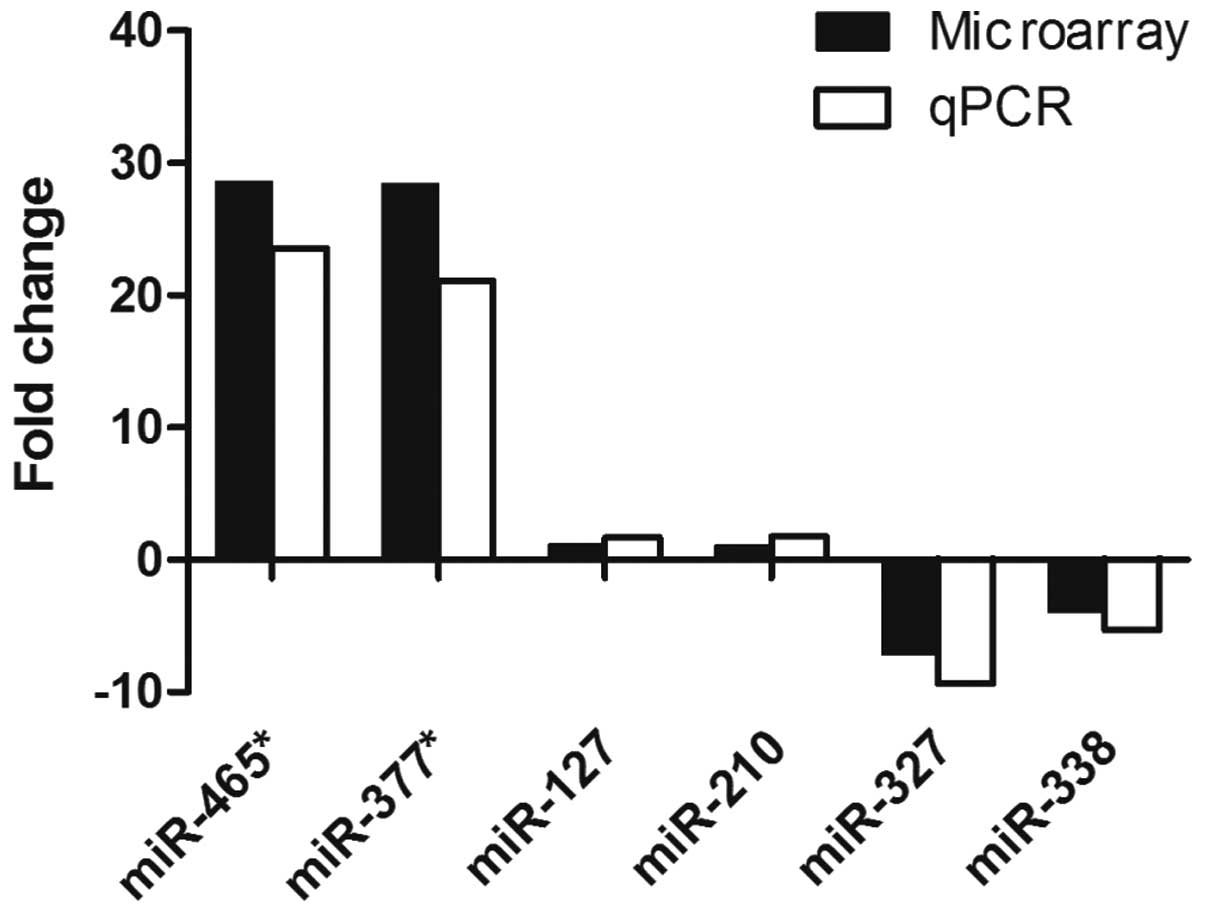
qPCR
qPCR was performed to validate the miRNA expression
levels in the neonatal rat lungs exposed to intrauterine hypoxia
for two days. miRNAs were selected based on high fold changes,
their expression in the lungs and functional studies in other
systems. miR-465* and miR-377* were selected
as the most upregulated miRNAs, while miR-327 and miR-338 exhibited
the most downregulated expression. miR-210 was selected as the
‘master hypoxamiR’, since this miRNA has been shown to exhibit high
and consistent upregulation under hypoxia in the majority of cell
types (17). Furthermore, miR-127
is an important miRNA in late lung development (5,8);
thus, was selected. The results from the qPCR analysis revealed
that all the selected miRNAs followed the same expression trend
observed in the microarray experiment (Fig. 3).
Discussion
In the present study, intrauterine hypoxia in late
gestation was shown to result in lung injury and marked changes in
miRNA expression in newborn rats. Hypoxia is required for fetal
development; however, excess hypoxia is detrimental. Complications
resulting from fetal hypoxia/anoxia are among the top ten causes of
fetal mortality (18). As
aforementioned, the lung development process begins early in
gestation and extends into adulthood, including five developmental
phases in rats: Embryonic (E0–E10), pseudoglandular (E11–E18),
canalicular (E19–E20), saccular (E21–P3) and alveolar (P4–P21)
(6). Each phase is critical and
indispensable. Hypoxia can produce temporary dysfunction or
permanent injury, depending on the duration, intensity of oxygen
deprivation and the age of the fetus (19). However, the mechanisms underlying
the effects of hypoxia on lung development remain unclear. Larson
and Thurlbeck (20) found that
pregnant rats exposed to hypoxia from early gestation (E14) until
near term (E21) produced offspring with decreased lung weight, DNA
and protein per lung when compared with a normoxic group (20). This long-term intrauterine hypoxia
was applied across three periods of lung development. However, the
present study investigated whether short-term hypoxic exposure in
the more mature lung lead to lung hypoplasia.
In the present study, rats were used as an animal
model to investigate hypertensive disorders complicating pregnancy
during the canalicular (E19–E20) stage. Newborn rats exposed to
intrauterine hypoxia between E19 and E20 exhibited a significant
reduction in body and lung wet weight, as well as a marked decrease
in the RAC and an increase in the Lm. In addition, the offspring
demonstrated fewer and larger alveoli, and the alveolar septum was
thicker. The canalicular phase is accompanied by the formation of
distal airway bronchioles and proximal to distal epithelial
differentiation. Mesenchymal cells begin to develop into
chondrocytes, fibroblasts and myofibroblasts (21). A previous study demonstrated that
during this stage, hypoxic impairment may irreversibly weaken
epithelial growth in the developing lung (22). Any factor affecting normal lung
development may disturb the balance between injury and repair,
leading to hypoplasia (21).
However, the underlying mechanisms are yet to be elucidated.
miRNAs are a large group of regulatory, noncoding,
small RNA molecules that are ~22 nucleotides in length (23). Up to a third of human genes
involved in the regulation of numerous biological processes,
including cellular differentiation, developmental timing, immune
responses, nerve system patterning and apoptosis, are regulated by
miRNAs (24). Previously, studies
have profiled the expression of different miRNAs at various stages
of lung development (8), with
several studies focusing on the canalicular stage (7,25).
However, to the best of our knowledge, no studies have investigated
intrauterine hypoxic lung development. In the present study, 69
differentially expressed miRNAs were identified between the hypoxic
and normoxic groups, including 55 upregulated miRNAs and 14
downregulated miRNAs, a number of which had not previously been
reported.
The expression levels of miR-465* and
miR-377* were the most significantly upregulated, while
the expression levels of miR-327 and miR-338 were the most
downregulated. It has previously been found that miR-465 families
are the most abundant X-linked miRNA molecules in newborn mouse
ovaries (26). In addition,
miR-377 has been shown to be upregulated in diabetic nephropathy
and lung tumors (27), and
abundantly expressed in transdifferentiated neuronal progenitors
(28). Furthermore, miR-327 is
upregulated in myocardial microvascular endothelial cells in
impaired angiogenesis of type 2 diabetic rats (29), while miR-338 has been previously
found to be downregulated in rats with pulmonary fibrosis (30). However, to the best of our
knowledge, the present study is the first to investigate these
miRNAs during lung development, and in particular, under hypoxic
exposure. miR-127 is important during the later stage of fetal lung
development (5); however, no
statistically significant difference in miR-127 expression was
identified between the hypoxic and normoxic groups in the present
study. The ‘master hypoxamiR’, miR-210, has been shown to exhibit
high and consistent upregulation under hypoxic conditions in the
majority of cell types (17);
however, the cells used in this study were mature. In the present
study, the expression of miR-210 did not change significantly in
the developing lung following intrauterine hypoxia exposure when
compared with the normoxic group. Studies have found that the
increased expression of a number of miRNAs was directly correlated
with the downregulation of predicted mRNA targets (31,32).
These observations indicated that inhibition of translation without
mRNA degradation may be the mechanism of miRNA-mediated gene
regulation during lung development. In the present study, more
miRNAs were found to be upregulated in the hypoxic group compared
with the normoxic group, which may constitute the mechanism
underlying lung hypoplasia resulting from intrauterine hypoxia
exposure.
In conclusion, the present study demonstrated that
intrauterine hypoxia results in lung hypoplasia and marked changes
in miRNA expression in newborn rats. However, the systematic
profiling of miRNA, mRNA and protein expression levels during
intrauterine hypoxic-exposed lung development requires further
investigation.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (nos. 81070522, 81000264 and
81370098) and the Open Fund Project of the University Innovation
Platform of Hunan Province (no. 2010.1-2012.12). The authors thank
members of the Department of Neonatology for their constructive
feedback in conducting the study and preparing the manuscript.
References
|
1
|
Habek D, Hodek B, Herman R and Habek JC:
Fetal hypoxia-etiology and pathophysiology of hypoxic damage. Lijec
Vjesn. 122:82–89. 2000.(In Croatian).
|
|
2
|
Jensen A and Berger R: Fetal circulatory
responses to oxygen lack. J Dev Physiol. 16:181–207. 1991.
|
|
3
|
de Grauw TJ, Myers RE and Scott WJ: Fetal
growth retardation in rats from different levels of hypoxia. Biol
Neonate. 49:85–89. 1986.PubMed/NCBI
|
|
4
|
Mach M, Dubovický M, Navarová J,
Brucknerová I and Ujházy E: Experimental modeling of hypoxia in
pregnancy and early postnatal life. Interdiscip Toxicol. 2:28–32.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kajekar R: Environmental factors and
developmental outcomes in the lung. Pharmacol Ther. 114:129–145.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Y, Kai G, Pu XD, Qing K, Guo XR and
Zhou XY: Expression profile of microRNAs in fetal lung development
of Sprague-Dawley rats. Int J Mol Med. 29:393–402. 2012.PubMed/NCBI
|
|
8
|
Khoshgoo N, Kholdebarin R, Iwasiow BM and
Keijzer R: MicroRNAs and lung development. Pediatr Pulmonol.
48:317–323. 2013. View Article : Google Scholar
|
|
9
|
Huang Y: National prevalence survey of
PIH. Chinese Journal of Obstetrics and Gynecology. 26:2–5.
1991.
|
|
10
|
Giles BL, Suliman H, Mamo LB, Piantadosi
CA, Oury TD and Nozik-Grayck E: Prenatal hypoxia decreases lung
extracellular superoxide dismutase expression and activity. Am J
Physiol Lung Cell Mol Physiol. 283:L549–L554. 2002.PubMed/NCBI
|
|
11
|
Mortola JP, Xu LJ and Lauzon AM: Body
growth, lung and heart weight, and DNA content in newborn rats
exposed to different levels of chronic hypoxia. Can J Physiol
Pharmacol. 68:1590–1594. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Al-Hasan YM, Evans LC, Pinkas GA,
Dabkowski ER, Stanley WC and Thompson LP: Chronic hypoxia impairs
cytochrome oxidase activity via oxidative stress in selected fetal
Guinea pig organs. Reprod Sci. 20:299–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Williams AE, Moschos SA, Perry MM, Barnes
PJ and Lindsay MA: Maternally imprinted microRNAs are
differentially expressed during mouse and human lung development.
Dev Dyn. 236:572–580. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sessa R and Hata A: Role of microRNAs in
lung development and pulmonary diseases. Pulm Circ. 3:315–328.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Truog WE, Xu D, Ekekezie II, et al:
Chronic hypoxia and rat lung development: analysis by morphometry
and directed microarray. Pediatr Res. 64:56–62. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang JR, Markham NE, Lin YJ, McMurtry IF,
Maxey A, Kinsella JP and Abman SH: Inhaled nitric oxide attenuates
pulmonary hypertension and improves lung growth in infant rats
after neonatal treatment with a VEGF receptor inhibitor. Am J
Physiol Lung Cell Mol Physiol. 287:L344–L351. 2004. View Article : Google Scholar
|
|
17
|
Bertero T, Robbe-Sermesant K, Le Brigand
K, et al: microRNAs target identification: lessons from hypoxamiRs.
Antioxid Redox Signal. Feb 3–2014.(Epub ahead of print).
|
|
18
|
Anderson RN: Deaths: leading causes for
2000. Natl Vital Stat Rep. 50:1–85. 2002.PubMed/NCBI
|
|
19
|
Waters KA and Machaalani R: Role of NMDA
receptors in development of respiratory control. Respir Physiol
Neurobiol. 149:123–130. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Larson JE and Thurlbeck WM: The effect of
experimental maternal hypoxia on fetal lung growth. Pediatr Res.
24:156–159. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi W, Xu J and Warburton D: Development,
repair and fibrosis: what is common and why it matters.
Respirology. 14:656–665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McQueston JA, Cornfield DN, McMurtry IF
and Abman SH: Effects of oxygen and exogenous L-arginine on EDRF
activity in fetal pulmonary circulation. Am J Physiol.
264:H865–H871. 1993.PubMed/NCBI
|
|
23
|
Grosshans H and Slack FJ: Micro-RNAs:
small is plentiful. J Cell Biol. 156:17–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu Y, Okubo T, Rawlins E and Hogan BL:
Epithelial progenitor cells of the embryonic lung and the role of
microRNAs in their proliferation. Proc Am Thorac Soc. 5:300–304.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ahn HW, Morin RD, Zhao H, et al: MicroRNA
transcriptome in the newborn mouse ovaries determined by massive
parallel sequencing. Mol Hum Reprod. 16:463–471. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Q, Wang Y, Minto AW, Wang J, Shi Q,
Li X and Quigg RJ: MicroRNA-377 is up-regulated and can lead to
increased fibronectin production in diabetic nephropathy. FASEB J.
22:4126–4135. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang SJ, Weng SL, Hsieh JY, Wang TY,
Chang MD and Wang HW: MicroRNA-34a modulates genes involved in
cellular motility and oxidative phosphorylation in neural
precursors derived from human umbilical cord mesenchymal stem
cells. BMC Med Gen. 4:652011. View Article : Google Scholar
|
|
29
|
Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM
and Hu RM: MicroRNA-320 expression in myocardial microvascular
endothelial cells and its relationship with insulin-like growth
factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol.
36:181–188. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Liu X, Chen S, et al:
Tectorigenin inhibits the in vitro proliferation and enhances
miR-338* expression of pulmonary fibroblasts in rats
with idiopathic pulmonary fibrosis. J Ethnopharmacol. 131:165–173.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chan SY and Loscalzo J: MicroRNA-210: a
unique and pleiotropic hypoxamir. Cell Cycle. 9:1072–1083. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan YC, Banerjee J, Choi SY and Sen CK:
miR-210: the master hypoxamir. Microcirculation. 19:215–223. 2012.
View Article : Google Scholar : PubMed/NCBI
|