Introduction
Periodontitis comprises a group of chronic
inflammatory diseases affecting the periodontium that can result in
the loosening and subsequent loss of teeth. Periodontitis is caused
by microorganisms that adhere to and grow on the surfaces of the
teeth, along with an overly aggressive immune response against
these microorganisms (1).
Consequently, an understanding of the bacterial flora in the human
oral cavity is essential for the diagnosis, prevention and therapy
of periodontal diseases; however, since the majority of oral
bacteria can not grow under artificial conditions (2,3), a
number of community-based microbiological methods are emerging as
more effective tools to study microbial ecology in the human oral
cavity.
Substrate utilization patterns, assessed using the
MicroResp™ method (4), are a means
of investigating the physiological diversity of microorganisms. The
MicroResp system is based on measuring the substrate-induced
respiration (SIR) of a range of different substrates to generate
patterns of potential sole carbon source utilization. Since
differences in utilization patterns are interpreted as differences
in the major active members of the microbial community, this method
has been widely used to examine communities from different
environments (4,5). Terminal restriction fragment length
polymorphism (TRFLP) is a molecular approach that can be
successfully used for the rapid assessment and comparison of
complex bacterial communities (6,7). The
application of TRFLP has led to the qualitative and quantitative
assessment of a significantly larger range of microbial communities
than the range that could previously be evaluated using traditional
culturing techniques (8).
In the study of the oral bacterial community, saliva
is considered to be the most suitable sample, as it is composed of
a variety of bacteria from different oral sites (9). Although TRFLP analysis has been used
for evaluating the oral bacterial community in a few studies
(7,9), the TRFLP method has not been compared
with other microbial ecology techniques. The aim of the present
study, therefore, was to measure colony-forming units (CFUs) in the
unstimulated saliva samples of patients with chronic periodontitis
and healthy subjects. The MicroResp and TRFLP methods were
additionally used to provide further understanding of the microbial
ecology of these saliva samples and to evaluate the hypothesis that
the oral community structure would be correlated with oral health
status.
Materials and methods
Sample collection
Saliva samples were collected in sterile containers
from 20 patients with chronic periodontitis (10 males and 10
females; mean age ± standard deviation, 46.3±10.8 years) who had
visited the First Affiliated Hospital (Hangzhou, China) and from 20
periodontally healthy subjects (10 males and 10 females; mean age ±
standard deviation, 43.3±12.5 years). All subjects who participated
in this study understood the nature of the research and exhibited
no serious illness. All subjects provided written informed consent.
They had not used any antibiotics in the last three months before
collection. Each sample contained 5-ml aliquots of saliva.
Plate counts of culturable
bacteria
Numbers of bacterial CFUs were estimated by the
dilution plate method. The saliva samples were serially diluted
with physiological salt solution and suspensions (0.1 ml) spread in
six replicates onto the blood agar plates (bioMérieux, Lyon,
France). The plates were incubated at 35°C and colonies counted
after 24 h.
Substrate utilization pattern
The MicroResp system (4) was used to measure the SIR pattern.
Briefly, the system utilizes a carbon dioxide detection microplate
attached to a 1.2-ml deepwell plate containing the saliva sample
and a range of carbon sources. The two plates are connected with a
rubber gasket, which forms a seal when the plates are clamped
together.
The saliva (100 µl) was placed in the deepwell plate
and all measurements were performed in triplicate. The MicroResp
system was incubated at 37°C for 6 h. Basal respiration was
measured in wells containing water (100 µl) only. SIR was measured
by adding one of 15 different carbon sources, which were
administered in aliquots of 100 µl, to generate a final
concentration of 20 gl−1. The carbon sources used were
alanine, galactose, glucosamine, arabinose, glucose, oxalic acid,
arginine, protocatechuic acid, citric acid, ketoglutaric acid,
malic acid, L-cysteine HCl, lysine, erythritol and xylitol (all
from Sigma-Aldrich Co., Ltd., Gillingham, UK).
TRFLP analysis
DNA was extracted using the DNeasy® kit from Qiagen
(Hilden, Germany), as per the manufacturer's instructions. For each
sample, internal regions of 16S rRNA genes were amplified using the
universal forward primer, 63F, labeled with caboxyfluorescein and
the universal reverse primer, 1087R (Sangon Biotech Co., Ltd,
Shanghai, China) (10). The labeled
polymerase chain reaction amplicons were checked by agarose gel
electrophoresis and purified using an UltraClean® DNA purification
kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). Approximately
l00 ng purified DNA products were digested using HhaI
(Sangon Biotech Co., Ltd) (11,12).
Following digestion, a 2-µl portion of each sample was mixed with
0.3 µl LIZ®-labeled internal size standard (Applied Biosystems,
Warrington, UK) and 12 µl formamide. The analysis of fragment size
was performed using an ABI PRISM® 3030×l genetic analyzer (Applied
Biosystems). The fragment analysis of the TRFLP data was conducted
between 35 and 1,200 bp. All terminal restriction fragments (TRFs)
with a fluorescence of <50 units were discarded and the relative
abundance of the TRFs were used. All peaks with heights that were
<2% of the total peak height were excluded from further analyses
to avoid potential artefacts.
Statistical analysis
Means and least significant differences at the 5%
level were calculated using a one-way analysis of variance. The SIR
and TRFLP data were also analyzed using principal component
analysis. The proportional color changes of individual carbon
sources in the MicroResp data set and the relative abundance of the
individual TRFs in the TRFLP data set were calculated and used for
principal component analysis, subsequent to the generation of a
correction matrix to transform the data to unit variance. The
correlation coefficients were made using simple linear regression.
All data analyses were performed using SPSS 16.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Colony-forming units and substrate
utilization patterns
The number of CFUs in the healthy subjects ranged
from 2.0×107 to 3.0×108 ml−1
(mean, 1.1×108 ml−1) saliva. Larger aerobic
bacterial populations were found in the patients with chronic
periodontitis compared with the control group, and the average CFU
count in the patient group was nearly three-fold higher
(P<0.001) than that in the control group. There was no
significant effect of age on the culturable bacteria. The bacterial
populations in the saliva were independent on the gender of
subjects, and no significant difference was found between the
female and male subjects.
The SIR rates for all the carbon sources were above
the basal respiration rate (water only). The highest SIR rate was
found with glucose and arabinose, and the SIR rates were 11- and
14-fold greater than the basal respiration rate, respectively. The
addition of amino acids, such as alanine and arginine, had the
smallest effect and only increased the SIR rate ∼1.5-fold.
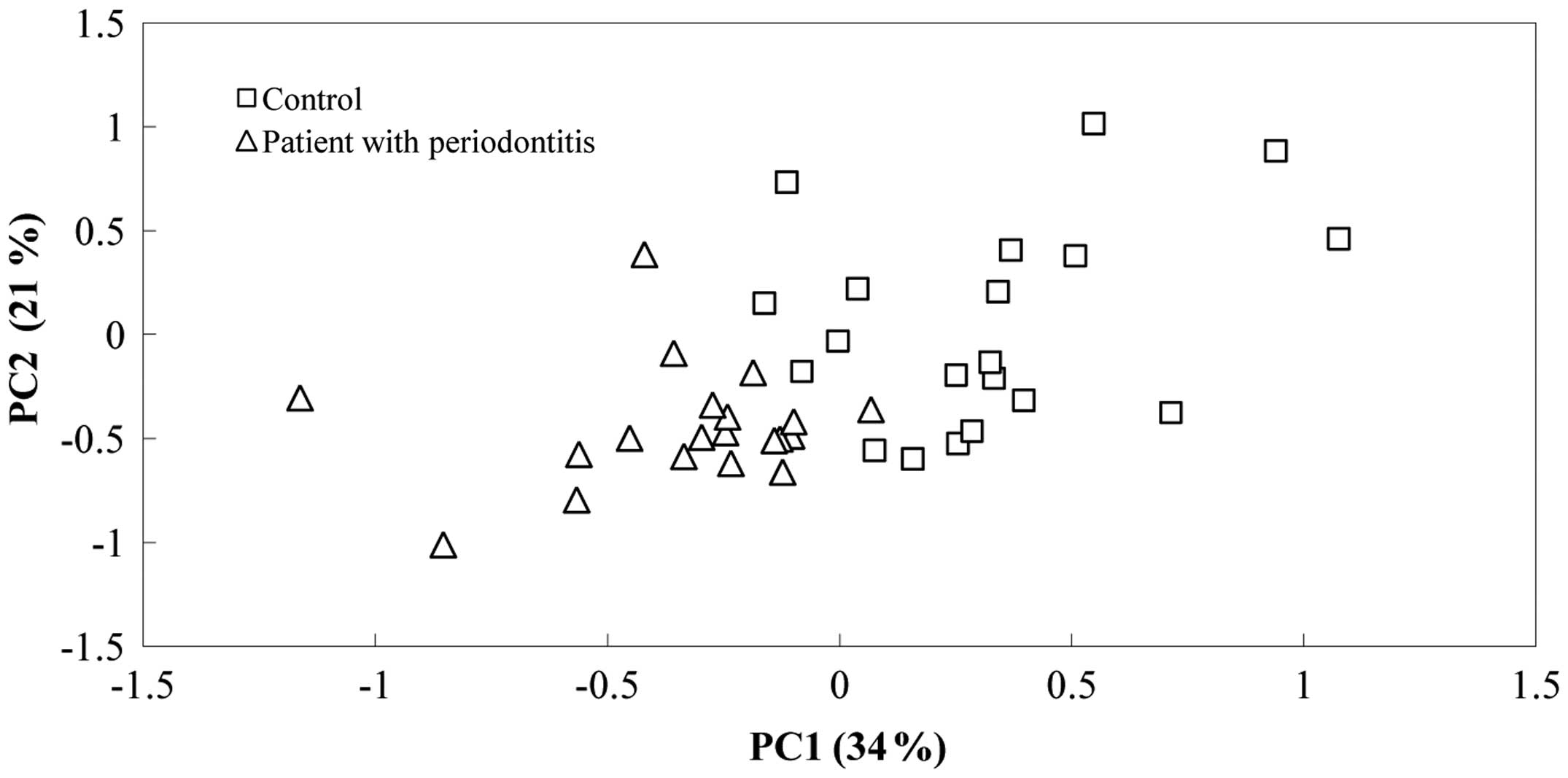
Principal component analysis (Fig.
1) using all 15 carbon sources revealed a separation of the
samples. The first principal component (PC1) of the MicroResp data
accounted for 34% of the variance, and the second (PC2) accounted
for 21% of the variation. The PC1 scores of the patient group were
significantly lower than those of the control group (Fig. 1). The oral microbial communities from
the patients with chronic periodontitis and the healthy subjects
differed in their preferred carbon sources (Table I; Fig.
1). Correlation analysis of the loadings of the most
influential individual carbon sources on the PC1 indicated that
arabinose, glucose and galactose were preferred by the patient
group, whereas erythritol, xylitol, malic acid and citric acid were
preferred by the control group. It was found, however, that the
principal components (PC1 and PC2) were not correlated with the
bacterial populations or age of the subjects (Table II).
 | Table I.Loading scores of the most influential
individual carbon sources on the first principal component. |
Table I.
Loading scores of the most influential
individual carbon sources on the first principal component.
| Substrate | Substrate class | Loading score |
|---|
| Arabinose | Carbohydrates | -0.433 |
| Glucose | Carbohydrates | -0.376 |
| Galactose | Carbohydrates | -0.312 |
| Citric acid | Carboxylic acids | 0.348 |
| Malic acid | Carboxylic acids | 0.413 |
| Xylitol | Sugar alcohols | 0.517 |
| Erythritol | Sugar alcohols | 0.621 |
 | Table II.Correlation coefficients (r) among
principal components and the bacterial populations and age of the
subjects. |
Table II.
Correlation coefficients (r) among
principal components and the bacterial populations and age of the
subjects.
|
| Substrate utilization
pattern | Bacterial TRFLP |
|---|
|
|
|
|
|---|
| Parameter | PC1 | PC2 | PC1 | PC2 |
|---|
| Colony forming
units | 0.288 | 0.149 | -0.506a | 0.023 |
| Age of the
subjects | 0.154 | 0.085 | 0.176 | 0.368a |
Terminal restriction fragment length
polymorphism analysis
Each of the saliva samples showed 15–29 TRFs
following digestion with HhaI. A total of 38 different TRFs
were detected. Overall, >90% of the TRFs were <600 bp. The
542-bp TRF was the most dominant in the TRFLP patterns of all the
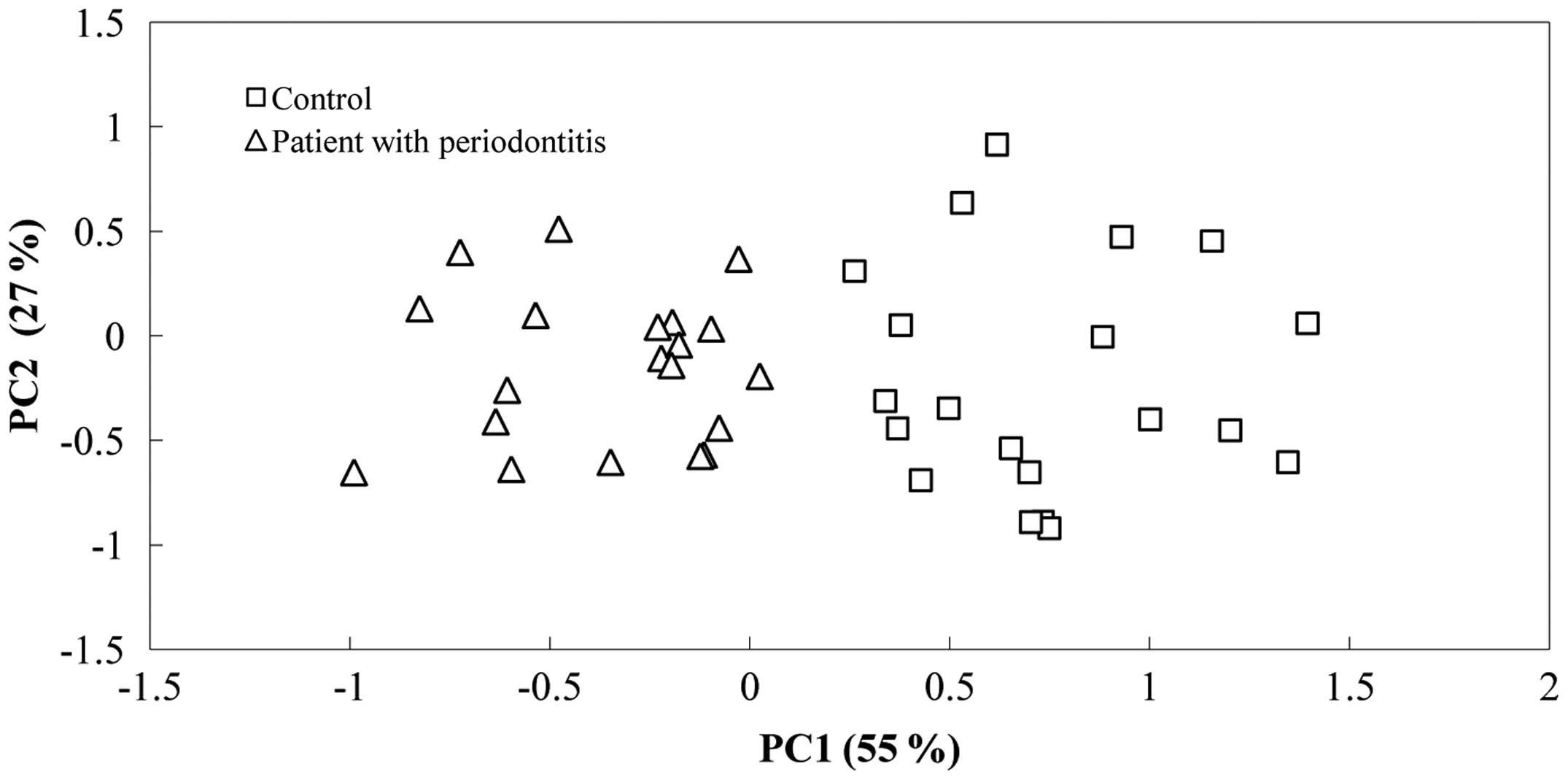
samples. Principal component analysis of the TRFLP data was used to
reveal shifts in the bacterial communities (Fig. 2). The PC1 of the TRFLP data accounted
for 55% of the variance and discriminated the patient group from
the control group, indicating that the two groups had different
microbial communities (Fig. 2). A
significant negative association was found between the PC1 scores
and CFUs (Table II). The PC2
accounted for 27% of the variation and there was a significant
positive correlation between the PC2 scores and age of the subjects
(Table II). Correlation analysis of
the loading scores of the most influential TRFs on the PC1
indicated that the TRFs of 68, 275, 330 and 436 bp were enriched in
the patient group, whereas the TRFs of 45, 192 and 424 bp showed
increases in relative abundance in the control group (Fig. 2; Table
III).
 | Table III.Loading scores of the most influential
individual TRFs on the first principal component. |
Table III.
Loading scores of the most influential
individual TRFs on the first principal component.
| TRF (bp) | Loading score |
|---|
| 68 | -0.473 |
| 275 | -0.386 |
| 330 | -0.352 |
| 436 | -0.311 |
| 424 | 0.334 |
| 192 | 0.375 |
| 45 | 0.448 |
Discussion
There is now considerable evidence documenting a
close association between bacterial community structure and oral
health status (7,13–15). The
bacterial composition in saliva is more similar to that of the soft
tissues than that of the supra- and subgingival plaque and can
reflect the periodontal health condition in the oral cavity
(16). As expected, significantly
larger aerobic bacterial populations were observed in the patients
with chronic periodontitis in the present study. The function and
composition of the bacterial community in saliva is highly
correlated with periodontal health.
The substrate utilization method is widely used due
to its simplicity and the fact that it yields considerable
information about an important functional attribute of microbial
communities (4). Since the ability
to utilize a range of carbon substrates is fundamental to the
ecological functions of microbes in different environments, the
difference in the substrate utilization pattern can reflect the
difference in the microbial community structure. In the present
study, a systematic change in the substrate utilization pattern was
associated with oral health status. Oral microbial communities from
the patients with chronic periodontitis preferred carbohydrates
while those from the control group preferred sugar alcohols and
carboxylic acids (Table I; Fig. 1). The results may suggest that the
shift in dominant microbial flora in the saliva is correlated with
periodontal disease, and that oral pathogenic bacteria may use
simple sugars as the preferred energy sources.
The TRFLP technique is commonly used for the
evaluation of microbial community composition and has been widely
used (10,17,18). The
TRFLP patterns allow the rapid comparison of the community
composition among patients with periodontitis, and can show
host-specific relatively stable oral bacterial flora (9). The present results indicated that the
TRFLP patterns derived from patients with periodontitis were
significantly different from the patterns of periodontally healthy
subjects. The data confirm findings from previous studies, which
reported that the saliva from patients with periodontitis contains
a special bacterial diversity (9,19).
Notably, the present TRFLP results also suggested that the age of
the subjects had a significant effect on the oral bacterial
flora.
The PC1 of the TRFLP data was significantly
correlated with the number of CFUs. This finding may suggest that a
change in oral microbial biomass is accompanied by a change in oral
microbial community composition, and that certain dominant species
determine the size of the microbial community. The 542-bp TRF was
found to be the most dominant in the saliva of the healthy subjects
and the patients with periodontitis. Compared with the 16S rDNA
clone library analysis of human saliva (20), the TRF was predominantly derived from
the members of the genus Streptococcus. The analysis of
traditional isolation and identification demonstrated that
Streptococcus was the main genus.
The three methods of conventional culture, substrate
utilization pattern and TRFLP analysis were simultaneously applied
to assess the size, function and composition of the oral microbial
community of healthy subjects and patients with periodontitis. The
methods all revealed that there was a systematic change in the
microbial ecological characteristics associated with oral health
status; however, the results obtained using the respective methods
were not in perfect agreement. Bacterial counts were highly
correlated with the TRFLP pattern but not with the substrate
utilization profile. It appears that the substrate utilization
pattern provides information about differences in community
functional diversity but they are not always clearly associated
with the size of the microbial community (5). Due to the high concentrations of the
carbon sources in the MicroResp system, the microbial species that
are able to use these carbon sources will grow and reproduce
quickly, and the initial microbial biomass may not be the major
factor affecting the substrate utilization pattern (21). In the present study the TRFLP method
showed more effective discrimination of the samples than the
substrate utilization pattern. Furthermore, the TRFLP analysis was
able to give more information and exhibited a higher sensitivity
than the substrate utilization. The age of the subjects was closely
correlated with the TRFLP pattern, but there was no significant
effect of age on the substrate utilization pattern (Table II). The present results suggest that
TRFLP analysis is useful in studying the composition of the oral
microbial community and in the diagnosis of chronic periodontitis,
particularly when used in conjunction with other methods.
Acknowledgements
This study was financially supported by grants from
the National Science Natural Foundation of China (no. 81101283) and
the Educational Commission of Zhejiang Province, China (no.
20061914).
References
|
1
|
Ehrlich GD, Hu FZ, Shen K, Stoodley P and
Post JC: Bacterial plurality as a general mechanism driving
persistence in chronic infections. Clin Orthop Relat Res. 20–24.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aas JA, Paster BJ, Stokes LN, Olsen I and
Dewhirst FE: Defining the normal bacterial flora of the oral
cavity. J Clin Microbiol. 43:5721–5732. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kumar PS, Griffen AL, Moeschberger ML and
Leys EJ: Identification of candidate periodontal pathogens and
beneficial species by quantitative 16S clonal analysis. J Clin
Microbiol. 43:3944–3955. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Campbell CD, Chapman SJ, Cameron CM,
Davidson MS and Potts JM: A rapid microtiter plate method to
measure carbon dioxide evolved from carbon substrate amendments so
as to determine the physiological profiles of soil microbial
communities by using whole soil. Appl Environ Microbiol.
69:3593–3599. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yao H, He Z, Wilson MJ and Campbell CD:
Microbial biomass and community structure in a sequence of soil
with increasing fertility and changing land use. Microb Ecol.
40:223–237. 2000.PubMed/NCBI
|
|
6
|
Liu WT, Marsh TL, Cheng H and Forney LJ:
Characterization of microbial diversity by determining terminal
restriction fragment length polymorphisms of genes encoding 16S
rRNA. Appl Environ Microbiol. 63:4516–4522. 1997.PubMed/NCBI
|
|
7
|
Takeshita T, Nakano Y, Kumagai T, Yasui M,
Kamio N, Shibata Y, Shiota S and Yamashita Y: The ecological
proportion of indigenous bacterial populations in saliva is
correlated with oral health status. ISME J. 3:65–78. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torsvik V and Øvreås L: Microbial
diversity and function in soil: from genes to ecosystems. Curr Opin
Microbiol. 5:240–245. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sakamoto M, Takeuchi Y, Umeda M, Ishikawa
I and Benno Y: Application of terminal RFLP analysis to
characterize oral bacterial flora in saliva of healthy subjects and
patients with periodontitis. J Med Microbiol. 52:79–89. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Singh BK, Nazaries L, Munro S, Anderson IC
and Campbell CD: Use of multiplex terminal restriction fragment
length polymorphism for rapid and simultaneous analysis of
different components of the soil microbial community. Appl Environ
Microbiol. 72:7278–7285. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Osborne CA, Rees GN, Bernstein Y and
Janssen PH: New threshold and confidence estimates for terminal
restriction fragment length polymorphism analysis of complex
bacterial communities. Appl Environ Microbiol. 72:1270–1278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horz HP, Haaf Ten A, Kessler O, Yekta Said
S, Seyfarth I, Hettlich M, Lampert F, Küpper T and Conrads G:
T-RFLP-based differences in oral microbial communities as risk
factor for development of oral diseases under stress. Environ
Microbiol Rep. 4:390–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Loesche WJ, Syed SA, Schmidt E and
Morrison EC: Bacterial profiles of subgingival plaques in
periodontitis. J Periodontol. 56:447–456. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ximénez-Fyvie LA, Haffajee AD and
Socransky SS: Microbial composition of supra- and subgingival
plaque in subjects with adult periodontitis. J Clin Periodontol.
27:722–732. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar PS, Leys EJ, Bryk JM, Martinez FJ,
Moeschberger ML and Griffen AL: Changes in periodontal health
status are associated with bacterial community shifts as assessed
by quantitative 16S cloning and sequencing. J Clin Microbiol.
44:3665–3673. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mager DL, Ximenez-Fyvie LA, Haffajee AD
and Socransky SS: Distribution of selected bacterial species on
intraoral surfaces. J Clin Periodontol. 30:644–654. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marsh TL, Saxman P, Cole J and Tiedje J:
Terminal restriction fragment length polymorphism analysis program,
a web-based research tool for microbial community analysis. Appl
Environ Microbiol. 66:3616–3620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saito D, Marsh TL, de Souza Cannavan F,
Höfling JF and Gonçalves RB: Assessment of intraradicular bacterial
composition by terminal restriction fragment length polymorphism
analysis. Oral Microbiol Immunol. 24:369–376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hutter G, Schlagenhauf U, Valenza G, Horn
M, Burgemeister S, Claus H and Vogel U: Molecular analysis of
bacteria in periodontitis: evaluation of clone libraries, novel
phylotypes and putative pathogens. Microbiology. 149:67–75. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sakamoto M, Umeda M, Ishikawa I and Benno
Y: Comparison of the oral bacterial flora in saliva from a healthy
subject and two periodontitis patients by sequence analysis of 16S
rDNA libraries. Microbiol Immunol. 44:643–652. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bååth E, Díaz-Raviña M, Frostegård S and
Campbell CD: Effect of metal-rich sludge amendments on the soil
microbial community. Appl Environ Microbiol. 64:238–245.
1998.PubMed/NCBI
|