Introduction
Osteoblasts are derived from undifferentiated
mesenchymal stem cells and play a key role in bone formation
(1). Therefore, investigation into
the expression levels and functions of master regulators is
essential in order to reveal the mechanisms underlying osteoblast
differentiation.
CCAAT-enhancer-binding homologous protein (CHOP),
also known as growth arrest and DNA damage-inducible gene 153 and
DNA damage inducible transcript 3, is a member of the basic leucine
zipper family of transcription factors (2). CHOP regulates mRNA transcription
indirectly by interacting with various nuclear proteins, including
CCAAT/enhancer-binding protein-β, activating transcription factor,
cAMP response element-binding protein and Fos/Jun (3–5). CHOP is
implicated in apoptosis, which may be induced by cellular stress,
including endoplasmic reticulum (ER) stress (6,7).
Transgenic mice overexpressing CHOP have shown reduced trabecular
bone volume due to decreased bone formation, resulting in
osteopenia (8). Furthermore, a
previous study demonstrated that hyperglycemia induced ER
stress-dependent CHOP expression in osteoblasts (9). CHOP expression in osteoblasts obtained
from diabetic rats or cultured in a high-glucose medium is elevated
(9), suggesting that abnormal
expression of CHOP may play an important role in the pathogenesis
of osteoporosis. However, the regulatory mechanisms underlying CHOP
expression remain largely unknown.
Signal transducer and activator of transcription 3
(STAT3) is a transcription factor that promotes cell survival and
differentiation (10,11). The aim of the present study was to
investigate the effects of STAT3 activation on the expression of
CHOP protein in osteoblasts. In addition, the effect of miRNA
(miR)-205 was analyzed in order to determine its role in the
STAT3-mediated regulation of CHOP protein expression. Potential
miRNAs that were able to regulate CHOP expression were analyzed by
bioinformatic software (data not shown) and (miR)-205 was selected
for further analysis.
Materials and methods
Cell culture
MC3T3-E1 cells were obtained from the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China), and were
maintained in a modified Eagles minimal essential medium
(Invitrogen Life Technologies, Carlsbad, CA, USA) containing 100
U/ml penicillin and 100 µg/ml streptomycin (Invitrogen Life
Technologies). Mouse bone marrow mesenchymal stem cells (MSCs) were
prepared from the bone marrow of femurs and tibias harvested from
seven-week-old male C57B/L6 mice (12). Interleukin (IL)-6 was purchased from
Merck Sharpe & Dohme (Shanghai, China). The present study was
approved by the Ethics Committee of the Qingpu Branch of Zhongshan
Hospital, Fudan University (Shanghai, China).
Small interfering RNA (siRNA) oligos
and miRNA
siRNA oligos targeting green fluorescent protein
(GFP) or STAT3 were designed and synthesized by GenePharm, Inc.
(Shanghai, China). siRNA targeting GFP was used as a negative
control for STAT3 siRNA. The negative control for miR-205 mimics
and antisense was purchased from Guangzhou RiboBio Co., Ltd.
(Guangdong, China).
Plasmids, transient transfections and
luciferase assays
An miR-205 promoter (420 bp) was amplified from the
mouse genomic DNA template and inserted into a pGL3 vector (Promega
Corporation, Madison, WI, USA). A mutant STAT3 binding site was
generated using a polymerase chain reaction (PCR) mutagenesis kit
(Toyobo Co., Ltd., Osaka, Japan) with the primer,
5-GATTCAGGGACATAAAACCAATAC-3′ (mutation site, AAAA), and a reverse
complementary primer. Reporter vectors carrying the miR-205 target
site were constructed by synthesizing a 3′-untranslated region
(UTR) fragment containing the predicted target sites (miRWalk)
(13) for CHOP cDNA, and
subsequently inserting the CHOP cDNA fragment into the multiple
cloning site of a pMIR-REPORT™ luciferase miRNA expression reporter
vector (Ambion Life Technologies, Carlsbad, CA, USA). Transient
transfection was performed using Lipofectamine® 2000 (Invitrogen
Life Technologies), according to the manufacturers instructions.
For the luciferase reporter assay, MC3T3-E1 cells were seeded in
24-well plates and transfected with the indicated plasmids. Cells
were harvested 36 h after transfection. Luciferase activity was
measured using the Dual-Luciferase® Reporter Assay System (Promega
Corporation).
RNA extraction, quantitative analysis
and western blot analysis
Total RNA was isolated from the tissues or cells
with TRIzol reagent (Invitrogen Life Technologies), and reverse
transcription was performed using a Takara RNA PCR kit (Takara
Biotechnology Co., Ltd., Dalian, China), following the
manufacturers instructions. In order to quantify the transcripts of
the genes of interest, quantitative PCR was performed using SYBR
Green Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) and a Light
Cycler 480 (Roche Diagnostics, Basel, Switzerland). The primer
sequences for the CHOP gene were as follows: forward 5′-
AAGCCTGGTATGAGGATCTGC-3′ and reverse:
5′-TTCCTGGGGATGAGATATAGGTG-3′. The PCR conditions included an
initial holding period at 94°C for 5 min, followed by a two-step
PCR program of 94°C for 10 sec and 60°C for 45 sec for 45 cycles.
For western blot analysis, tissues and cells were lysed in
radioimmunoprecipitation buffer containing 50 mM Tris-HCl, 150 mM
NaCl, 5 mM MgCl2, 2 mM EDTA, 1 mM NaF, 1% NP40 and 0.1%
SDS. The antibodies used were monoclonal rabbit anti-STAT3 (D3Z2G;
1:1,000; #12640; Cell Signaling Technology, Inc., Danvers, MA,
USA), monoclonal mouse anti-CHOP (1:2,000; #ab11419; Abcam,
Cambridge, UK) and monoclonal mouse anti-GAPDH (1:5,000;
#sc-365062; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Chromatin immunoprecipitation (ChIP)
assays
ChIP assay kits were purchased from Upstate
Biotechnology, Inc. (New York, NY, USA). MC3T3-E1 cells were fixed
with formaldehyde. DNA was sheared into 200–1,000-bp fragments
using sonication. Chromatin was incubated and precipitated with the
STAT3 antibody or IgG (Santa Cruz Biotechnology, Inc.).
Statistical analysis
Values are expressed as the mean ± standard error of
the mean. Statistical analyses were performed with Graphpad
software, version 5.0 (La Jolla, CA, USA). The two-tailed Students
t-test was used to evaluate the statistical significance of the
differences between the two groups. Values of *P<0.05,
**P<0.01 or ***P<0.001 were considered to indicate a
statistically significant difference.
Results
IL-6 treatment and STAT3
overexpression downregulate CHOP protein levels
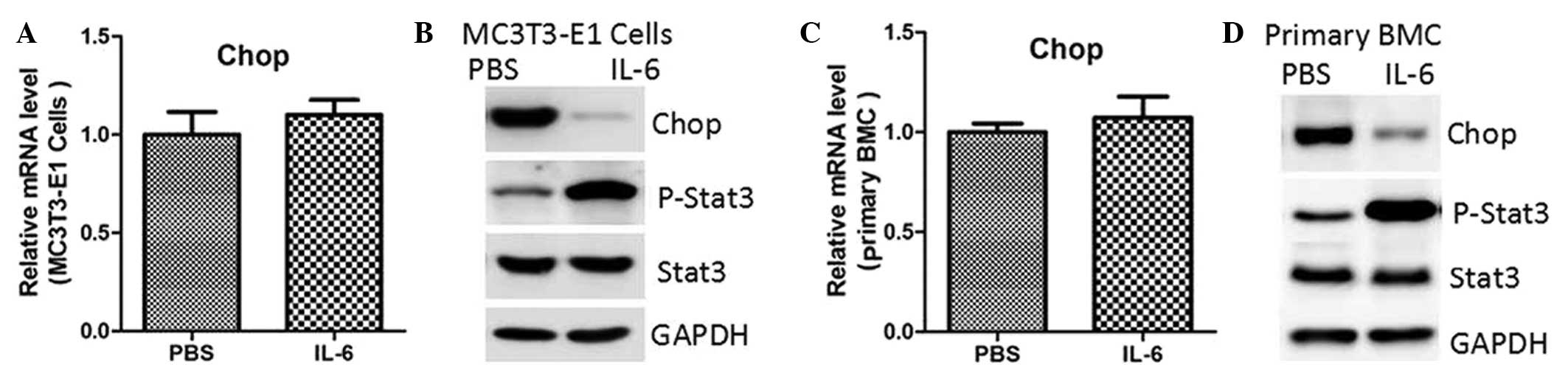
To assess whether STAT3 regulated CHOP expression,
MC3T3-E1 cells were treated with IL-6, a known STAT3 agonist. As
shown in Fig. 1A, IL-6 treatment did
not affect CHOP mRNA expression. However, the protein expression
levels were notably reduced, accompanied by enhanced
phosphorylation of STAT3 (Fig. 1B).
Similar results were observed in the primary mouse bone marrow MSCs
(Fig. 1C and D). To rule out the
potential non-specific effects of IL-6, these two cell types were
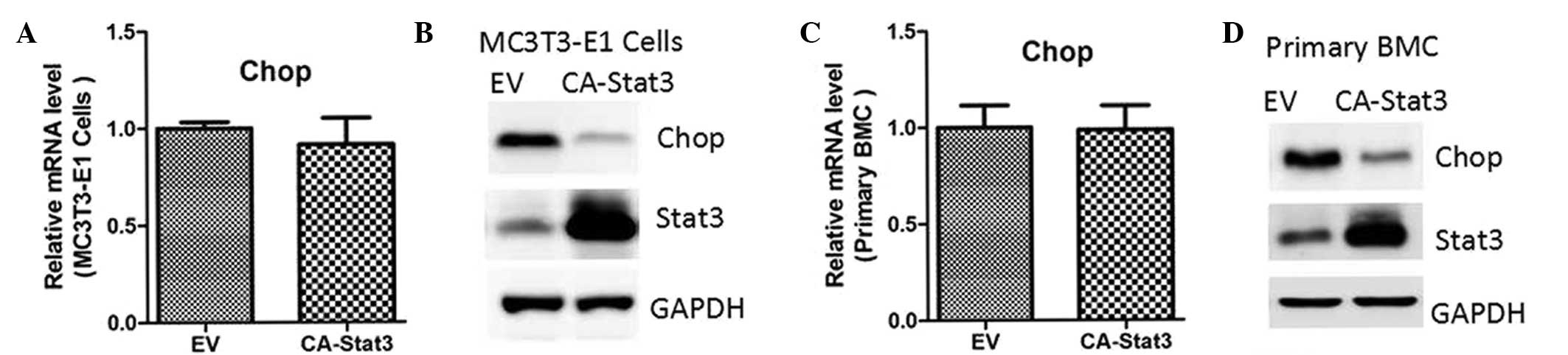
transfected with lentiviruses containing an empty vector or
constitutively-activated (CA)-STAT3 (14). The results indicated that CA-STAT3
also inhibited protein expression of CHOP, while the mRNA
expression levels remained unaffected (Fig. 2).
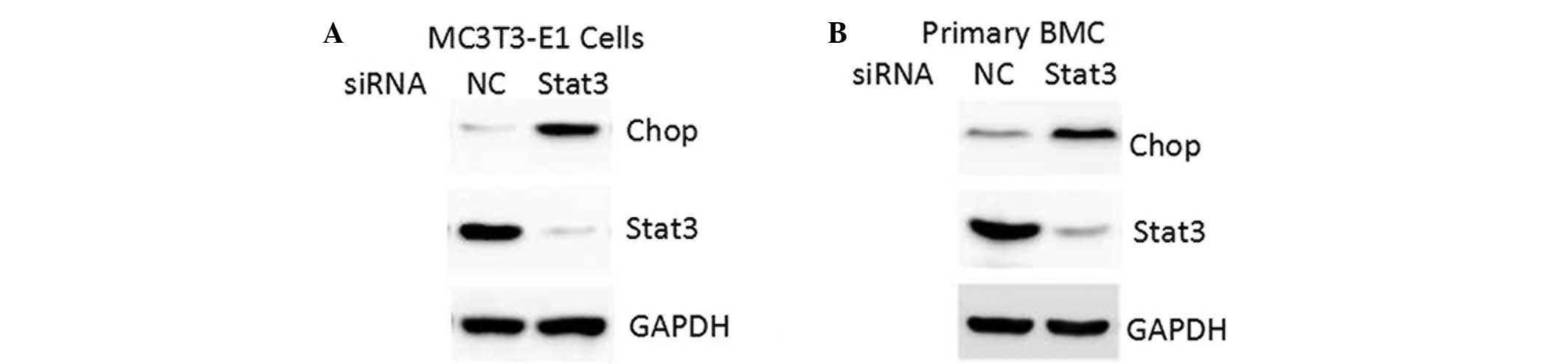
STAT3 inhibition by siRNA oligos
increases CHOP expression
Endogenous STAT3 expression was inhibited by siRNA
oligos. Knockdown of STAT3 increased CHOP protein levels in the
MC3T3-E1 and primary mouse bone marrow MSCs (Fig. 3). These results demonstrated that
STAT3 may be a negative regulator in the control of CHOP expression
in osteoblasts.
Regulation of miR-205 by STAT3
activation
STAT3 activation was found to regulate CHOP protein
expression, but not mRNA expression. Thus, it was hypothesized that
STAT3 regulates CHOP expression at a translational level. miRNAs
are known to recognize and bind to the target 3-UTR of mRNAs, which
leads to mRNA degradation or translational inhibition of the target
mRNA and downregulation of target gene expression (15,16).
Using the miRWalk algorithm based on seed recognition, a number of
miRNAs were identified that potentially interacted with the CHOP
transcript (data not shown). However, only miR-205 was notably
elevated in the MC3T3-E1 cells treated with IL-6 or transfected
with CA-STAT3 (Fig. 4A and B).
Targeted knockdown of endogenous STAT3 also reduced miR-205
expression (Fig. 4C), suggesting
that STAT3 activation increases the levels of miR-205.
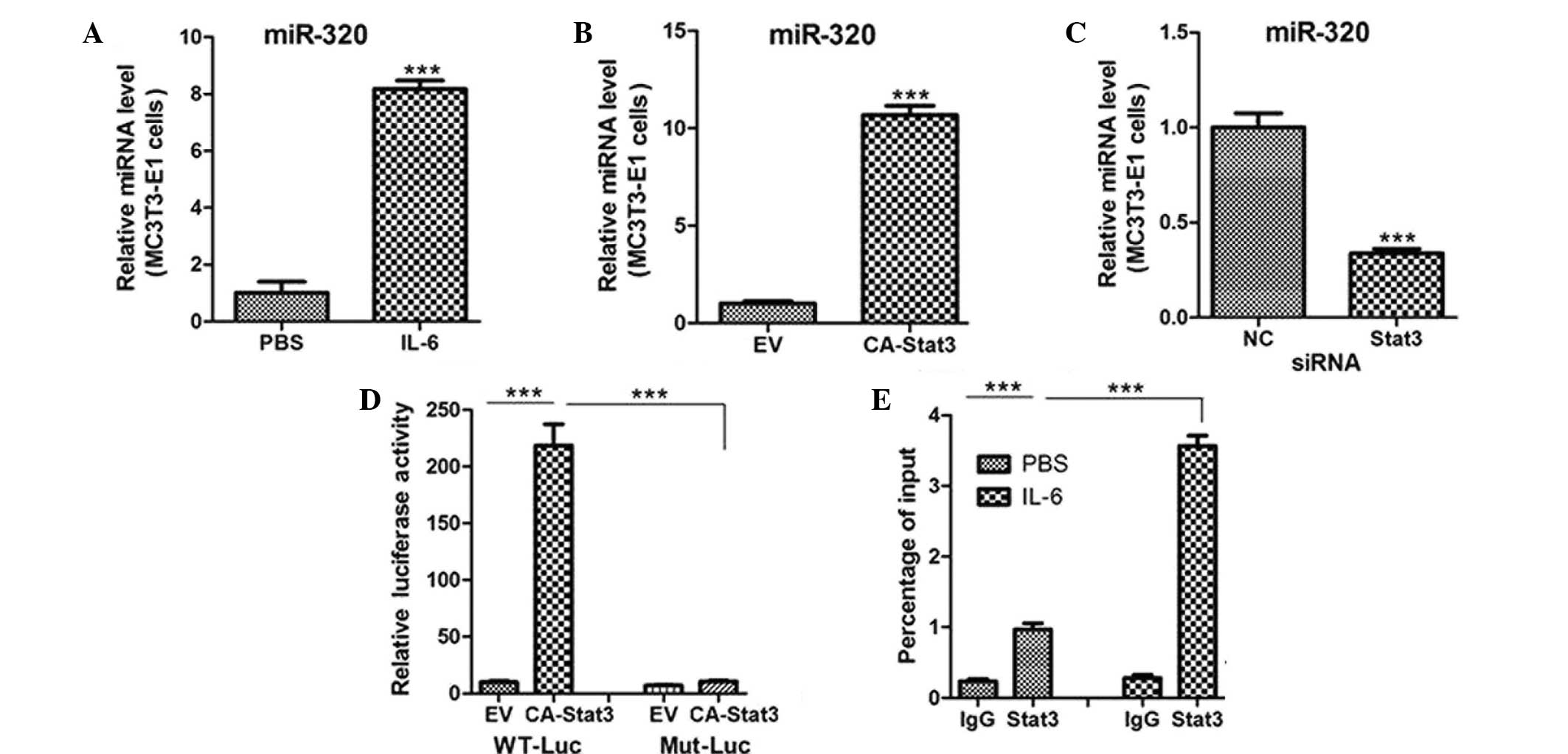 | Figure 4.STAT3 activation transcriptionally
regulates miR-205 expression. Quantitative polymerase chain
reaction (PCR) analysis of miR-205 expression in MC3T3-E1 cells
treated with (A) PBS or IL-6 (10 ng/ml) for 12 h, (B) transfected
with an EV or CA-STAT3 and (C) transfected with siRNA oligos
targeting STAT3 or NC. (D) Relative luciferase activity of MC3T3-E1
cells cotransfected with WT or Mut miR-205 promoters and EV or
CA-STAT3. (E) Quantitative PCR analysis of chromatin
immunoprecipitation assays showing STAT3 binding to the miR-205
promoter. Chromatins were prepared from miR-205 cells treated with
PBS or IL-6 (10 ng/ml). ***P<0.001. miR, microRNA; siRNA, small
interfering RNA; PBS, phosphate-buffered saline; IL-6,
interleukin-6; EV, empty vector; CA-STAT3, constitutively
active-signal transducer and activator of transcription 3; NC,
negative control; WT, wild type; Mut, mutant. |
In order to investigate whether STAT3 is a
transcriptional activator of miR-205, MC3T3-E1 cells were
transfected with a reporter vector encoding luciferase, under the
control of the miR-205 promoter. Concurrent expression of CA-STAT3
with the miR-205 reporter construct enhanced miR-205 promoter
activity (Fig. 4D), which was
inhibited by mutation of the STAT3 DNA-binding site in the miR-205
promoter (Fig. 4D). In addition,
ChIP assays were performed to assess whether STAT3 directly binds
to the miR-205 promoter. Levels of STAT3 protein bound to the
miR-205 promoter were significantly increased in the MC3T3-E1 cells
treated with IL-6 (Fig. 4E).
Therefore, the results indicated that miR-205 may be a
transcriptional target of STAT3 in osteoblasts.
miR-205 directly targets the 3-UTR of
the CHOP gene in osteoblasts
To confirm CHOP as the target gene of miR-205, a
fragment of the CHOP gene containing the binding sites for miR-205
was engineered into a luciferase reporter vector and luciferase
reporter activity assays were conducted. Transfection of miR-205
caused a substantial reduction in luciferase activity in the
luciferase expression constructs carrying the target fragment
(Fig. 5A). Furthermore, the
repressive effect of miR-205 on the CHOP 3-UTR was inhibited by
point mutations in the miR-205-binding site region of the CHOP
3-UTR (Fig. 5A).
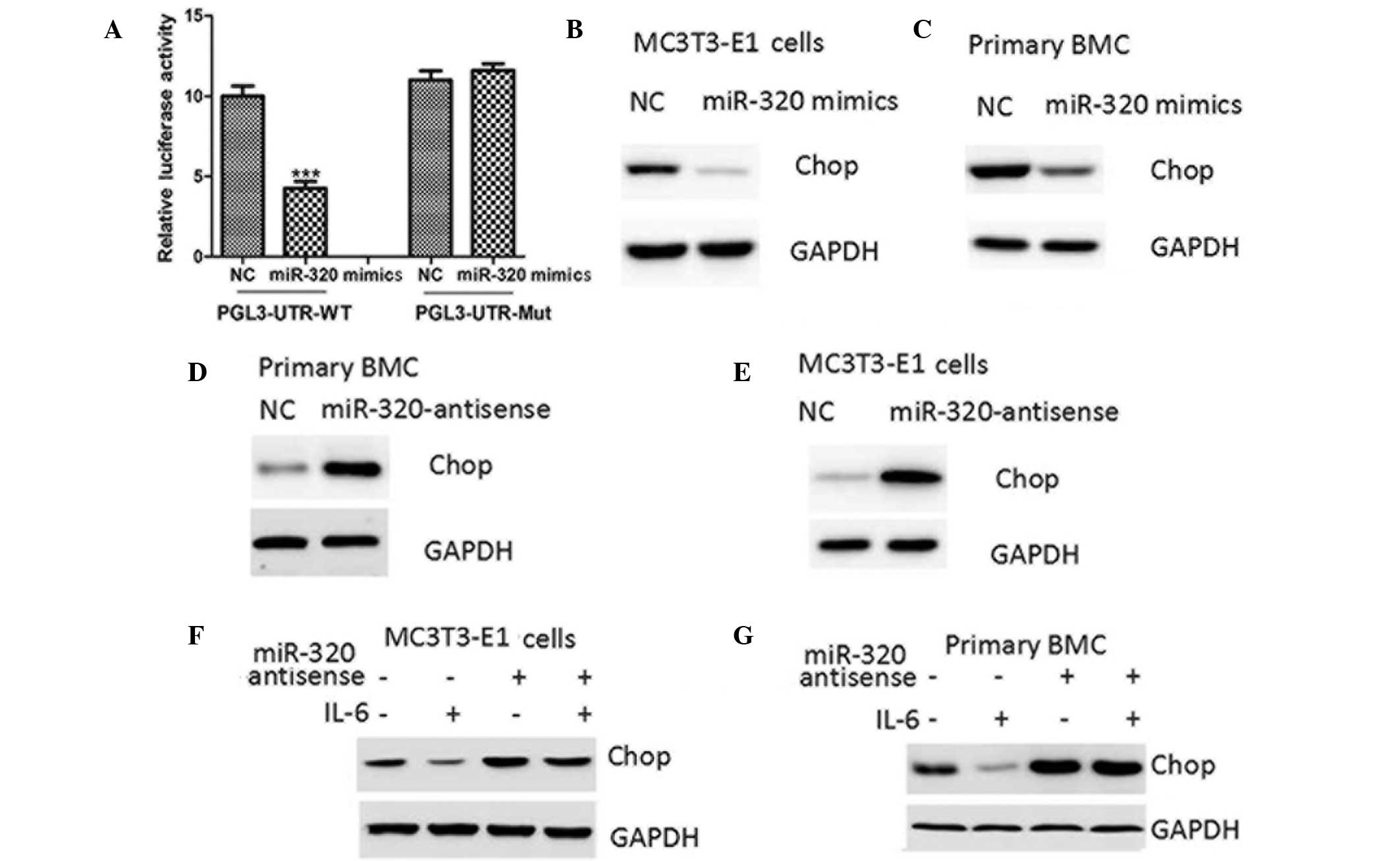 | Figure 5.miR-205 negatively regulates CHOP
protein expression through targeting its 3-UTR. (A) Relative
luciferase activity of WT or Mut CHOP 3-UTR in MC3T3-E1 cells
transfected with NC or miR-205 mimics. Western blot analysis of
CHOP protein expression in (B) MC3T3-E1 cells and (C) mouse primary
BMCs transfected with NC or miR-205 mimics, (D) MC3T3-E1 and (E)
mouse primary BMCs transfected with NC or miR-205 antisense and in
(F) MC3T3-E1 and (G) mouse primary BMCs pretransfected with NC or
miR-205 antisense for 24 h and then treated with PBS or IL-6 (10
ng/ml) for 24 h. ***P<0.001, vs. NC. NC, negative control; miR,
microRNA; UTR, untranslated region; CHOP, CCAAT-enhancer binding
homologous protein; IL-6, interleukin-6; WT, wild type; Mut,
mutant; PBS, phosphate-buffered saline; BMC, bone marrow cell. |
Ectopic expression of miR-205 decreased the protein
expression levels of CHOP in MC3T3-E1 and mouse primary bone marrow
cells (Fig. 5B and C), while the
inhibition of miR-205 was observed to upregulate CHOP protein
levels (Fig. 5D and E). Furthermore,
miR-205 antisense negated the inhibitory effect of STAT3 activation
on CHOP protein levels (Fig. 5F).
Therefore, the results suggested that STAT3 negatively regulated
CHOP protein expression through the upregulation of miR-205 in
osteoblasts.
Discussion
In the present study, STAT3 was shown to repress
CHOP protein expression in osteoblasts. At a molecular level,
miR-205 was found to be a transcriptional target of STAT3. miR-205
directly targeted the CHOP 3-UTR, suggesting that CHOP is regulated
by miR-205 in osteoblasts. Collectively, the present observations
indicate that activation of STAT3 may inhibit CHOP expression
through upregulation of miR-205. STAT3 activation has been reported
to promote osteoblast formation and protect against ethanol-induced
bone loss (17,18). Therefore, the present study proposes
a novel mechanism for the protective roles of STAT3 in
osteoblasts.
A previous study showed that miR-205 negatively
regulates androgen receptors and is inversely correlated to the
occurrence of metastases and shortened overall survival in prostate
cancer patients (19). However,
miR-205 expression is significantly upregulated in human
endometrial endometrioid carcinoma tissues, which promotes tumor
proliferation and invasion by targeting estrogen-related receptors
(20). Therefore, miR-205 may
function as a tumor suppressor or as an oncogene, depending on the
cellular context. Furthermore, miR-205 is expressed in a
lineage-related pattern in mesenchymal cell types and is regulated
by runt-related transcription factor-2, a master regulator of
osteoblast differentiation (21).
Therefore, the function of miR-205 in osteoblasts and bone
formation should be investigated further in future studies.
Timofeeva et al (22) demonstrated that STAT3 suppresses mRNA
transcription of the CHOP gene (22). The authors demonstrated that the
STAT3-binding region of the CHOP promoter was localized in the
DNase I hypersensitive site of chromatin in cancer cells, but not
in non-transformed cells, indicating that STAT3 binding and
suppressive action may be chromatin structure-dependent (22). In accordance with these observations,
the regulation of CHOP by STAT3 has been hypothesized to be cell-
or tissue-specific.
In summary, the current study demonstrated that
STAT3 activation reduces CHOP protein levels in osteoblasts, which
is partly dependent on the upregulation of miR-205. Further
investigation into this osteoblast signaling pathway may aid the
understanding of the pathogenic mechanisms underlying
osteoblast-related diseases, including osteoporosis.
Acknowledgements
This study was supported by grants from the Shanghai
Key Medical Specialty Construction, Orthopedic Trauma (no. ZK
2012A36) and the Leading Discipline Program of the Qingpu Branch of
Zhongshan Hospital, Fudan University, (no. WL 2011-01).
References
|
1
|
DiGirolamo DJ, Clemens TL and Kousteni S:
The skeleton as an endocrine organ. Nat Rev Rheumatol. 8:674–683.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramji DP and Foka P:
CCAAT/enhancer-binding proteins: structure, function and
regulation. Biochem J. 365:561–575. 2002.PubMed/NCBI
|
|
3
|
Tabas I and Ron D: Integrating the
mechanisms of apoptosis induced by endoplasmic reticulum stress.
Nat Cell Biol. 13:184–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malhi H and Kaufman RJ: Endoplasmic
reticulum stress in liver disease. J Hepatol. 54:795–809. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scull CM and Tabas I: Mechanisms of ER
stress-induced apoptosis in atherosclerosis. Arterioscler Thromb
Vasc Biol. 31:2792–2797. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schönthal AH: Pharmacological targeting of
endoplasmic reticulum stress signaling in cancer. Biochem
Pharmacol. 85:653–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eizirik DL, Miani M and Cardozo AK:
Signalling danger: endoplasmic reticulum stress and the unfolded
protein response in pancreatic islet inflammation. Diabetologia.
56:234–241. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pereira RC, Stadmeyer LE, Smith DL,
Rydziel S and Canalis E: CCAAT/Enhancer-binding protein homologous
protein (CHOP) decreases bone formation and causes osteopenia.
Bone. 40:619–626. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu W, Zhu X, Wang Q and Wang L:
Hyperglycemia induces endoplasmic reticulum stress-dependent CHOP
expression in osteoblasts. Exp Ther Med. 5:1289–1292.
2013.PubMed/NCBI
|
|
10
|
Taub R: Liver regeneration: from myth to
mechanism. Nat Rev Mol Cell Biol. 5:836–847. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu H, Pardol D and Jove R: STATs in cancer
inflammation and immunity: a leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edgar CM, Chakravarthy V, Barnes G, et al:
Autogenous regulation of a network of bone morphogenetic proteins
(BMPs) mediates the osteogenic differentiation in murine marrow
stromal cells. Bone. 40:1389–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
miRWalk, . The database on predicted and
validated microRNA targets. http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/Accessed.
March 20–2012
|
|
14
|
Lee H, Herrmann A, Deng JH, et al:
Persistently activated Stat3 maintains constitutive NF-kappaB
activity in tumors. Cancer Cell. 15:283–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nicolaidou V, Wong MM, Redpath AN, et al:
Monocytes induce STAT3 activation in human mesenchymal stem cells
to promote osteoblast formation. PLoS One. 7:e398712012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JR, Shankar K, Nagarajan S, Badger TM
and Ronis MJ: Protective effects of estradiol on ethanol-induced
bone loss involve inhibition of reactive oxygen species generation
in osteoblasts and downstream activation of the extracellular
signal-regulated kinase/signal transducer and activator of
transcription 3/receptor activator of nuclear factor-kappaB ligand
signaling cascade. J Pharmacol Exp Ther. 324:50–59. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hagman Z, Haflidadóttir BS, et al: miR-205
negatively regulates the androgen receptor and is associated with
adverse outcome of prostate cancer patients. Br J Cancer.
108:1668–1676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su N, Qiu H, Chen Y, Yang T, Yan Q and Wan
X: miR-205 promotes tumor proliferation and invasion through
targeting ESRRG in endometrial carcinoma. Oncol Rep. 29:2297–2302.
2013.PubMed/NCBI
|
|
21
|
Zhang Y, Xie RL, Croce CM, et al: A
program of microRNAs controls osteogenic lineage progression by
targeting transcription factor Runx2. Proc Natl Acad Sci USA.
108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Timofeeva OA, Tarasova NI, Zhang X, et al:
STAT3 suppresses transcription of proapoptotic genes in cancer
cells with the involvement of its N-terminal domain. Proc Natl Acad
Sci USA. 110:1267–1272. 2013. View Article : Google Scholar : PubMed/NCBI
|