Introduction
Drug resistance is a phenomenon that has received
serious attention in recent years in everyday medical practice, and
this may also be described as responsiveness or non-responsiveness
as patients respond partially to medical treatment or have no
response at all (1). Cardiovascular
diseases (CVDs) are the most common cause of mortality globally
(2). An estimated 17.3 million
people succumbed to CVDs in 2008, representing 30% of all global
mortalities (2). It is estimated
that 7.3 million of these mortalities were due to coronary heart
disease and 6.2 million were due to stroke (3). Low and middle income countries are
highly affected, as >80% of CVD mortalities occur in such
countries, with almost equal incidence in men and women (2). Clopidogrel is an antiplatelet drug that
is administered in the form of a prodrug to cardiac patients with
acute coronary syndrome (ACS) and patients undergoing percutaneous
coronary intervention (PCI). It has been reported that clopidogrel
non-responsiveness varies between 5 and 44% among different
populations; therefore, clopidogrel and aspirin are recommended in
combination as the two therapies inhibit the aggregation of
platelets via different mechanisms (4). As a monotherapy, clopidogrel does not
effectively inhibit platelet aggregation; therefore, it is
preferentially used with aspirin, which provides additive
advantages in the reduction of atherothrombotic events as compared
with either drug alone (5).
The non-responsiveness to clopidogrel in cardiac
patients of different populations is due to genetic variations in
the cytochrome P450 (CYP) gene (6,7). During
its action, clopidogrel is converted into its active metabolite
(via 2-oxo-clopidogrel) by hepatic CYP isoenzymes in the liver.
Clopidogrel functions as an inhibitor of adenosine
diphosphate-induced platelet activation and aggregation by blocking
the P2Y12 receptor selectively and irreversibly (1). The pharmacokinetics and
pharmacodynamics of clopidogrel are modulated by genetic
polymorphism of CYP2C19 in healthy volunteers and in patients
(6,8–11). In
comparison with subjects without a CYP2C19 variant allele, subjects
carrying one or two CYP2C19 loss-of-function alleles have been
found to have lower plasma concentrations of the active metabolite
of clopidogrel and a reduction in the antiplatelet effect of
clopidogrel as evaluated by ex vivo platelet aggregation
tests (8). These CYP isoforms are
involved in the inter-individual response variability (6,7). In the
response to clopidogrel therapy, polymorphisms in CYP2C19 are
considered to affect both steps of the hepatic metabolism of
clopidogrel. Carriers of at least one ‘poor metabolizer allele’ of
CYP2C19 (either *2 or *3) have lower levels of the active
metabolite of clopidogrel and have reduced platelet inhibition
(12). Furthermore, the significant
inter-ethnic variability in the allelic frequencies of CYP2C19*2
has been associated with differential clopidogrel resistance
(13). The significance of screening
for the loss of function allelic variants of CYP2C19*2 and their
association with clopidogrel resistance has been discussed in
detail in a review (14). Such
mutations in this variant allele are responsible for the inability
of the CYP enzyme to convert clopidogrel into its active
metabolite, which results in the increased risk of mortality, heart
attack or stroke among patients who have undergone PCI. Thus, the
genotyping of allelic variant CYP2C19*2 is recommended as a
suitable and affordable strategy for the identification of patients
at risk of thrombosis (15).
In the present study, the aim was to determine the
association of genetic polymorphism of the CYP2C19 isoform
CYP2C19*2 with clopidogrel resistance in Pakistani cardiac
patients. A total of 100 cardiac patients with PCI or ACS who were
on clopidogrel therapy were analyzed for the CYP2C19*2 variant
using an allele-specific primer extension polymerase chain reaction
(PCR) technique. The allele frequency distribution of CYP2C19*2
variants was determined. Furthermore, the gel-based single
nucleotide polymorphism (SNP) identification technique was
validated using Sanger sequencing of CYP2C19*2 variant
amplicons.
Materials and methods
Methodology
Blood samples of 100 cardiac patients on clopidogrel
therapy were collected in K3 ethylenediamine tetraacetic acid
(EDTA) vials from the Punjab Institute of Cardiology and Mayo
Hospital (both Lahore, Pakistan). The samples were processed for
molecular analysis. The present study was approved by the ethical
committee of the Department of Zoology, University of Punjab
(Lahore, Pakistan). Informed consent was obtained from either the
patients or the patients' families.
Molecular analysis
Extraction of DNA
The DNA was extracted using a Genomic DNA Extraction
kit (Invitrogen Life Technologies, Carlsbad, CA, USA). The quality
of DNA was determined by agarose gel electrophoresis. The DNA was
quantified using a spectrophotometer (NanoDrop™ 2000; Thermo
Scientific, Wilmington, DE, USA).
Design of primers
The allele-specific and amplification primers for
allelic variants of CYP2C19*2 were designed using Primer 3
(http://frodo.wi.mit.edu) and their sequences are
shown in Table I. Genomic DNA
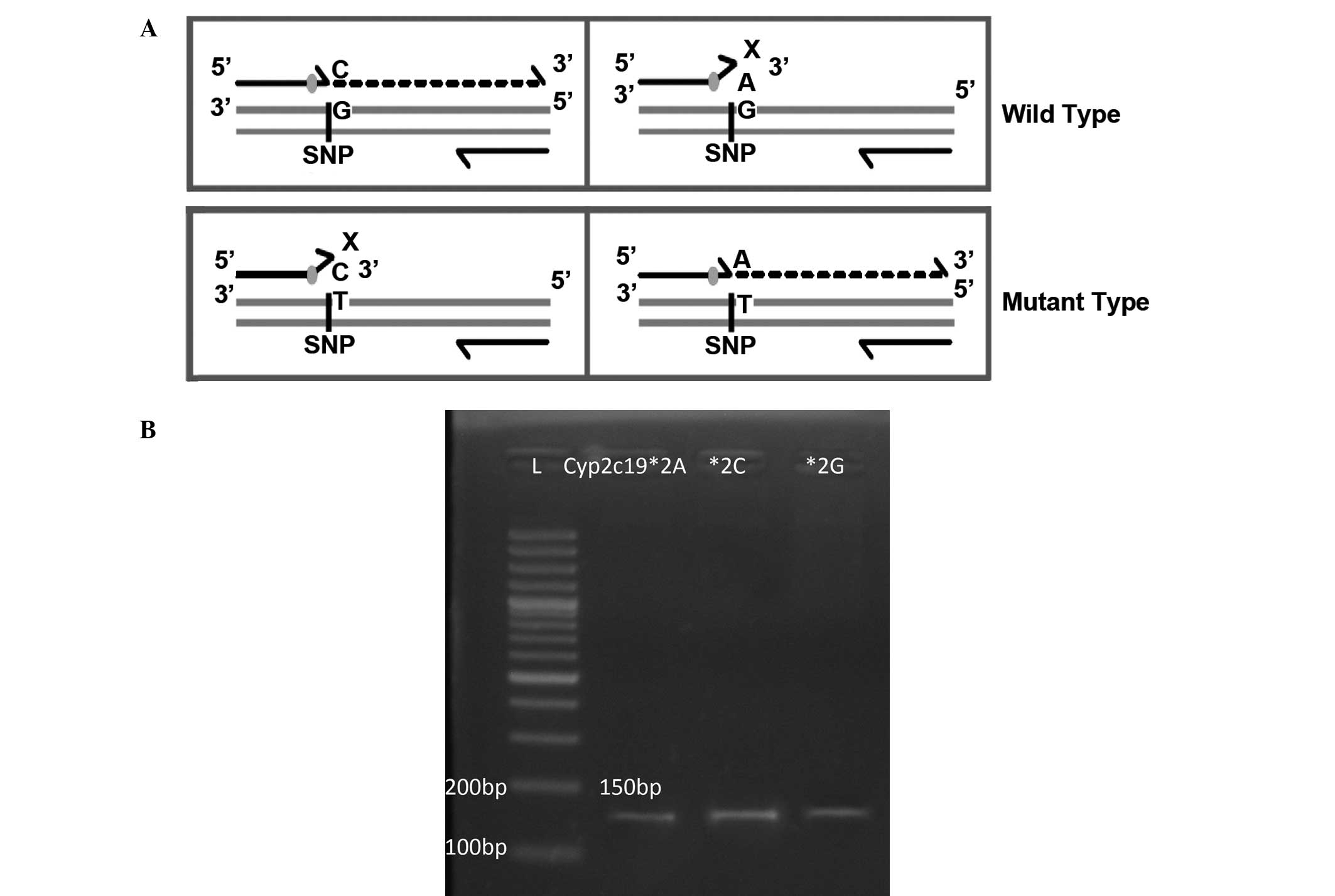
flanking the SNP was amplified with allele-specific primers. Three
different pairs of primers were used for SNP amplification, one
with wild-type allele-specific primer and the other two with mutant
allele-specific primer; the reverse primer was non-allele-specific
and identical in wild and mutant genotypes (Fig. 1A). The strategy for the design of
allele-specific primers followed that of Hirotsu et al
(16).
 | Table I.Allele specific primer sequences for
CYP2C19*2. |
Table I.
Allele specific primer sequences for
CYP2C19*2.
| CYP2C19 allele | Name of primer | Sequence of
primer | Annealing temperature
(°C) |
|---|
| CYP2C19*2 | A forward |
5′-CCACTATCATTGATTATTTCCCA-3′ | 53 |
|
| C forward |
5′-CCACTATCATTGATTATTTCCCC-3′ | 55 |
|
| G forward |
5′-CCACTATCATTGATTATTTCCCG-3′ | 55 |
|
| Universal
reverse |
5′-TAAAGTCCCGAGGGTTGTTG-3′ | 58 |
PCR amplification
PCR was performed in a 20-µl reaction volume
containing 10 ng genomic DNA, 0.4 pM of each oligonucleotide
primer, 1X PCR Buffer (Fermentas, Thermo Fisher Scientific,
Waltham, MA, USA), 200 µM dNTPs (Fermentas), 2 mM MgCl2
and 2 U Taq Polymerase (Fermentas). The following PCR
cycling conditions were used: 5 min at 95°C for 1 cycle, 32 cycles
at 95°C for 30 sec, with various annealing temperatures (as given
in Table I) for 30 sec and 72°C for
30 sec, followed by 1 cycle at 72°C for 5 min. PCR was carried out
using a 2720 Thermal Cycler (Applied Biosystems Life Technologies,
Foster City, CA, USA).
Gel electrophoresis
Amplified SNP products were electrophoresed on 2%
agarose gel stained with ethidium bromide (EtBr) and visualized
with a UV transilluminator (Benchtop 3UV Transilluminator; UVP,
LLC, Upland, CA, USA). The CYP2C19 variant CYP2C19*2 was genotyped
by the gel-based genotyping method (Fig.
1B).
Sanger sequencing
In order to validate the gel-based method of SNP
identification, Sanger sequencing of the purified PCR products of
selected samples was performed to confirm the different allelic
variants of CYP2C19*2. Sequencing of the purified products using
reverse primers was conducted with the BigDye® Terminator v3.1
Cycle Sequencing kit according to the instructions of the
manufacturer (Applied Biosystems, Foster City, CA, USA). The
amplification consisted of pre-denaturation at 96°C for 1 min,
followed by 35 cycles of denaturing at 96°C for 15 sec, annealing
at 55°C for 15 sec, and extension at 72°C for 1 min, and a final
extension at 72°C for 5 min. Sequencing products were resuspended
in 10 µl formamide and denatured at 95°C for 5 min. The DNA
sequencing was performed on an ABI PRISM 3100 Genetic Analyzer
(Applied Biosystems). The sequencing results were assembled using
ABI PRISM sequencing analysis software version 3.7 (Applied
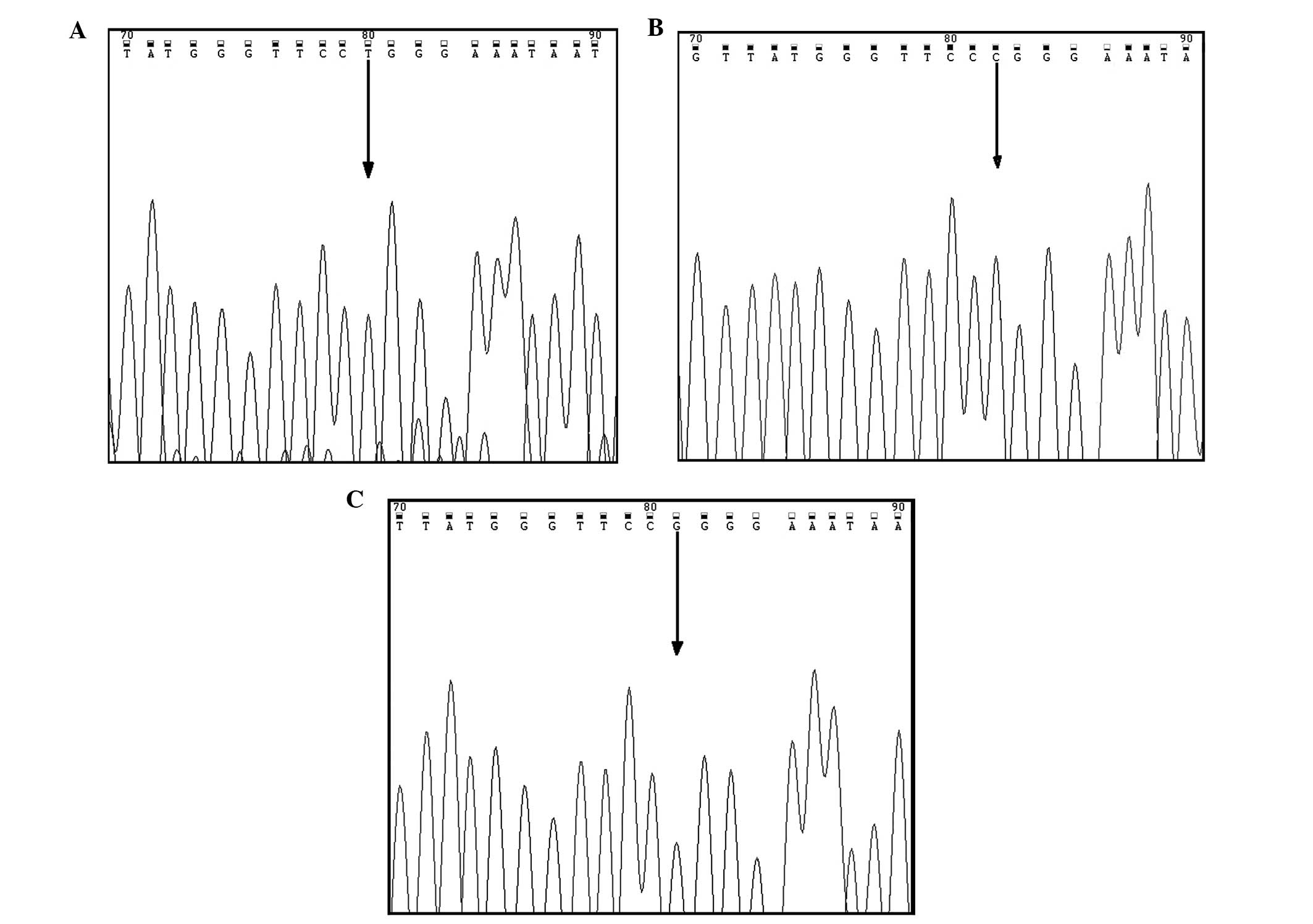
Biosystems) and analyzed with Chromas software (http://www.technelysium.com.au/chromas.html). The
chromatograms of different SNP variants using reverse primers for
sequencing are presented in Fig. 2A
(CYP2C19*2A), Fig. 2B (CYP2C19*2G)
and Fig. 2C (CYP2C19*2C).
Results and Discussion
The present study screened Pakistani cardiac
patients enrolled for clopidogrel therapy for loss-of-function
alleles of CYP2C19*2, which have been observed to be involved in
clopidogrel resistance in numerous ethnic groups (17–20). A
total of 100 cardiac patients on clopidogrel therapy were analyzed
by an allele-specific extension-based SNP identification method
with gel electrophoresis in this study. The amplified PCR products
for three different allelic variants were confirmed by the Sanger
sequencing method. The results of sequencing chromatograms
demonstrated 100% concordance with the results of the gel
electrophoresis method in the selected samples. The allele
frequency distribution of CYP2C19*2 variants was calculated in the
Pakistani cardiac patients. For the first time, the present study
screened out the C variant of the CYP2C19*2 allele in a Pakistani
population; this variant, to the best of our knowledge has not been
reported before in any other population. Among the 100 cardiac
patients analyzed in this study, 18% were heterozygous for
CYP2C19*2 A/C/G variants, 35% were heterozygous for A/G variants,
13% were heterozygous for C/G variants, 6% were heterozygous for
A/C variants, 7% were homozygous for A variant, 5% were homozygous
for C variant and 16% were homozygous for G wild-type (Table II). The complete list of allelic
variants of CYP2C19*2 found in the Pakistani cardiac patients is
shown in Table III.
 | Table II.Comparison of the allele frequency
distribution (%) of variants of CYP2C19*2 (SNP accession number,
rs4244285) associated with clopidogrel resistance among different
populations. |
Table II.
Comparison of the allele frequency
distribution (%) of variants of CYP2C19*2 (SNP accession number,
rs4244285) associated with clopidogrel resistance among different
populations.
|
| Population
(ref.) |
|---|
|
|
|
|---|
| Allelic variant of
CYP2C19*2 | Pakistani | Tunisian (15) | Thai (21) | Malaysian (13) | Indian (13) | Chinese (13) | Caucasian (13) |
|---|
| A | 7 | – | 7.32 | 9 | 13 | 6 | 2–3 |
| A/G | 35 | 11.5 | 5.61 | – | – | – | – |
| C | 5 | – | – | – | – | – | – |
| A/C | 6 | – | – | – | – | – | – |
| C/G | 13 | – | – | – | – | – | – |
| A/C/G | 18 | – | – | – | – | – | – |
 | Table III.Complete list of allelic variants of
CYP2C19*2 (SNP accession number, rs4244285) in Pakistani cardiac
patients. |
Table III.
Complete list of allelic variants of
CYP2C19*2 (SNP accession number, rs4244285) in Pakistani cardiac
patients.
| Sample ID | Allele A | Allele C | Allele G | Genotype | Sample ID | Allele A | Allele C | Allele G | Genotype |
|---|
| 1 | A |
| G | A/G | 51 | A |
| G | A/G |
| 2 |
|
| G | C/G | 52 | A |
|
| A/A |
| 3 |
|
| G | G/G | 53 | A |
| G | A/G |
| 4 |
|
| G | G/G | 54 | A |
|
| A/A |
| 5 | A | C | G | A/C/G | 55 | A | C |
| A/C |
| 6 | A |
| G | A/G | 56 | A |
| G | A/G |
| 7 |
|
| G | G/G | 57 |
| C | G | C/G |
| 8 |
| C | G | C/G | 58 | A |
| G | A/G |
| 9 | A |
|
| A/A | 59 | A |
| G | A/G |
| 10 | A |
| G | A/G | 60 |
| C |
| C/C |
| 11 | A |
| G | A/G | 61 |
| C | G | C/G |
| 12 |
| C | G | C/G | 62 | A | C | G | A/C/G |
| 13 | A | C | G | A/C/G | 63 | A |
| G | A/G |
| 14 | A |
| G | A/G | 64 | A |
| G | A/G |
| 15 |
|
| G | G/G | 65 | A |
|
| A/A |
| 16 |
| C | G | C/G | 66 | A | C | G | A/C/G |
| 17 |
| C | G | C/G | 67 | A |
| G | A/G |
| 18 |
|
| G | G/G | 68 | A | C | G | A/C/G |
| 19 | A |
|
| A/A | 69 | A | C |
| A/C |
| 20 | A | C | G | A/C/G | 70 |
| C | G | C/G |
| 21 |
| C |
| C/C | 71 | A | C |
| A/C |
| 22 | A |
| G | A/G | 72 |
|
| G | G/G |
| 23 |
| C | G | C/G | 73 |
|
| G | G/G |
| 24 |
|
| G | G/G | 74 | A | C | G | A/C/G |
| 25 | A |
| G | A/G | 75 | A |
| G | A/G |
| 26 | A | C |
| A/C | 76 |
| C |
| C/C |
| 27 | A |
| G | A/G | 77 | A | C | G | A/C/G |
| 28 | A |
|
| A/A | 78 |
|
| G | G/G |
| 29 | A |
| G | A/G | 79 |
|
| G | G/G |
| 30 | A | C | G | A/C/G | 80 |
| C | G | C/G |
| 31 | A |
| G | A/G | 81 | A |
| G | A/G |
| 32 | A |
|
| A/A | 82 |
| C | G | C/G |
| 33 | A |
| G | A/G | 83 | A |
| G | A/G |
| 34 |
| C | G | C/G | 84 |
|
| G | G/G |
| 35 | A |
| G | A/G | 85 |
| C |
| C/C |
| 36 | A |
| G | A/G | 86 |
|
| G | G/G |
| 37 | A | C | G | A/C/G | 87 | A |
| G | A/G |
| 38 | A |
| G | A/G | 88 | A | C | G | A/C/G |
| 39 |
|
| G | G/G | 89 | A |
| G | A/G |
| 40 | A |
| G | A/G | 90 |
|
| G | G/G |
| 41 | A | C |
| A/C | 91 | A | C |
| A/C |
| 42 | A |
| G | A/G | 92 | A | C | G | A/C/G |
| 43 |
|
| G | G/G | 93 | A |
| G | A/G |
| 44 | A |
| G | A/G | 94 |
| C |
| C/C |
| 45 | A | C | G | A/C/G | 95 |
| C | G | C/G |
| 46 | A |
| G | A/G | 96 | A | C | G | A/C/G |
| 47 | A |
| G | A/G | 97 | A | C | G | A/C/G |
| 48 | A |
| G | A/G | 98 | A | C | G | A/C/G |
| 49 | A | C | G | A/C/G | 99 | A | C | G | A/C/G |
| 50 | A |
| G | A/G | 100 |
|
| G | G/G |
Tri-allelic SNPs in the CYP2C19*2 allele (CYP2C19*2
A/C/G SNPs) were identified for the first time, to the best of our
knowledge, in Pakistani cardiac patients with an incidence of 18%.
There is no evidence of tri-allelic variants in any other
populations, including the Asian population, in the literature. The
association of bi-allelic variants has been observed in many other
populations (13,15,20,21).
However, the present study observed a significant frequency (42%)
of new allelic variant C, both in homozygous and heterozygous
forms, in Pakistani cardiac patients. The allelic variant CYP2C19*2
is secondary to G<A and G<C nucleotide substitution at
position 681 at the junction of intron 4 and exon 5 with loss of
function as confirmed by the Sanger sequencing method (Fig. 2). The mechanisms responsible for
inducing mutations on the two strands of DNA duplex can be easily
understood if it is considered that if a base mismatch is present,
it may create instability. Thus, following the mutation of a G-C
base pair to G-A, further mutation to C-A may occur; if DNA
replication reads through this mismatch, the G allele will have
mutated to both C and T. Alternatively, mutations may occur
concurrently across the two strands of the duplex, due to a
chemical or radiation event, for example. Although bi-allelic SNPs
have a reasonably low density in the human genome, some tri-allelic
and tetra-allelic sites are present in the human population
(22). There are considered to be
approximately twice as many tri-allelic sites than are expected by
chance (23,24). Three different mutational mechanisms
that are reported to be responsible for generating such an excess
of tri-allelic sites are: i) Hyper-mutable regions in DNA; ii) the
simultaneous generation of two of the alleles at a tri-allelic site
within a single individual; and iii) subsequent mutations induced
by a single SNP by the process of base mismatching in heteroduplex
DNA during recombination (23,24).
Certain sites may be hypermutable, and an elevation of the mutation
rate of at least two pathways at such sites will result in an
excess of tri-allelic sites. The mutation rate of a site depends
upon the nucleotides adjacent to it; the best known example is the
CpG dinucleotide at which transition and transversion mutations
occur at increased frequency (23,24).
Other adjacent nucleotides are also known to affect the mutation
rate (25–27). However, the role of this new allelic
variant C of CYP2C19*2 in clopidogrel resistance remains to be
determined.
There is significant inter-ethnic variability in the
allelic frequencies of CYP2C19*2. As shown in the present study,
the prevalence of bi-allelic heterozygous CYP2C19*2 variants was
very high (54%) compared with that in other studied populations
such as Tunisian (11.5%) (15) and
Thai populations (5.61%) (21). The
prevalence of the bi-allelic homozygous CYP2C19*2A variant was 7%
in the population of the present study compared with 2–3% in
Caucasian, 6% in Chinese, 7.32% in Thai, 9% in Malaysian and 13% in
Indian populations (13).
Furthermore, the prevalence of a bi-allelic homozygous CYP2C19*2C
variant was observed for the first time with a frequency of 5% in
the present study population; this has not been found in any other
population. Such differences in the frequencies of CYP2C19*2
variants are responsible for the differential dose response in
cardiac patients during treatment. Therefore, the present study is
further supports that the genotyping of allelic variant CYP2C19*2
as a more suitable and affordable strategy for identifying patients
at risk of thrombosis than repeatedly performing platelet
monitoring (15).
Despite providing informative genotyping data of
CYP2C19*2 variants in a Pakistani population, there were several
limitations to the present study. Only patients admitted to
hospital were enrolled in the study. Therefore, some selection bias
is likely. The small sample size of patients is insufficient to
provide significant allele frequency distribution of CYP2C19*2
variants. A large-scale study is required to strengthen the present
findings regarding the allele frequency distribution of CYP2C19*2
variants that contribute as a risk factor for stent thrombosis. It
has been demonstrated in previous studies that CYP2C19*2
polymorphisms inhibit the antiplatelet effect of clopidogrel
(28,29). However, platelet aggregation was not
assessed in the present study population. Therefore, it is
suggested that it is necessary to investigate the role of the newly
reported allelic variant CYP2C19*2C in the inhibition of the
antiplatelet response to clopidogrel.
In conclusion, allelic variants of CYP2C19*2 have
been screened out for the first time, to the best of our knowledge,
and new variant CYP2C19*2C has been identified in Pakistani cardiac
patients with ACS and PCI. Due to the new variant C, tri-allelic
SNPs in CYP2C19*2 allele (CYP2C19*2 A/C/G SNPs) were observed in
Pakistani cardiac patients, in contrast with observations in other
studied populations. Significant inter-ethnic variability has been
observed in the allelic frequencies of CYP2C19*2, providing
evidence that genetic polymorphism of CYP2C19*2 is important in the
response to clopidogrel. Hence, the genetic screening of CYP2C19*2
allele variants is emphasized as a more suitable tool for selecting
the appropriate anti-platelet agent to treat a cardiac patient, and
would be a step towards personalized medicine. However, further
studies are required to investigate other likely factors involved
in clopidogrel resistance, and there is also a requirement for a
larger study to better assess the role of genotyping in the
evaluation of the phenomenon of clopidogrel resistance.
Acknowledgements
The authors thank Punjab University Research
Committee 2012/2013 (University of the Punjab, Lahore-Pakistan) for
providing financial assistance. The laboratory facility provided by
Vital Molecular Diagnostic Laboratory (Lahore, Pakistan) and the
sequencing facility provided by the Center for Applied Molecular
Biology (CAMB), Lahore are also highly acknowledged.
References
|
1
|
Ding Z, Kim S, Dorsam RT, Jin J and
Kunapuli SP: Inactivation of the human P2Y12 receptor by thiol
reagents requires interaction with both extracellular cysteine
residues, Cys17 and Cys270. Blood. 101:3908–3914. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alwan A: Global status report on
noncommunicable diseases 2010. World Health Organization; Geneva:
2010
|
|
3
|
Mendis S, Puska P and Norrving B: Global
Atlas on cardiovascular disease prevention and control. World
Health Organization in collaboration with the World Heart
Federation and the World Stroke Organization; Geneva: 2011
|
|
4
|
Angiolillo DJ, Fernandez-Ortiz A, Bernardo
E, et al: Identification of low responders to a 300-mg clopidogrel
loading dose in patients undergoing coronary stenting. Thrombosis
Res. 115:101–108. 2005. View Article : Google Scholar
|
|
5
|
Antman EM, Hand M, Armstrong PW, et al:
2007 focused update of the ACC/AHA 2004 guidelines for the
management of patients with ST-elevation myocardial infarction: a
report of the American College of Cardiology/American Heart
Association Task Force on Practice Guidelines: developed in
collaboration with the Canadian Cardiovascular Society endorsed by
the American Academy of Family Physicians: 2007 Writing Group to
review new evidence and update the ACC/AHA 2004 guidelines for the
management of patients with ST-elevation myocardial infarction,
writing on behalf of the 2004 Writing Committee. Circulation.
117:296–329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hulot JS, Bura A, Villard E, et al:
Cytochrome P450 2C19 loss-of-function polymorphism is a major
determinant of clopidogrel responsiveness in healthy subjects.
Blood. 108:2244–2247. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sibbing D, Stegherr J, Latz W, et al:
Cytochrome P450 2C19 loss-of-function polymorphism and stent
thrombosis following percutaneous coronary intervention. Eur Heart
J. 30:916–922. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brandt JT, Close SL, Iturria SJ, et al:
Common polymorphisms of CYP2C19 and CYP2C9 affect the
pharmacokinetic and pharmacodynamic response to clopidogrel but not
prasugrel. J Throm Haemost. 5:2429–2436. 2007. View Article : Google Scholar
|
|
9
|
Giusti B, Gori AM, Marcucci R, et al:
Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4
IVS10 + 12 G/A and P2Y12 T744C polymorphisms, is associated with
response variability to dual antiplatelet treatment in high-risk
vascular patients. Pharmacogenet Genom. 17:1057–1064. 2007.
View Article : Google Scholar
|
|
10
|
Trenk D, Hochholzer W, Fromm MF, et al:
Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel
platelet reactivity associated with adverse 1-year clinical outcome
of elective percutaneous coronary intervention with drug-eluting or
bare-metal stents. J Am Coll Cardiol. 51:1925–1934. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fontana P, Senouf D and Mach F: Biological
effect of increased maintenance dose of clopidogrel in
cardiovascular outpatients and influence of the cytochrome P450
2C19*2 allele on clopidogrel responsiveness. Thromb Res.
121:463–468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferreiro JL and Angiolillo DJ: Clopidogrel
response variability: current status and future directions. Thromb
Haemost. 102:7–14. 2009.PubMed/NCBI
|
|
13
|
Chan MY: Clopidogrel pharmacogenetics of
east, south and other Asian populations. Euro Heart J Suppl.
14:A41–A42. 2012. View Article : Google Scholar
|
|
14
|
Rehman K, Tariq MA and Akhtar T:
Clopidogrel resistance and its genetics. Pak Heart J. 46:151–158.
2013.
|
|
15
|
Abid L, Laroussi L, Bahloul A, Siala A,
Abdelhédi R, Kharrat N, Hentati M and Kammoun S: Impact of
cytochrome P450 2C19*2 polymorphism on the clinical cardiovascular
events after stent implantation in patients receiving clopidogrel
of a southern Tunisian region. World J Cardiovasc Dis. 3:4–10.
2013. View Article : Google Scholar
|
|
16
|
Hirotsu N, Murakami N, Kashiwagi T, Ujiie
K and Ishimaru K: Protocol: a simple gel-free method for SNP
genotyping using allele-specific primers in rice and other plant
species. Plant Methods. 6:122010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aynacioglu AS, Sachsc C, Bozkurt A, et al:
Low frequency of defective alleles of cytochrome P450 enzymes 2C19
and 2D6 in the Turkish population. Clin Pharmacol Ther. 66:185–192.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lamba JK, Dhiman RK and Kohli KK: Genetic
polymorphism of the hepatic cytochrome P450 2C19 in north Indian
subjects. Clin Pharmacol Ther. 63:422–427. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ozawa S, Soyama A, Saeki M, et al: Ethnic
differences in genetic polymorphisms of CYP2D6, CYP2C19, CYP3As and
MDR1/ABCB1. Drug Metabol Pharmacokinet. 19:83–95. 2004. View Article : Google Scholar
|
|
20
|
Zand N, Tajik N, Moghaddam AS and Milanian
I: Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in
a healthy Iranian population. Clin Exp Pharmacol Physiol.
34:102–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sukasem C, Tunthong R, Chamnanphon M, et
al: CYP2C19 polymorphisms in the Thai population and the clinical
response to clopidogrel in patients with atherothrombotic-risk
factors. Pharmgenomics Pers Med. 6:85–91. 2013.PubMed/NCBI
|
|
22
|
Hodgkinson A and Eyre-Walker A: Human
triallelic sites: evidence for a new mutational mechanism?
Genetics. 184:233–241. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Coulondre C, Miller JH, Farabaugh PJ and
Gilbert W: Molecular-basis of base substitution hotspots in
Escherichia coli. Nature. 274:775–780. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blake RD, Hess ST and Nicholson-Tuell J:
The influence of nearest neighbors on the rate and pattern of
spontaneous point mutations. J Mol Evol. 34:189–200. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Z, Fu YX, Hewett-Emmett D and
Boerwinkle E: Investigating single nucleotide polymorphism (SNP)
density in the human genome and its implications for molecular
evolution. Gene. 312:207–213. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hwang DG and Green P: Bayesian Markov
chain Monte Carlo sequence analysis reveals varying neutral
substitution patterns in mammalian evolution. Proc Natl Acad Sci
USA. 101:13994–14001. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bird AP: DNA methylation and the frequency
of CpG in animal DNA. Nucleic Acids Res. 8:1499–1504. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mega JL, Close SL, Wiviott SD, et al:
Cytochrome P-450 polymorphisms and response to clopidogrel. New
Engl J Med. 360:354–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jinnai T, Horiuchi H, Makiyama T, et al:
Impact of CYP2C19 polymorphisms on the antiplatelet effect of
clopidogrel in an actual clinical setting in Japan. Circ J.
73:1498–1503. 2009. View Article : Google Scholar : PubMed/NCBI
|