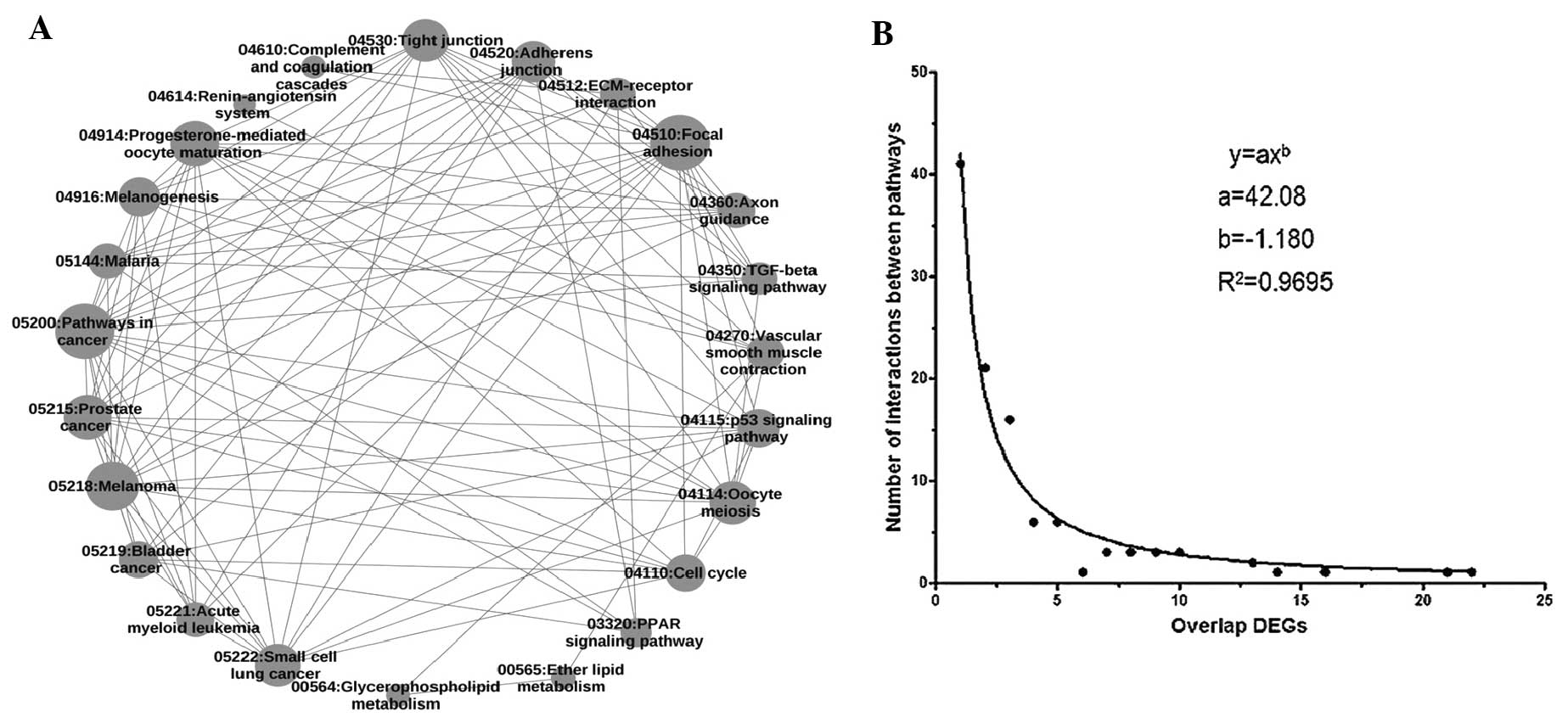
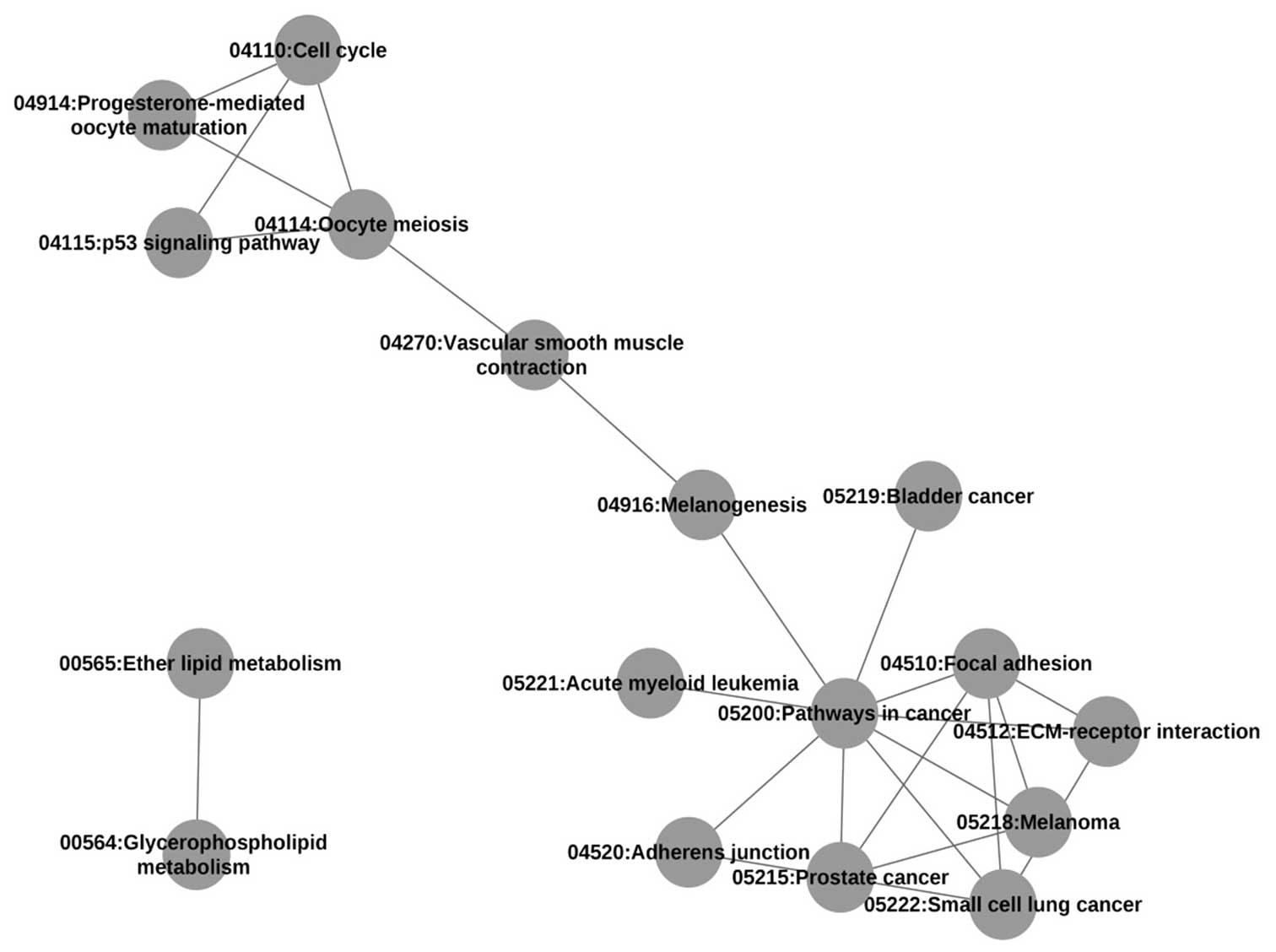
|
1
|
Igene H: Global health inequalities and
breast cancer: An impending public health problem for developing
countries. Breast J. 14:428–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chia S, Norris B, Speers C, et al: Human
epidermal growth factor receptor 2 overexpression as a prognostic
factor in a large tissue microarray series of node-negative breast
cancers. J Clin Oncol. 26:5697–5704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Planas-Silva MD, Bruggeman RD, Grenko RT
and Smith JS: Overexpression of c-Myc and Bcl-2 during progression
and distant metastasis of hormone-treated breast cancer. Exp Mol
Pathol. 82:85–90. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ota D, Mimori K, Yokobori T, et al:
Identification of recurrence-related microRNAs in the bone marrow
of breast cancer patients. Int J Oncol. 38:955–962. 2011.PubMed/NCBI
|
|
5
|
Ohira M and Nakagawara A: Global genomic
and RNA profiles for novel risk stratification of neuroblastoma.
Cancer Sci. 101:2295–2301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fujita T, Ikeda H, Kawasaki K, Taira N,
Ogasawara Y, Nakagawara A and Doihara H: Clinicopathological
relevance of UbcH10 in breast cancer. Cancer Sci. 100:238–248.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berlingieri MT, Pallante P, Sboner A, et
al: UbcH10 is overexpressed in malignant breast carcinomas. Eur J
Cancer. 43:2729–2735. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang S, Chen C, Meng Y, et al: Effective
suppression of breast tumor growth by an anti-EGFR/ErbB2 bispecific
antibody. Cancer Lett. 325:214–219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tinnikov AA, Yeung KT, Das S and Samuels
HH: Identification of a novel pathway that selectively modulates
apoptosis of breast cancer cells. Cancer Res. 69:1375–1382. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Donato M, Xu Z, Tomoiaga A, et al:
Analysis and correction of crosstalk effects in pathway analysis.
Genome Res. 23:1885–1893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bradley EW, Ruan MM, Vrable A and Oursler
MJ: Pathway crosstalk between Ras/Raf and PI3K in promotion of
M-CSF-induced MEK/ERK-mediated osteoclast survival. J Cell Biochem.
104:1439–1451. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gerits N, Kostenko S, Shiryaev A,
Johannessen M and Moens U: Relations between the mitogen-activated
protein kinase and the cAMP-dependent protein kinase pathways:
Comradeship and hostility. Cell Signal. 20:1592–1607. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pham H, Chong B, Vincenti R and Slice LW:
Ang II and EGF synergistically induce COX-2 expression via CREB in
intestinal epithelial cells. J Cell Physiol. 214:96–109. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang D, Xia D and Dubois RN: The crosstalk
of PTGS2 and EGF signaling pathways in colorectal cancer. Cancers
(Basel). 3:3894–3908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Krysan K, Reckamp KL, Dalwadi H, Sharma S,
Rozengurt E, Dohadwala M and Dubinett SM: Prostaglandin E2
activates mitogen-activated protein kinase/Erk pathway signaling
and cell proliferation in non-small cell lung cancer cells in an
epidermal growth factor receptor-independent manner. Cancer Res.
65:6275–6281. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang C, Cigliano A, Delogu S, Armbruster
J, et al: Functional crosstalk between AKT/mTOR and Ras/MAPK
pathways in hepatocarcinogenesis: Implications for the treatment of
human liver cancer. Cell Cycle. 12:1999–2010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong FX, Breitling R, McEntee CW, Wittner
BS, Nemhauser JL and Chory J: A bioconductor package for detecting
differentially expressed genes in meta-analysis. Bioinformatics.
22:2825–2827. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mootha VK, Lindgren CM, Eriksson KF, et
al: PGC-1alpha-responsive genes involved in oxidative
phosphorylation are coordinately downregulated in human diabetes.
Nat Genet. 34:267–273. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suárez-Fariñas M, Lowes MA, Zaba LC and
Krueger JG: Evaluation of the psoriasis transcriptome across
different studies by gene set enrichment analysis (GSEA). PLoS ONE.
5:e102472010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Subramanian A, Tamayo P, Mootha VK, et al:
Gene set enrichment analysis: A knowledge-based approach for
interpreting genome-wide expression profiles. Proc Natl Acad Sci
USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lisowska KM, Dudaladava V, Jarzab M, et
al: BRCA1-related gene signature in breast cancer: The role of ER
status and molecular type. Front Biosci (Elite Ed). 3:125–136.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Richardson AL, Wang ZC, De Nicolo A, et
al: X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Efron B and Tibshirani R: Empirical Bayes
methods and false discovery rates for microarrays. Genet Epidemiol.
23:70–86. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cui X and Churchill GA: Statistical tests
for differential expression in cDNA microarray experiments. Genome
Biol. 4:2102003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yekutieli D and Benjamini Y:
Resampling-based false discovery rate controlling multiple test
procedures for correlated test statistics. J Stat Plan Inference.
82:171–196. 1999. View Article : Google Scholar
|
|
26
|
Breitling R and Herzyk P: Rank-based
methods as a non-parametric alternative of the T-statistic for the
analysis of biological microarray data. J Bioinform Comput Biol.
3:1171–1189. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Long JM, Bell CW, Fagg WS IV, et al:
Microarray and pathway analysis reveals decreased CDC25A and
increased CDC42 associated with slow growth of BCL2 overexpressing
immortalized breast cell line. Cell Cycle. 7:3062–3073. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bissell MJ, Radisky DC, Rizki A, Weaver VM
and Petersen OW: The organizing principle: Microenvironmental
influences in the normal and malignant breast. Differentiation.
70:537–546. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huan J, Wang L, Xing L, et al: Insights
into significant pathways and gene interaction networks underlying
breast cancer cell line MCF-7 treated with 17β-estradiol (E2).
Gene. 533:346–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
del Sol A, Balling R, Hood L and Galas D:
Diseases as network perturbations. Curr Opin Biotechnol.
21:566–571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miller JA, Oldham MC and Geschwind DH: A
systems level analysis of transcriptional changes in Alzheimer's
disease and normal aging. J Neurosci. 28:1410–1420. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shao L, Wang L, Wei Z, et al: Dynamic
network of transcription and pathway crosstalk to reveal molecular
mechanism of MGd-treated human lung cancer cells. PLoS ONE.
7:e319842012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian M, Chen X, Xiong Q, et al:
Phosphoproteomic analysis of protein phosphorylation networks in
Tetrahymena thermophila, a model single-celled organism. Mol
Cell Proteomics. 13:503–519. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bartlett TE, Olhede SC and Zaikin A: A DNA
methylation network interaction measure, and detection of network
oncomarkers. PLoS ONE. 9:e845732014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Remon J, Morán T, Reguart N, Majem M,
Carcereny E and Lianes P: Beyond EGFR TKI in EGFR-mutant non-small
cell lung cancer patients: Main challenges still to be overcome.
Cancer Treat Rev. 40:723–729. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Z, Wang Z, Liang Z, et al: Expression
and clinical significance of IGF-1, IGFBP-3, and IGFBP-7 in serum
and lung cancer tissues from patients with non-small cell lung
cancer. Onco Targets Ther. 6:1437–1444. 2013.PubMed/NCBI
|
|
37
|
Bilke S, Schwentner R, Yang F, et al:
Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma
and prostate cancer. Genome Res. 23:1797–1809. 2013. View Article : Google Scholar : PubMed/NCBI
|