Introduction
The vacuolar H+-ATPase (V-ATPase) is a
transmembrane enzyme that actively pumps protons to the
extracellular matrix or intracellular compartments (e.g. endosomes,
lysosomes and secretory vesicles), in order to regulate the
alkalinization of the cytosol and acidification of cellular
compartments (1–3). The V-ATPase is widely distributed in
eukaryotic cells and exhibits particularly high levels of activity
in cancer cells (4). The
significance of the excessive activity of the V-ATPase in cancer
cells is to maintain the slightly alkaline intracellular pH, in
order to promote the survival of tumor cells when excess acidosis
is produced due to the ‘Warburg effect’; furthermore, the acidified
extracellular environment facilitates metastasis (5,6). High
levels of V-ATPase activity therefore promote the malignancy of
tumors. Functions of the V-ATPase have been discovered over many
years and include regulating signal transduction (7), glucose metabolism (8), lysosome functions (9), endosomal trafficking (10,11) and
the endoplasmic reticulum stress response (12). Considerable evidence supports the
suggestion that the V-ATPase represents a potential target of
anti-tumor therapy (13–16). Bafilomycin A1 is a specific inhibitor
of the c subunit of the V-ATPase and has been found to inhibit the
proliferation and metastasis of cancer cells (17).
MicroRNAs (miRNAs), the intrinsic, small,
non-protein-coding RNAs that effectively regulate gene expression,
play important roles in determining the proliferation or apoptosis
of cancer cells (18,19). The aim of the present study was to
investigate the effects of bafilomycin A1 on the BEL-7402
hepatocellular carcinoma and HO-8910 ovarian cancer cell lines and
to use microarray and reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) techniques to explore the altered pathways
and miRNA expression induced by bafilomycin A1 in these cell
lines.
Materials and methods
Cells and chemicals
The human BEL-7402 hepatocellular carcinoma and
HO-8910 ovarian cancer cell lines were purchased from the Cell Bank
of the Chinese Academy of Science (Shanghai, China) and maintained
in RPMI-1640 medium (Gibco-BRL, Rockville, MD, USA) supplemented
with 10% fetal calf serum (FCS; Hangzhou Sijiqing Bioengineering
Material Co., Ltd., Hangzhou, China), 100 U/ml penicillin and 100
µg/ml streptomycin. Cells were incubated at 37°C in a 5%
CO2 and 95% air atmosphere and were subcultured at ~80%
confluence. Bafilomycin A1 was purchased from Sigma-Aldrich (St.
Louis, MO, USA).
Water-soluble tetrazolium salt (WST)-1
cell proliferation assay
Cell proliferation and viability were analyzed using
the WST-1 Cell Proliferation Assay kit (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
instructions. Briefly, cells were harvested using 0.05% trypsin and
suspended in culture medium containing 10% FCS at a concentration
of 5×104 cells/ml, and 200 µl suspension was added to
each well of a 96-well plate. Cells were cultured for 20 h for
adhesion. Bafilomycin A1 was added to the wells at the final
concentrations of 200, 400 and 800 nM, in triplicate. At 24, 48 and
72 h, 20 µl WST-1 was added to the cells. Following incubation at
37°C for 4 h, the plates were read to determine the optical density
(OD) at 435 nm with 675 nm reference using a spectrophotometer.
Soft-agar colony formation assay
Soft agar assays were performed in a 6-well plate.
Each well contained 2 ml 0.5% agarose (Sigma-Aldrich) at the bottom
and 2 ml 0.35% agarose containing 1×104 cells on top.
The medium used was the aforementioned culture media with serial
concentrations of bafilomycin A1. Each concentration was set in
triplicate. Cells were grown at 37°C in a 5% CO2 and 95%
air atmosphere for 14 days. Colonies were counted under a
phase-contrast microscope (XD-202; Nanjing Jiangnan Novel Optics,
Co., Ltd., Nanjing, China). The relative proliferation ratio was
calculated using the following formula: Relative proliferation
ratio = Number of treated/Number of control.
Capsase-3 and −9 assays
A total of 1×106 BEL-7402 and HO-8910
cells, respectively, were seeded in a 6-well plate (Corning, Inc.,
Corning, NY, USA) and cultured with the serial concentrations of
bafilomycin A1 in RPMI-1640 medium with 10% FCS at 37°C in a 5%
CO2 and 95% air atmosphere for 24, 48 and 72 h. The
activities of caspase-3 and −9 in the cellular extracts were
determined by colorimetric assay, using commercial Caspase-3 and −9
Activity kits (Beyotime Institute of Biotechnology) according to
the manufacturer's instructions. The protein concentrations of the
samples were determined using a Bicinchoninic Acid Assay kit
(Beyotime Institute of Biotechnology). The relative enzyme activity
was calculated using the following formula: Relative enzyme
activity = enzyme activity of treated cells per µg protein/enzyme
activity of control cells per µg protein.
Transmission electron microscopy
(TEM)
A total of 1×106 BEL-7402 and HO-8910
cells, respectively, were seeded and cultured with the serial
concentrations of bafilomycin A1 in RPMI-1640 medium with 10% FCS
at 37°C in a 5% CO2 and 95% air atmosphere for 48 h. The
cells were washed with phosphate-buffered saline (PBS), harvested
using 0.03% trypsin, re-washed with PBS to remove the trypsin and
fixed in 2.5% glutaraldehyde at 4°C overnight. The cells were then
centrifuged at 500 × g for 5 min to remove the glutaraldehyde and
postfixed in 1% OsO4 for 2 h. Following postfixation,
the cells were washed with PBS, dehydrated in ethanol and embedded
in Epon 812 (Sigma-Aldrich). Ultrathin sections were sliced using a
Leica-Reichert Ultracut ultramicrotome (Leica Microsystems GmbH,
Wetzlar, Germany), stained with 2% aqueous uranyl acetate
(Sigma-Aldrich) and lead citrate (Sigma-Aldrich) and observed with
an HT-7700 electron microscope (Hitachi, Ltd., Tokyo, Japan).
Cell invasion assay
The cells were cultured in the aforementioned
complete culture medium until reaching 80% confluence and then
starved in serum-free medium for 8 h. The cells were subsequently
washed with PBS, harvested, diluted to the concentration of
1×105 cells/ml in 0.1% serum RPMI-1640 medium with
serial concentrations of bafilomycin A1 and then plated in an
invasion chamber (1×104 cells in 100 µl medium per well)
of a Cell Invasion Assay kit (Chemicon International, Inc.,
Temecula, CA, USA). The invasion chamber was inserted into the
feeder tray, which contained 150 µl 10% serum RPMI-1640 medium per
well. The plate was incubated for 20 h at 37°C in 5% CO2
and 95% air. The cells that had invaded through the membrane were
washed with detachment solution and stained with lysis buffer/dye
solution, and the fluorescence absorbance was measured in a
luminescence spectrometer at 480/520 nm.
Total RNA extraction
Total RNA of the control cells and cells that had
been exposed to 400 nM bafilomycin A1 for 48 h was extracted using
TRIzol® reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA)
in accordance with the manufacturer's instructions. RNA
concentration and purity were determined via ultraviolet
spectrometry, measuring the OD at 260 and 280 nm.
Microarray of mRNA and miRNA
Total extracted RNA was subjected to mRNA and miRNA
microarray using the Affymetrix GeneChip® Human Gene and miRNA
Arrays (Affymetrix, Inc., Santa Clara, CA, USA), respectively, at
Genminix Informatics Ltd., Co. (Shanghai, China) according to the
instructions provided with the Affymetrix arrays. Briefly, RNA was
extracted from cells following treatment with 400 nM bafilomycin A1
for 48 h using TRIzol reagent (Invitrogen, Life Technologies). The
quantity and purity of the RNA were assessed using a NanoDrop
ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington,
DE, USA). The purity and integrity of each extracted RNA met the
following requirements: A260/A280 >1.6, A260/A230 >1 and RNA
Integrity Number >5. The small RNA fraction indicated an
abundance of RNA <200 nt, compared with total RNA using Agilent
RNA 6000 Nano assay (Agilent Technologies, Inc., Santa Clara, CA,
USA), and was acceptable for miRNA assay. The possibility of
genomic DNA contamination was excluded using gel electrophoresis. A
total of 2 µg RNA from each group was respectively converted into
cyanine-5-labeled target cRNA, hybridized to the Affymetrix
GeneChip Human Gene and miRNA Arrays, respectively, using the
Affymetrix GeneChip Fluidics Station 450 (Affymetrix, Inc.), and
scanned with an Affymetrix GeneChip Scanner 3000 7G (Affymetrix,
Inc.). Following normalization, differentially expressed mRNAs or
miRNAs were established at log2 (fold change) >1 and
P<0.05.
RT-qPCR analysis of miRNA
RNA was transcribed into cDNA using the miScript II
RT kit (Qiagen, Hilden, Germany). The reaction components were as
follows: Total RNA, 1 µg; miScript HiSpec Buffer, 4 µl; Nucleic
Acid Mix, 2 µl; miScript Reverse Transcriptase Mix, 2 µl;
RNase-free H2O, <20 µl. The reaction was performed at
37°C for 60 min and 95°C for 5 min on an ABI PCR 9700 system
(Applied Biosystems, Foster City, CA, USA). cDNA was diluted in 80
µl nuclease-free H2O for the addition of LightCycler®
480 SYBR Green I Master (Roche Diagnostics, Indianapolis, IN, USA).
The reaction system for the qPCR was as follows: LightCycler 480
SYBR Green I Master, 5 µl; forward primer, 0.2 µl; reverse primer,
0.2 µl (provided by RiboBio Co., Ltd., Guangzhou, China); cDNA, 1
µl; nuclease-free H2O, 3.6 µl. The PCR was run using an
ABI 7500 Fast system (Applied Biosystems), as follows: 94°C for 2
min, followed by 40 cycles of 94°C for 15 sec, 62°C for 40 sec and
70°C for 40 sec. The mRNA expression was normalized against the
expression of U6. All samples were analyzed in triplicate. Relative
expression was calculated using the comparative threshold cycle
(Ct) method and was indicated as the fold change, as follows: Fold
change = 2−(ΔCttreated - ΔCtcontrol).
Data analysis
Microarray data were analyzed by Genminix
Informatics Ltd., Co. The integrated analysis of the differential
mRNA and miRNA expression due to treatment with bafilomycin A1 was
performed to reveal the target genes of the drug via miRNA
regulation. The target genes of the differential miRNA were
predicted using bioinformatics [TargetScan (http://www.targetscan.org/) and MicroCosm Targets
(http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets)].
The overlapping altered genes in the mRNA microarray and the miRNA
predicted genes were subjected to Gene Ontology (GO) term and
pathway analyses, in order to organize the genes into hierarchical
categories and uncover the altered pathways and miRNA-gene
regulatory network upon which bafilomycin A1 likely has an
effect.
Statistical analysis
Statistical analysis was carried out using Microsoft
Excel software (Microsoft Corp., Redmond, WA, USA). Data were
compared using a two-tailed Student's t-test, and P<0.05 was
considered to indicate a statistically significant difference.
Results
Bafilomycin A1 inhibits BEL-7402 and
HO-8910 cell proliferation
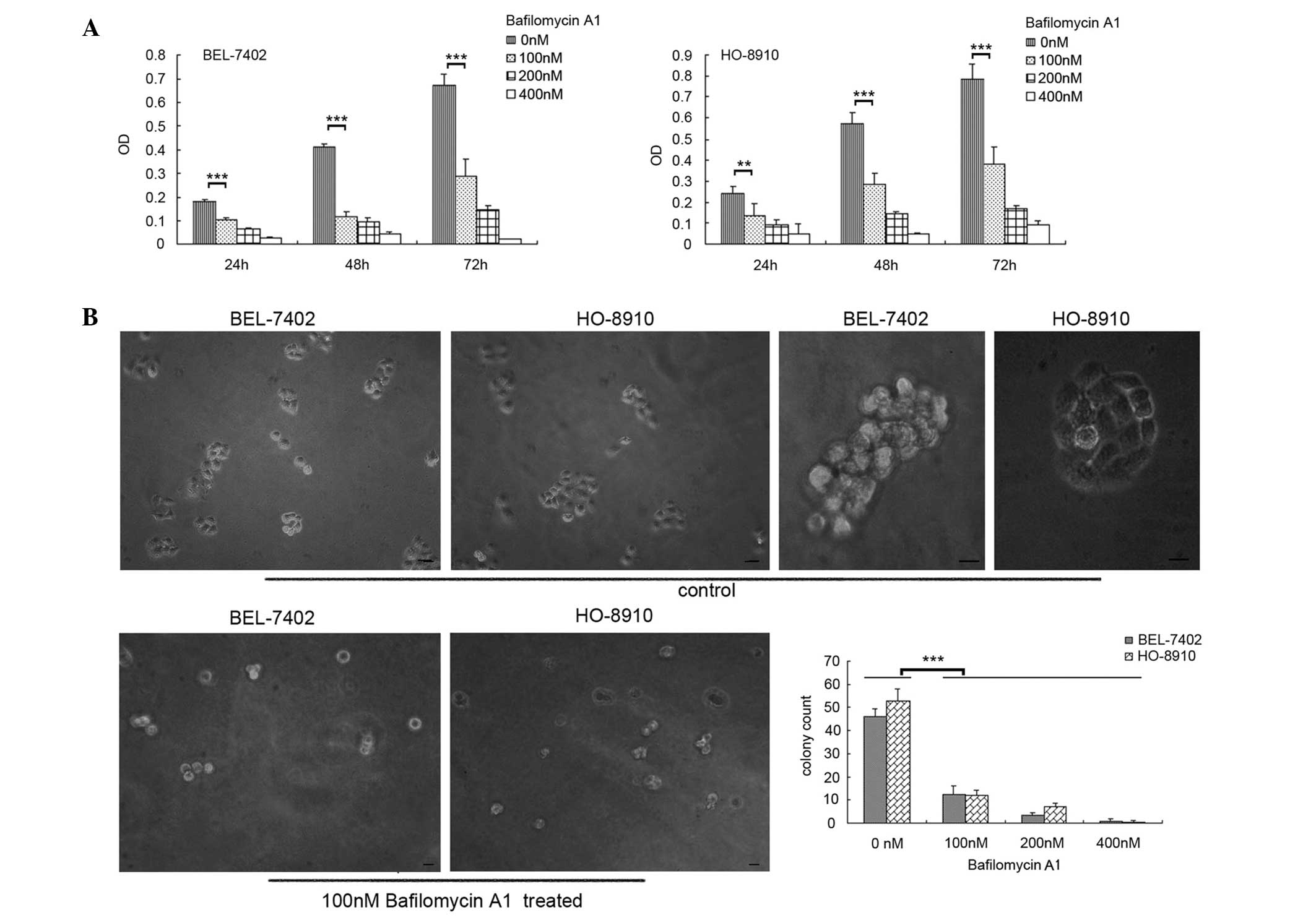
WST-1 cell proliferation and soft-agar colony
formation assays were performed to determine the effect of
bafilomycin A1 on BEL-7402 and HO-8910 cells. The results indicated
that the proliferation of the two cell lines was inhibited by
bafilomycin A1, as shown in Fig. 1A.
The inhibitory effect was also evident in the soft-agar colony
formation assay (Fig. 1B). Compared
with the control cells, few micro- or middle-colonies (containing
3–5 or 6–10 cells/colony, respectively) and non-macro-colonies
(containing >10 cells) could be observed among the bafilomycin
A1-treated cells, and the total numbers of colonies were
significantly reduced.
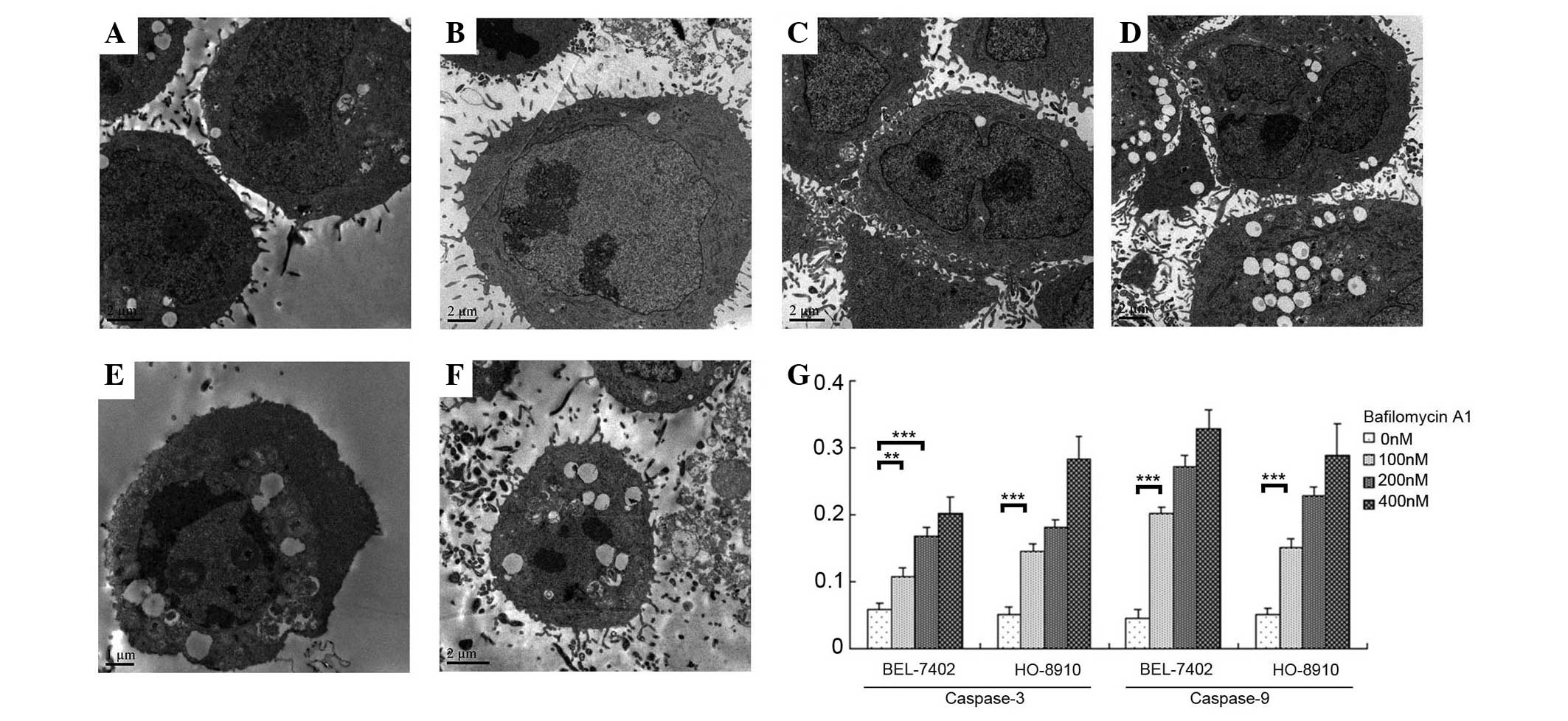
TEM observations of bafilomycin
A1-treated BEL-7402 and HO-8910 cells
TEM was used to capture images of the cells treated
with various concentrations of bafilomycin A1 at 48 h.
Non-bafilomycin A1-treated BEL-7402 and HO-8910 cells are shown in
Fig. 2A and B, respectively.
BEL-7402 and HO-8910 cells treated with 100 nM bafilomycin A1
(Fig. 2C and D, respectively)
exhibited indentations and folding of the nuclear membrane, whereas
BEL-7402 and HO-8910 cells treated with 200 nM bafilomycin A1
(Fig. 2E and F, respectively)
exhibited chromosome condensation at the periphery of the nuclear
membrane, condensed nuclei and vacuolar cytoplasms. The images
suggested an apoptotic response to the cellular toxicity.
Furthermore, the assays for capsase-3 and −9, which are two
important enzymes involved in apoptosis, showed significant
increases following bafilomycin A1-treatment (Fig. 2G).
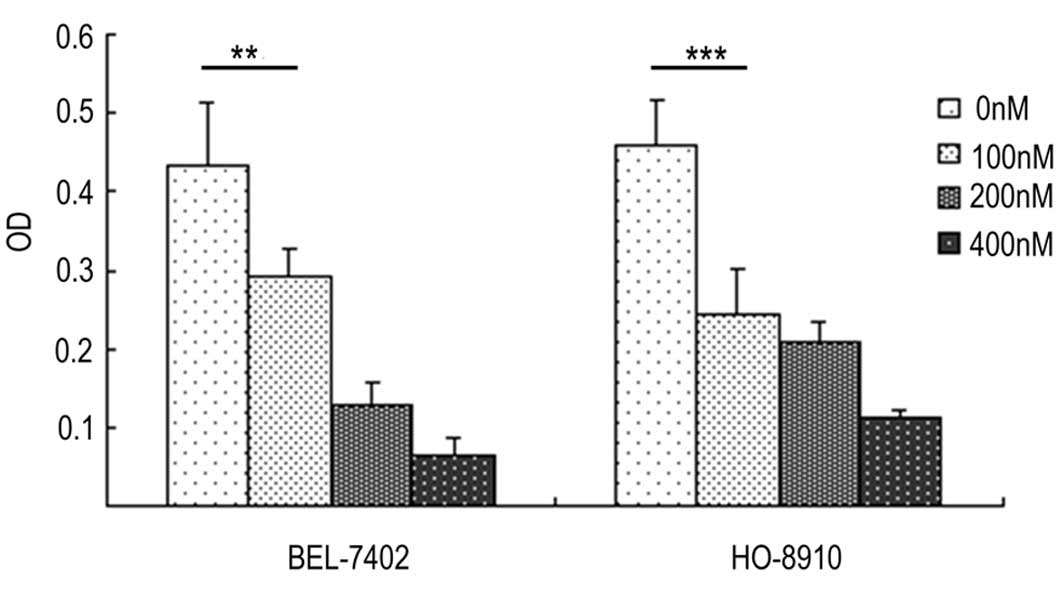
Bafilomycin A1 suppresses the invasion
of BEL-7402 and HO-8910 cells
According to the cell invasion assays, the invasive
potential of the BEL-7402 and HO-8910 cells was significantly
suppressed with the bafilomycin A treatment, as shown in Fig. 3.
Bafilomycin A1 induces alterations in
certain pathways
The numbers of altered mRNAs and miRNAs in BEL-7402
and HO-8910 cells 48 h after exposure to 400 nM bafilomycin A1 are
shown in Table I. The overlapping
altered genes in the mRNA microarray and the miRNA predicted genes
were determined, and GO analysis was performed. The altered
pathways are shown in Fig. 4A. After
treatment with 400 nM bafilomycin A1 for 48 h, the following miRNAs
were altered in the two cell lines: miR-923, miR-1246, miR-149*,
miR-638 and miR-210 were significantly upregulated, whereas
miR-99a, miR-181a-2* and miR-339-5p were significantly
downregulated. These results were confirmed by qPCR (Fig. 4B).
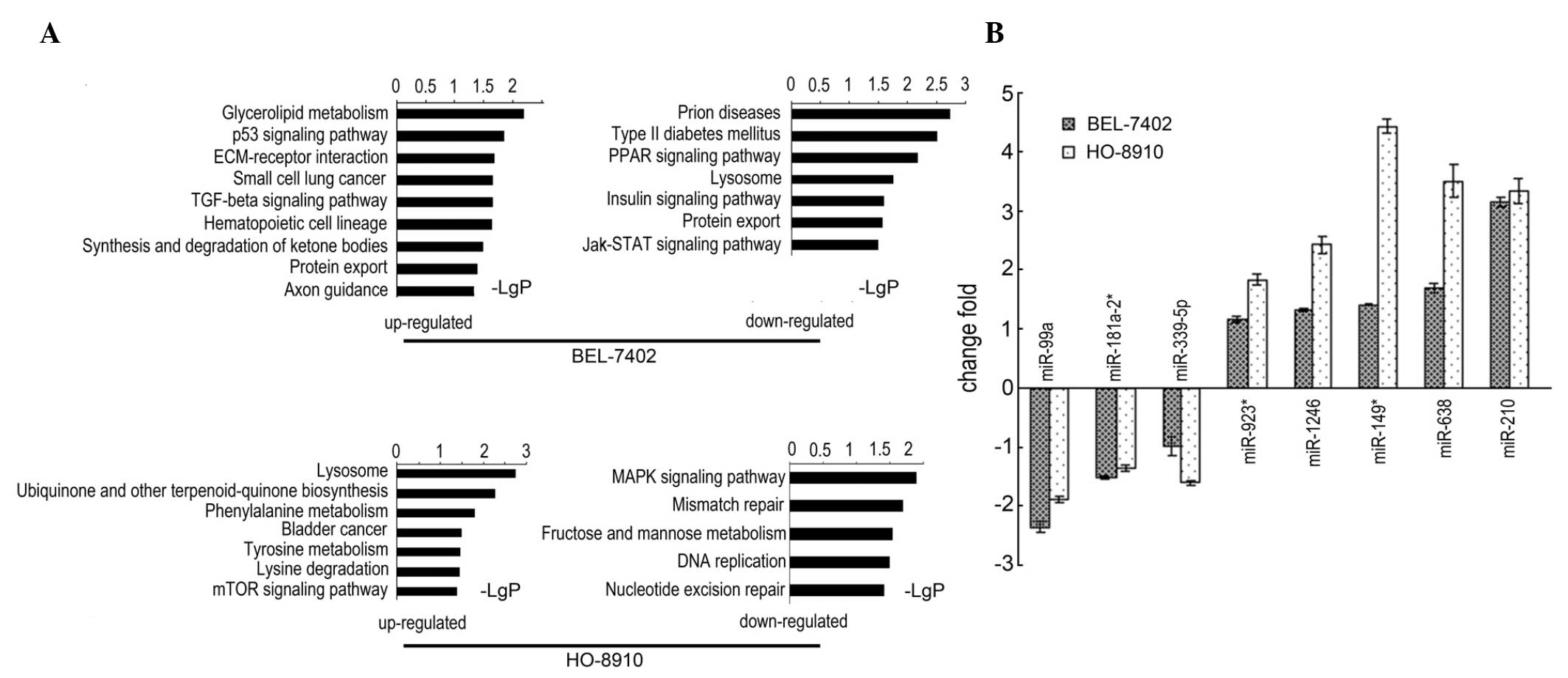 | Figure 4.Bafilomycin A1-induced altered
pathways and miRNAs. (A) miRNA-mRNA integrity analysis demonstrated
the significantly altered pathways in the BEL-7402 and HO-8910 cell
lines. (B) Quantitative polymerase chain reaction assay confirmed
the eight common significantly altered miRNAs following treatment
with bafilomycin A1, log2 fold change is shown (each of the eight,
P<0.001 vs. the control). miRNA, microRNA; ECM, extracellular
matrix; TFG, transforming growth factor; PPAR, peroxisome
proliferator-activated receptor; Jak, Janus kinase; STAT, signal
transducer and activator of transcription; mTOR, mammalian target
of rapamycin; MAPK, mitogen-activated protein kinase. |
 | Table I.Numbers of altered genes and
miRNAs. |
Table I.
Numbers of altered genes and
miRNAs.
|
| Number of altered
genes | Number of altered
miRNAs |
|---|
|
|
|
|
|---|
| Cell line | Upregulated | Downregulated | Upregulated | Downregulated |
|---|
| BEL-7402 | 252 | 127 | 12 | 8 |
| HO-8910 | 126 | 57 | 18 | 20 |
Discussion
The V-ATPase is typically highly activated in cancer
cells, in which it has the following functions: Regulating cell
proliferation, apoptosis and autophagy; facilitating cancer
metastasis; contributing to the acquirement of drug resistance; and
affecting signal transduction (20–23).
Inhibitors of the V-ATPase, therefore, are promising anti-cancer
chemicals. In the present study, the inhibitory effects of
bafilomycin A1 on two cell lines, BEL-7402 and HO-8910, were
investigated, and it was found that bafilomycin A1 suppressed the
proliferation, induced the apoptosis and attenuated the invasive
potential of the cells.
Following exposure to 400 nM bafilomycin A1 for 48
h, an mRNA-miRNA microarray integrity analysis was performed to
reveal the altered pathways, among which several were associated
with glucose or lipid metabolism, including glycerolipid
metabolism, insulin signaling, fructose and mannose metabolism,
phenylalanine metabolism and the synthesis and degradation of
ketone bodies (Fig. 4); this finding
may result from the coupling of glucose metabolism and regulation
of the cytosolic/extracellular pH gradient by the V-ATPase. By
dysregulating the V-ATPase, bafilomycin A1 thus affected the
metabolism of glucose (8,24). Pathways associated with DNA repair or
duplication were also found to be altered, including the p53
signaling pathway, nucleotide excision repair, and DNA replication
and mismatch repair, which suggested the toxicity of bafilomycin A1
and was consistent with the retarded proliferation. A number of
altered signal pathways induced by bafilomycin A1, which were
suggested to be highly related to the functions of V-ATPase, have
been previously reported, such as mammalian target of rapamycin
(25), AMP-activated protein kinase
(26) and transforming growth
factor-β (27). The present study is
the first to additionally report Janus kinase-signal transducer and
activator of transcription and peroxisome proliferator-activated
receptor (Fig. 4).
Notably, the lysosome pathway was altered in both
cell lines, but in a contrasting manner, i.e., in BEL-7402 cells
the pathway was downregulated, whereas in HO-8910 cells the pathway
was upregulated. Previous studies have suggested that the functions
of lysosomes and the V-ATPase are closely associated and that their
interactions are involved in the regulations of autophagy and
apoptosis (9,28).
Liver cancer and ovarian cancer are different types
of solid cancer; men are more susceptible to the former, while the
latter is female specific. This is one of the reasons that these
two cell lines were selected for the present study; in order to
expand their contrasts. However, bafilomycin A1 displayed a high
efficiency of inhibitory effects on both tumor types in
vitro. Although the inhibitory effects of bafilomycin A1 in the
two cancer cell lines were similar, the cellular pathways involved
in the action of bafilomycin A1 were not identical, as an miRNA
microarray showed that 8 miRNAs were altered in both cell lines.
miRNAs serve an advanced regulatory function in cellular pathways,
with one miRNA typically impacting numerous mRNAs. The miRNAs that
are commonly altered by bafilomycin A1 in the two cell lines may
represent promising targets for anti-cancer therapies. However,
further studies are required to determine whether the miRNAs
exhibit similar effects in other types of solid tumor.
Acknowledgements
This study was supported by grants from the State
Key Laboratory of Oncogenes and Related Genes (no. 90-10-02), the
Clinical Medicine Science and Technology Project of Jiangsu
Province China (no. BL2013024) and the National Nature Science
Foundation of China (no. 81372404).
References
|
1
|
Nishi T and Forgac M: The vacuolar
(H+)-ATPases-nature's most versatile proton pumps. Nat
Rev Mol Cell Biol. 3:94–103. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marshansky V, Rubinstein JL and Grüber G:
Eukaryotic V-ATPase: Novel structural findings and functional
insights. Biochimica Biophy Acta. 1837:857–879. 2014. View Article : Google Scholar
|
|
3
|
Breton S and Brown D: Regulation of
luminal acidification by the V-ATPase. Physiology (Bethesda).
28:318–329. 2013.PubMed/NCBI
|
|
4
|
Sun-Wada GH and Wada Y: Vacuolar-type
proton pump ATPases: Acidification and pathological relationships.
Histol Histopathol. 28:805–815. 2013.PubMed/NCBI
|
|
5
|
Barar J and Omidi Y: Dysregulated pH in
tumor microenvironment checkmates cancer therapy. BioImpacts.
3:149–162. 2013.PubMed/NCBI
|
|
6
|
Chung C, Mader CC, Schmitz JC, Atladottir
J, Fitchev P, Cornwell ML, Koleske AJ, Crawford SE and Gorelick F:
The vacuolar-ATPase modulates matrix metalloproteinase isoforms in
human pancreatic cancer. Lab Invest. 91:732–743. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sennoune SR and Martinez-Zaguilan R:
Vacuolar H(+)-ATPase signaling pathway in cancer. Curr Protein Pept
Sci. 13:152–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fogarty FM, O'Keeffe J, Zhadanov A,
Papkovsky D, Ayllon V and O'Connor R: HRG-1 enhances cancer cell
invasive potential and couples glucose metabolism to
cytosolic/extracellular pH gradient regulation by the vacuolar-H(+)
ATPase. Oncogene. 33:4653–4663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakashima S, Hiraku Y, Tada-Oikawa S,
Hishita T, Gabazza EC, Tamaki S, Imoto I, Adachi Y and Kawanishi S:
Vacuolar H+-ATPase inhibitor induces apoptosis via
lysosomal dysfunction in the human gastric cancer cell line MKN-1.
J Biochem. 134:359–364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan Y, Denef N and Schüpbach T: The
vacuolar proton pump, V-ATPase, is required for notch signaling and
endosomal trafficking in drosophila. Dev Cell. 17:387–402. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Marshansky V and Futai M: The V-type
H+-ATPase in vesicular trafficking: Targeting,
regulation and function. Curr Opin Cell Biol. 20:415–426. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee GH, Kim DS, Kim HT, Lee JW, Chung CH,
Ahn T, Lim JM, Kim IK, Chae HJ and Kim HR: Enhanced lysosomal
activity is involved in Bax inhibitor-1-induced regulation of the
endoplasmic reticulum (ER) stress response and cell death against
ER stress: Involvement of vacuolar H+-ATPase (V-ATPase).
J Biol Chem. 286:24743–24753. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Avnet S, Di Pompo G, Lemma S, Salerno M,
Perut F, Bonuccelli G, Granchi D, Zini N and Baldini N: V-ATPase is
a candidate therapeutic target for Ewing sarcoma. Biochim Biophys
Acta. 1832:1105–1116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hernandez A, Serrano-Bueno G,
Perez-Castineira JR and Serrano A: Intracellular proton pumps as
targets in chemotherapy: V-ATPases and cancer. Curr Pharm Des.
18:1383–1394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graham RM, Thompson JW and Webster KA:
Inhibition of the vacuolar ATPase induces Bnip3-dependent death of
cancer cells and a reduction in tumor burden and metastasis.
Oncotarget. 5:1162–1173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luciani F, Spada M, De Milito A, Molinari
A, Rivoltini L, Montinaro A, Marra M, Lugini L, Logozzi M, Lozupone
F, et al: Effect of proton pump inhibitor pretreatment on
resistance of solid tumors to cytotoxic drugs. J Nat Cancer Inst.
96:1702–1713. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morimura T, Fujita K, Akita M, Nagashima M
and Satomi A: The proton pump inhibitor inhibits cell growth and
induces apoptosis in human hepatoblastoma. Pediat Surg Int.
24:1087–1094. 2008. View Article : Google Scholar
|
|
18
|
Baer C, Claus R and Plass C: Genome-wide
epigenetic regulation of miRNAs in cancer. Cancer Res. 73:473–477.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y and Taniguchi T: MicroRNAs and DNA
damage response: Implications for cancer therapy. Cell Cycle.
12:32–42. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
von Schwarzenberg K, Wiedmann RM, Oak P,
Schulz S, Zischka H, Wanner G, Efferth T, Trauner D and Vollmar AM:
Mode of cell death induction by pharmacological vacuolar
H+-ATPase (V-ATPase) inhibition. J Biol Chem.
288:1385–1396. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ohta T, Arakawa H, Futagami F, Fushida S,
Kitagawa H, Kayahara M, Nagakawa T, Miwa K, Kurashima K, Numata M,
et al: Bafilomycin A1 induces apoptosis in the human pancreatic
cancer cell line Capan-1. J Pathol. 185:324–330. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hendrix A, Sormunen R, Westbroek W,
Lambein K, Denys H, Sys G, Braems G, Van den Broecke R, Cocquyt V,
Gespach C, et al: Vacuolar H+ ATPase expression and
activity is required for Rab27B-dependent invasive growth and
metastasis of breast cancer. Int J Cancer. 133:843–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu Q, Lu S, Huang L, Wang T, Wan Y, Zhou
CX, Zhang C, Zhang Z and Li X: The expression of V-ATPase is
associated with drug resistance and pathology of non-small-cell
lung cancer. Diagn Pathol. 8:1452013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dechant R, Binda M, Lee SS, Pelet S,
Winderickx J and Peter M: Cytosolic pH is a second messenger for
glucose and regulates the PKA pathway through V-ATPase. EMBO J.
29:2515–2526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Valapala M, Wilson C, Hose S, Bhutto IA,
Grebe R, Dong A, Greenbaum S, Gu L, Sengupta S, Cano M, et al:
Lysosomal-mediated waste clearance in retinal pigment epithelial
cells is regulated by CRYBA1/βA3/A1-crystallin via V-ATPase-MTORC1
signaling. Autophagy. 10:480–496. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang CS, Jiang B, Li M, Zhu M, Peng Y,
Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al: The lysosomal
v-ATPase-ragulator complex is a common activator for AMPK and
mTORC1, acting as a switch between catabolism and anabolism. Cell
Metab. 20:526–540. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao X, Yang Q, Qin J, Zhao S, Li X, Fan J,
Chen W, Zhou Y, Mao H and Yu X: V-ATPase promotes transforming
growth factor-β-induced epithelial-mesenchymal transition of rat
proximal tubular epithelial cells. Am J Physiol Renal Physiol.
302:F1121–F1132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jung JY and Robinson CM: Interleukin-27
inhibits phagosomal acidification by blocking vacuolar ATPases.
Cytokine. 62:202–205. 2013. View Article : Google Scholar : PubMed/NCBI
|