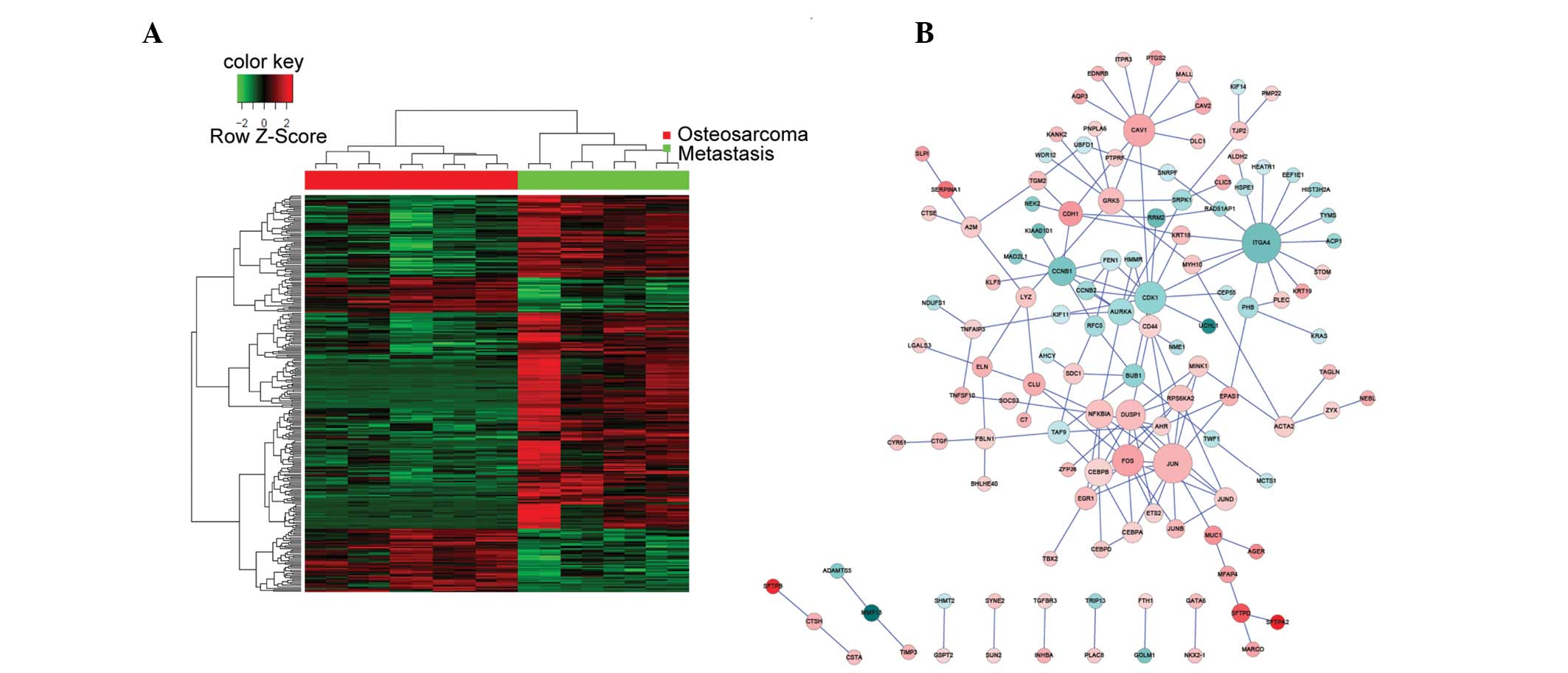
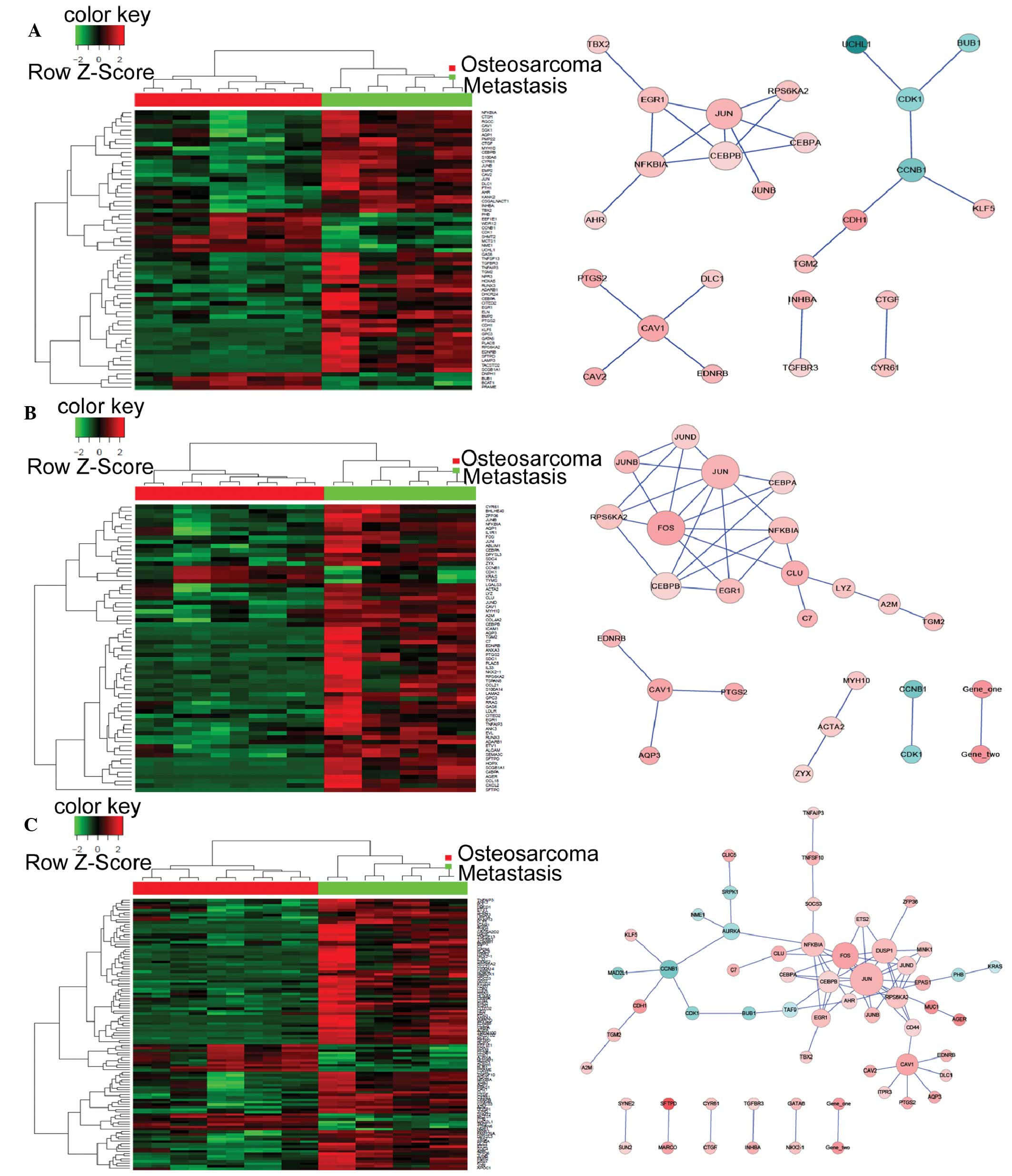
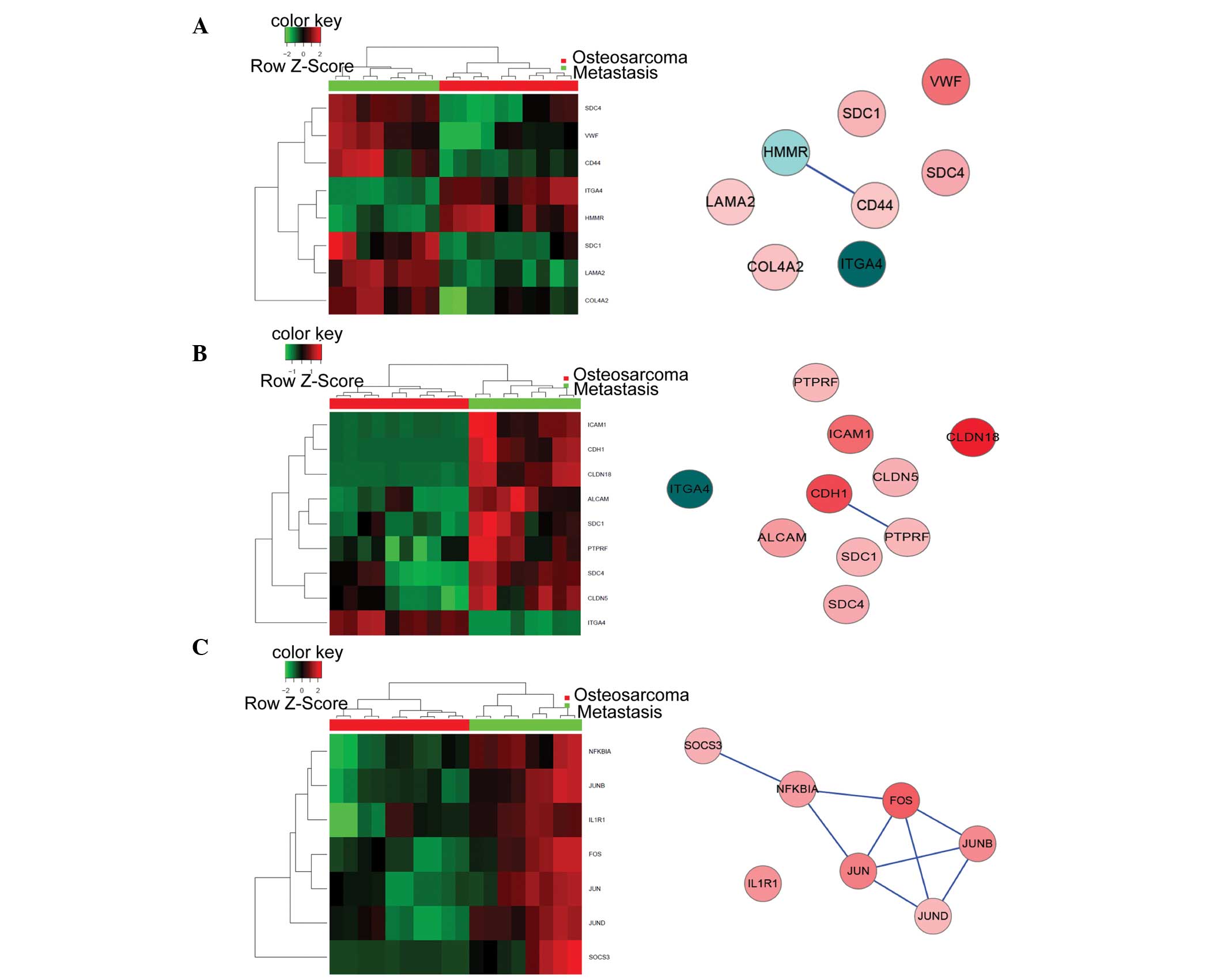
|
1
|
Whelan J, McTiernan A, Cooper N, Wong YK,
Francis M, Vernon S and Strauss SJ: Incidence and survival of
malignant bone sarcomas in England 1979-2007. Int J Cancer.
131:E508–E517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bielack SS, Carrle D, Hardes J, Schuck A
and Paulussen M: Bone tumors in adolescents and young adults. Curr
Treat Options Oncol. 9:67–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hegyi M, Semsei AF, Jakab Z, Antal I, Kiss
J, Szendroi M, Csoka M and Kovacs G: Good prognosis of localized
osteosarcoma in young patients treated with limb-salvage surgery
and chemotherapy. Pediatr Blood Cancer. 57:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Botter SM, Neri D and Fuchs B: Recent
advances in osteosarcoma. Curr Opin Pharmacol. 16:15–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mialou V, Philip T, Kalifa C, Perol D,
Gentet JC, Marec-Berard P, Pacquement H, Chastagner P, Defaschelles
AS and Hartmann O: Metastatic osteosarcoma at diagnosis: Prognostic
factors and long-term outcome-the French pediatric experience.
Cancer. 104:1100–1109. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Flores RJ, Li Y, Yu A, Shen J, Rao PH, Lau
SS, Vannucci M, Lau CC and Man TK: A systems biology approach
reveals common metastatic pathways in osteosarcoma. BMC Syst Biol.
28:50–67. 2012. View Article : Google Scholar
|
|
7
|
Zhang Y, Zhang L, Zhang G, Li S, Duan J,
Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al: Osteosarcoma
metastasis: Prospective role of ezrin. Tumour Biol. 35:5055–5059.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rao-Bindal K, Rao CK, Yu L and Kleinerman
ES: Expression of c-FLIP in pulmonary metastases in osteosarcoma
patients and human xenografts. Pediatr Blood Cancer. 60:575–579.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagao-Kitamoto H, Nagata M, Nagano S,
Kitamoto S, Ishidou Y, Yamamoto T, Nakamura S, Tsuru A, Abematsu M,
Fujimoto Y, et al: GLI2 is a novel therapeutic target for
metastasis of osteosarcoma. Int J Cancer. 136:1276–1284. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang G, Nishimoto K, Zhou Z, Hughes D and
Kleinerman ES: miR-20a encoded by the miR-17-92 cluster increases
the metastatic potential of osteosarcoma cells by regulating Fas
expression. Cancer Res. 72:908–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: MicroRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
12
|
Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu
LH, Bu J, Wang ZH and Xiao T: Screening for metastatic osteosarcoma
biomarkers with a DNA microarray. Asian Pac J Cancer Prev.
15:1817–1822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu J, Irizarry R, MacDonald J and Gentry
J: Gcrma: Background adjustment using sequence information. R
package. version 2.36.0.
|
|
14
|
Gentleman R, Carey V, Huber W and Hahne F:
Genefilter: Methods for filtering genes from microarray
experiments. R package. version 1.46.1.
|
|
15
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. 397–420. 2005. View Article : Google Scholar
|
|
16
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 57:289–300.
1995.
|
|
17
|
Tavazoie S, Hughes JD, Campbell MJ, Cho RJ
and Church GM: Systematic determination of genetic network
architecture. Nat Genet. 22:281–285. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warnes GR, Bolker B, Bonebakker L, et al:
gplots: Various R programming tools for plotting data. R package
version 2. 2009.
|
|
19
|
Carlson M: GOdb: A set of annotation maps
describing the entire Gene Ontology. R package. version 2.14.0.
|
|
20
|
Tenenbaum D: KEGGREST: Client-side REST
access to KEGG. R package. version 1.4.0.
|
|
21
|
Prasad Keshava TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:D767–D772. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chatr-Aryamontri A, Breitkreutz BJ,
Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas
N, O'Donnell L, et al: The BioGRID interaction database: 2013
update. Nucleic Acids Res. 41:D816–D823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McDowall MD, Scott MS and Barton GJ: PIPs:
Human protein-protein interaction prediction database. Nucleic
Acids Res. 37:D651–D656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Y and Ma L: The emerging molecular
machinery and therapeutic targets of metastasis. Trends Pharmacol
Sci. 36:349–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamashita K, Katoh H and Watanabe M: The
homeobox only protein homeobox (HOPX) and colorectal cancer. Int J
Mol Sci. 14:23231–23243. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kovárová D, Plachy J, Kosla J, Trejbalová
K, Čermák V and Hejnar J: Downregulation of HOPX controls
metastatic behavior in sarcoma cells and identifies genes
associated with metastasis. Mol Cancer Res. 11:1235–1247. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ozaki K, Nagata M, Suzuki M, Fujiwara T,
Ueda K, Miyoshi Y, Takahashi E and Nakamura Y: Isolation and
characterization of a novel human lung-specific gene homologous to
lysosomal membrane glycoproteins 1 and 2: Significantly increased
expression in cancers of various tissues. Cancer Res. 58:3499–3503.
1998.PubMed/NCBI
|
|
28
|
Nagelkerke A, Bussink J, Mujcic H, Wouters
BG, Lehmann S, Sweep FC and Span PN: Hypoxia stimulates migration
of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded
protein response. Breast Cancer Res. 15:R22013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanao H, Enomoto T, Kimura T, Fujita M,
Nakashima R, Ueda Y, Ueno Y, Miyatake T, Yoshizaki T, Buzard GS, et
al: Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in
uterine cervical cancer. Cancer Res. 65:8640–8645. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Yao Y, Gong C, Yu F, Su S, Liu B,
Deng H, Wang F, Lin L, et al: CCL18 from tumor-associated
macrophages promotes breast cancer metastasis via PITPNM3. Cancer
Cell. 19:541–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li HY, Cui XY, Wu W, Yu FY, Yao HR, Liu Q,
Song EW and Chen JQ: Pyk2 and Src mediate signaling to
CCL18-induced breast cancer metastasis. J Cell Biochem.
115:596–603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuespert K, Pils S and Hauck CR: CEACAMs:
Their role in physiology and pathophysiology. Curr Opin Cell Biol.
18:565–571. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Beauchemin N and Arabzadeh A:
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs)
in cancer progression and metastasis. Cancer Metastasis Rev.
32:643–671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Govaere O, Komuta M, Berkers J, Spee B,
Janssen C, de Luca F, Katoonizadeh A, Wouters J, van Kempen LC,
Durnez A, et al: Keratin 19: A key role player in the invasion of
human hepatocellular carcinomas. Gut. 63:674–685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Barlow M, Edelman M, Glick RD, Steinberg
BM and Soffer SZ: Celecoxib inhibits invasion and metastasis via a
cyclooxygenase 2-independent mechanism in an in vitro model of
Ewing sarcoma. J Pediatr Surg. 47:1223–1227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu X, Cai M, Ji F and Lou LM: The impact
of COX-2 on invasion of osteosarcoma cell and its mechanism of
regulation. Cancer Cell Int. 14:272014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee EJ, Choi EM, Kim SR, Park JH, Kim H,
Ha KS, Kim YM, Kim SS, Choe M, Kim JI and Han JA: Cyclooxygenase-2
promotes cell proliferation, migration and invasion in U2OS human
osteosarcoma cells. Exp Mol Med. 39:469–476. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naruse T, Nishida Y, Hosono K and Ishiguro
N: Meloxicam inhibits osteosarcoma growth, invasiveness and
metastasis by COX-2-dependent and independent routes.
Carcinogenesis. 27:584–592. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pucci S, Mazzarelli P, Nucci C, Ricci F
and Spagnoli LG: CLU “in and out”: Looking for a link. Adv Cancer
Res. 105:93–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang C, Jiang K, Kang X, Gao D, Sun C, Li
Y, Sun L, Zhang S, Liu X, Wu W, et al: Tumor-derived secretory
clusterin induces epithelial-mesenchymal transition and facilitates
hepatocellular carcinoma metastasis. Int J Biochem Cell Biol.
44:2308–2320. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marshall JC, Collins J, Marino N and Steeg
P: The Nm23-H1 metastasis suppressor as a translational target. Eur
J Cancer. 46:1278–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Takács-Vellai K: The metastasis suppressor
Nm23 as a modulator of Ras/ERK signaling. J Mol Signal. 9:42014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jarrett SG, Novak M, Dabernat S, Daniel
JY, Mellon I, Zhang Q, Harris N, Ciesielski MJ, Fenstermaker RA,
Kovacic D, et al: Metastasis suppressor NM23-H1 promotes repair of
UV-induced DNA damage and suppresses UV-induced melanomagenesis.
Cancer Res. 72:133–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ouatas T, Salerno M, Palmieri D and Steeg
PS: Basic and translational advances in cancer metastasis: Nm23. J
Bioenerg Biomembr. 35:73–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McCorkle JR, Leonard MK, Kraner SD,
Blalock EM, Ma D, Zimmer SG and Kaetzel DM: The metastasis
suppressor NME1 regulates expression of genes linked to metastasis
and patient outcome in melanoma and breast carcinoma. Cancer
Genomics Proteomics. 11:175–194. 2014.PubMed/NCBI
|
|
46
|
Price JT and Thompson EW: Mechanisms of
tumour invasion and metastasis: Emerging targets for therapy.
Expert Opin Ther Targets. 6:217–233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vogt PK: Fortuitous convergences: The
beginnings of JUN. Nat Rev Cancer. 2:465–469. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sze KM, Chu GK, Lee JM and Ng IO:
C-terminal truncated hepatitis B virus x protein is associated with
metastasis and enhances invasiveness by C-Jun/matrix
metalloproteinase protein 10 activation in hepatocellular
carcinoma. Hepatology. 57:131–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vleugel MM, Greijer AE, Bos R, van der
Wall E and van Diest PJ: c-Jun activation is associated with
proliferation and angiogenesis in invasive breast cancer. Hum
Pathol. 37:668–674. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Burgermeister E, Liscovitch M, Röcken C,
Schmid RM and Ebert MP: Caveats of caveolin-1 in cancer
progression. Cancer Lett. 268:187–201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu H, Shen H, Zhang Y, Zhong F, Liu Y, Qin
L and Yang P: CAV1 promotes HCC cell progression and metastasis
through Wnt/β-catenin pathway. PLoS One. 9:e1064512014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sáinz-Jaspeado M, Lagares-Tena L, Lasheras
J, Navid F, Rodriguez-Galindo C, Mateo-Lozano S, Notario V, Sanjuan
X, Del Garcia Muro X, Fabra A and Tirado OM: Caveolin-1 modulates
the ability of Ewing's sarcoma to metastasize. Mol Cancer Res.
8:1489–1500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Crépieux P, Kwon H, Leclerc N, Spencer W,
Richard S, Lin R and Hiscott J: I kappaB alpha physically interacts
with a cytoskeleton-associated protein through its signal response
domain. Mol Cell Biol. 17:7375–7385. 1997.PubMed/NCBI
|
|
54
|
Torabian SZ, de Semir D, Nosrati M,
Bagheri S, Dar AA, Fong S, Liu Y, Federman S, Simko J, Haqq C, et
al: Ribozyme-mediated targeting of IkappaBgamma inhibits melanoma
invasion and metastasis. Am J Pathol. 174:1009–1016. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kummer C and Ginsberg MH: New approaches
to blockade of alpha4-integrins, proven therapeutic targets in
chronic inflammation. Biochem Pharmacol. 72:1460–1468. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Park J, Song SH, Kim TY, Choi MC, Jong HS,
Kim TY, Lee JW, Kim NK, Kim WH and Bang YJ: Aberrant methylation of
integrin alpha4 gene in human gastric cancer cells. Oncogene.
23:3474–3480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Qian F, Vaux DL and Weissman IL:
Expression of the integrin alpha 4 beta 1 on melanoma cells can
inhibit the invasive stage of metastasis formation. Cell.
77:335–347. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gosslar U, Jonas P, Luz A, Lifka A, Naor
D, Hamann A and Holzmann B: Predominant role of alpha 4-integrins
for distinct steps of lymphoma metastasis. Proc Natl Acad Sci USA.
93:4821–4826. 1996. View Article : Google Scholar : PubMed/NCBI
|