Introduction
Currently, ~33.2 million people are living with a
human immunodeficiency virus (HIV) infection worldwide, and 2.5
million new cases occur annually, as reported by the Joint United
Nations Programme on HIV and acquired immunodeficiency syndrome
(AIDS) in 2011 (1). The HIV/AIDS
pandemic has become a global health threat and a great deal of
effort has been focused on improving the HIV treatment.
Highly-active antiretroviral therapy (HAART) is one
of the greatest successes of modern medicine and brings hope to
HIV-positive patients by improving their life quality and
postponing AIDS; however, emergence of drug resistant variants,
side effects and poor adherence demonstrates the necessity for
novel therapeutic approaches (2).
RNA interference (RNAi) is a highly conserved and sequence-specific
post-transcriptional gene silencing mechanism in eukaryotes, which
has shown the most promising results in repressing viral
replication (3–5). RNAi-based gene therapy has been widely
used to inhibit HIV-1 replication in cell culture systems and
animal models (6,7). However, emerging HIV-1 escape mutants
due to the error-prone nature of viral reverse transcriptase is a
major obstacle to therapy (8,9).
Numerous efforts have been made to minimize the
emergence of escape mutants. Song et al (10) first demonstrated the inhibition of
HIV replication using synergistic small interfering RNA (siRNA)
targeting cellular C-C chemokine receptor type 5 and viral p24
transcripts (10). In addition, the
emergence of viral escape mutants was shown to be delayed by using
a multiple short-hairpin RNA (shRNA) approach from a single vector
(11). An alternative approach has
been applied using the simultaneous expression of multiple siRNAs
from polycistronic microRNAs (miRNAs or miRs) (12,13).
Although predominantly focused on combinatorial RNAi to reduce
HIV-1 viral escape mutations, the multiple RNAi approach may induce
side effects, such as cell toxicity and off-target effects. It has
been demonstrated that high expression levels of shRNA can induce
cell toxicity due to over-saturation of RNAi (14). Furthermore, off-target effects have
been shown to increase the occurrence of unwanted cellular mRNA
cleavage or suppression (15).
In line with previous studies (16,17), the
present study investigated a specific miRNA expression vector
targeting 3-untranslated regions (3-UTRs), which is highly
conserved among the HIV subtypes A, B and C, and present in all
HIV-1 transcripts. The main aim of the present study was to design
an artificial miRNA with maximum efficacy and minimal side effects
(18). Furthermore, the present
study may strengthen the evidence in support of controlled clinical
trials targeting viral replication in patients infected with
HIV.
Materials and methods
Target sequence design
HIV-1 3-UTR mRNA reference sequences (accession no.
AF033819.3) were used to identify candidate target sequences using
BLOCK-iT™ RNAi Designer (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA USA). Candidate target sequences were
subsequently analyzed with all HIV-1 subtypes in the Los Alamos
National Laboratory Database (http://www.hiv.lanl.gov) in order to identify the
target sequence with the optimal complete identity among the major
HIV-1 subtypes, as shown in Table I.
The final target sequence (8418–8438 nt) was conserved (100%
identity) among HIV-1 clades A-C. Target sequence was then
incorporated into the 5′ and 3′ arm of hsa-miR30a containing a
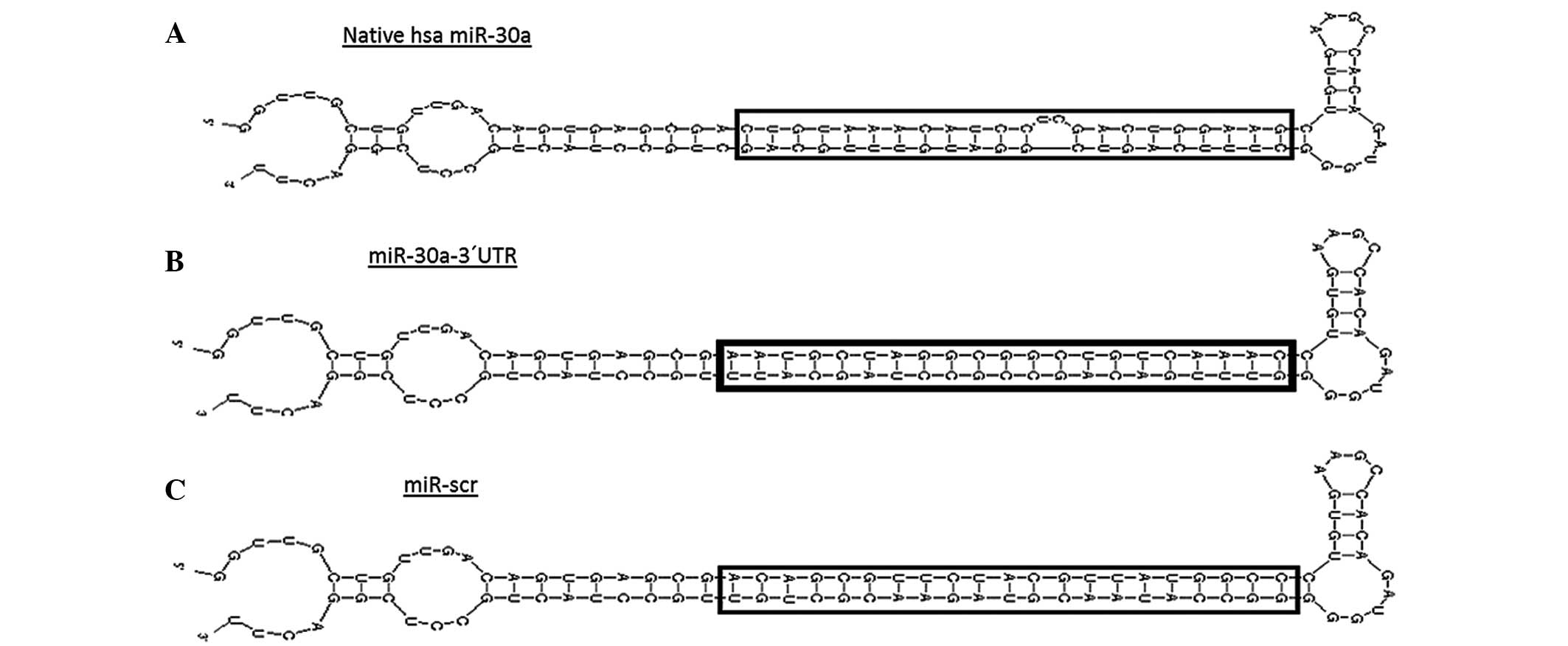
natural loop, and the 5′ and 3′ flank sequences (Fig. 1A) together with HIV-1 3′UTR target
sequence (Fig. 1B), which was
referred to as miR-3′UTR in the present study.
 | Table I.Basic local alignment search tool of
target sequence between clades A, B and C. |
Table I.
Basic local alignment search tool of
target sequence between clades A, B and C.
| GenBank accession
no. | Subtype | Query:
GTTTGACAGCCGCCTAGCATT | Identities |
|---|
| AF538657 | A1 |
549–5′-GTTTGACAGCCGCCTAGCATT-3′-569 | 100% |
| JN944919 | B |
8418––5′-GTTTGACAGCCGCCTAGCATT-3′-8438 | 100% |
| JX985439 | C |
555––5′-GTTTGACAGCCGCCTAGCATT-3′-575 | 100% |
UNAFold online software (http://unafold.rna.albany.edu/) was used to predict
target accessibility and artificial miR-3′UTR thermodynamic
stability. In order to demonstrated that miR-3′UTR was capable of
acting directly on HIV sequence, scrambled sequence was designed by
Genescrip Bioanformatic online tools (https://www.genscript.com). The scramble sequence was
designed according to the target sequence and incorporated into the
5′ and 3′ arm of hsa-miR30a according to the GC percentage and the
melting temperature of the target sequence (Fig. 1C). hsa-miR-29a containing HIV-1 3′UTR
seed site was also used as a positive control.
Construction of plasmids
To generate an artificial miR-3′-UTR expression
plasmids were generated by Metabion International AG (Steinkirchen,
Germany) by annealing two forward and reverse synthetic
oligonucleotide strands and introducing them into the
HindIII/BamHI sites of pCDNA 3 and pAc enhanced green
fluorescent protein (EGFP) plasmids (Promega Corporation, Madison,
WI, USA). The strands were as follows: Forward,
AGCTTGGTTGCTGTTGACAGTGAGCGAATGCTAGGCGGCTGTCAAACCTGTGAAGCCACAGATGGGGTTTGACAGCCGCCTAGCATTTGCCTACTGCCTCGGACTTG
and reverse,
GATCCAAGTCCGAGGCAGTAGGCAAATGCTAGGCGGCTGTCAAACCCCATCTGTGGCTTCACAGGTTTGACAGCCGCCTAGCATTCGCTCACTGTCAACAGCAACCAA.
A pre-miR-scrambled expression plasmid was constructed by inserting
the annealed forward and reverse strand scrambled oligonucleotides.
Target sequence was then synthesized chemically by adding 50 extra
nucleotides to the 5 and 3 sites, followed by insertion into the
cloning XhoI/NotI site of the psi-CHECK-2 vector
(Promega Corporation). This construct was referred to as
psi-CHECK-HIV 3′-UTR in the present study.
Cell culture and transfection
Human embryonic kidney (HEK) 293T cells were
purchased from the National Cell Bank of Iran (Tehran, Iran) and
maintained in Dulbeccos modified Eagles medium (DMEM) supplemented
with 10% fetal bovine serum (FBS), penicillin (100 U/ml) and
streptomycin (100 µg/ml; all Thermo Fisher Scientific, Inc.) at
37°C. The cells were trypsinized (Thermo Fisher Scientific, Inc.) 1
day prior to transfection, resuspended in DMEM without antibiotics
and seeded in 24-well plates at a density of 1.2×105
cells/well.
miR30a, miR-scr and pNL4-3 plasmids were transfected
with Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) into cells according to the manufacturers
protocol. The transfection efficiency of EGFP expression was
measured by fluorescence microscopy (BX51; Olympus Corporation,
Tokyo, Japan). Co-transfection with various plasmids was performed
as follows. Two groups were used to test target gene expression;
one was treated with pNL4-3+ miR-30a, whereas the other was treated
with pNL4-3+ miR-scr. Untreated cells served as the control group.
Three groups were used to assess viral production: Group 1,
pNL4-3+miR-3a experimental group; group 2, pNL4-3+miR29a positive
control; and group 3, pNL4-3+miR-scr negative control.
Stem-loop reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
RT-qPCR
Cells were lysed via the freeze/thaw method and
total RNA was extracted from the HEK-293T cells expressing
artificial miRNAs using TRI reagent (Sigma-Aldrich, St. Louis, MO,
USA). Nanodrop 2000 spectrophotometer was used for RNA
quantification (Nanodrop Technologies; Thermo Fisher Scientific,
Inc.). To digest unwanted DNA, 1 µg RNA was treated with DNase
(Thermo Fisher Scientific, Inc.) according to the manufacturers
protocol. Specific artificial miR-3-UTR stem-loop
(5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTTTGA-3′) and
small nucleolar RNA stem loop primers
(5′GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCTTCCT-3′) were
designed using Primer 3 (version 4.0.0; http://bioinfo.ut.ee/primer3–0.4.0/). Using these
specific primers, 100 ng of total RNA obtained from the DNase
treatment was reverse transcribed into cDNA using avian
myeloblastosis virus reverse transcriptase (Roche Diagnostics GmbH,
Mannheim, Germany). To detect artificial miR-3-UTR, stem-loop qPCR
was applied using 2 µl cDNA, specific primers (0.5 µm miR-3-UTR
forward and stem-loop reverse primers) and iQ SYBR Green Supermix
(Bio-Rad Laboratories, Inc., Hercules, USA) at a final volume of 20
µl. The PCR conditions were as follows: 94°C for 15 sec, 58°C for
30 sec and 72°C for 30 sec for 45 cycles, followed by melting curve
analysis (Rotor-Gene Q; Qiagen GmbH, Hilden, Germany) to assess the
specificity of SYBR amplification. Small nucleolar RNA was used as
the endogenous control.
RT-qPCR was conducted in order to measure mRNA
expression. Total RNA was extracted from the transfected cells and
DNase treatment was performed as described above. cDNA was
synthesized using 200 ng DNase-treated total RNA and 5 µm random
primer with avian myeloblastosis virus-reverse transcriptase (Roche
Diagnostics GmbH) in 20 µl reaction mixture according to the
manufacturers protocol. iQ SYBR Green Supermix (Bio-Rad
Laboratories, Inc.) was used to measure the mRNA expression levels
with an iQ master containing dNTPs, 50 U/ml iTaq DNA polymerase, 6
mM MgCl2, SYBR Green I, 20 nM fluorescein, 0.5µl cDNA
and 0.5 µM of each primer (forward, 3′UTR-GCCGCCTAGCATTTCATCAC-5′
and reverse 3′UTR-GAAAGTCCCCAGCGGAAAGT-5′). PCR conditions were
94°C for 5 min enzyme activation, followed by 94°C for 30 sec, 57°C
for 30 sec and 72°C for 30 sec with a melting curve analysis. The
2−∆∆Cq method (19) was
used to compare the HIV gene expression levels in cells exposed and
unexposed to artificial miR-3′UTR. GAPDH was used to normalize the
expression levels of target genes.
Dual luciferase reporter assay
Following the addition of 500 ng psi-CHECK-HIV 3-UTR
and 500 ng artificial miR-3-UTR to Lipofectamine® 2000,
plasmids were co-transfected into HEK-293T cells. To measure
luciferase activity, the transfected cells were lysed 24 h
post-transfection using passive lysis buffer (Promega Corporation).
Firefly and Renilla luciferase activities were determined using a
FB 12 tube luminometer (Berthold Detection Systems GmbH, Pforzheim,
Germany) according to the manufacturers protocol. Briefly, firefly
Luciferase expression was used to normalize target specific
Renilla luciferase expression. Relative luciferase activity
levels were calculated from the ratio of Firefly to Renilla
luciferase activity.
HIV challenge and p24 antigen
assay
To assess the inhibition of artificial miR-3-UTR on
virus production, HEK-293T cells were co-transfected with 500 ng
HIV-1 infectious pNL4-3 plasmid clone (Pasteur Institute of Iran,
Tehran, Iran), 500 ng artificial miR-3-UTR and 500 ng miR-scr
constructs, using Lipofectamine® 2000. Virus production
was measured by Elecsys 2010 (Roche Diagnostics GmbH) 2 days
post-transfection. Briefly, 200 µl cell culture supernatant was
collected and centrifuged at 300 × g for 10 min to monitor CA-P24
levels using an HIV Combi assay kit (Roche Diagnostics GmbH)
according to the manufacturers protocol.
Statistical analysis
Statistical analyses were performed using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA). Luciferase activity was
measured using ratio comparison and the results of the Combi assay
were determined using repeated analysis of variance.
Results
Assessment of expression and
processing of artificial miR-3-UTR
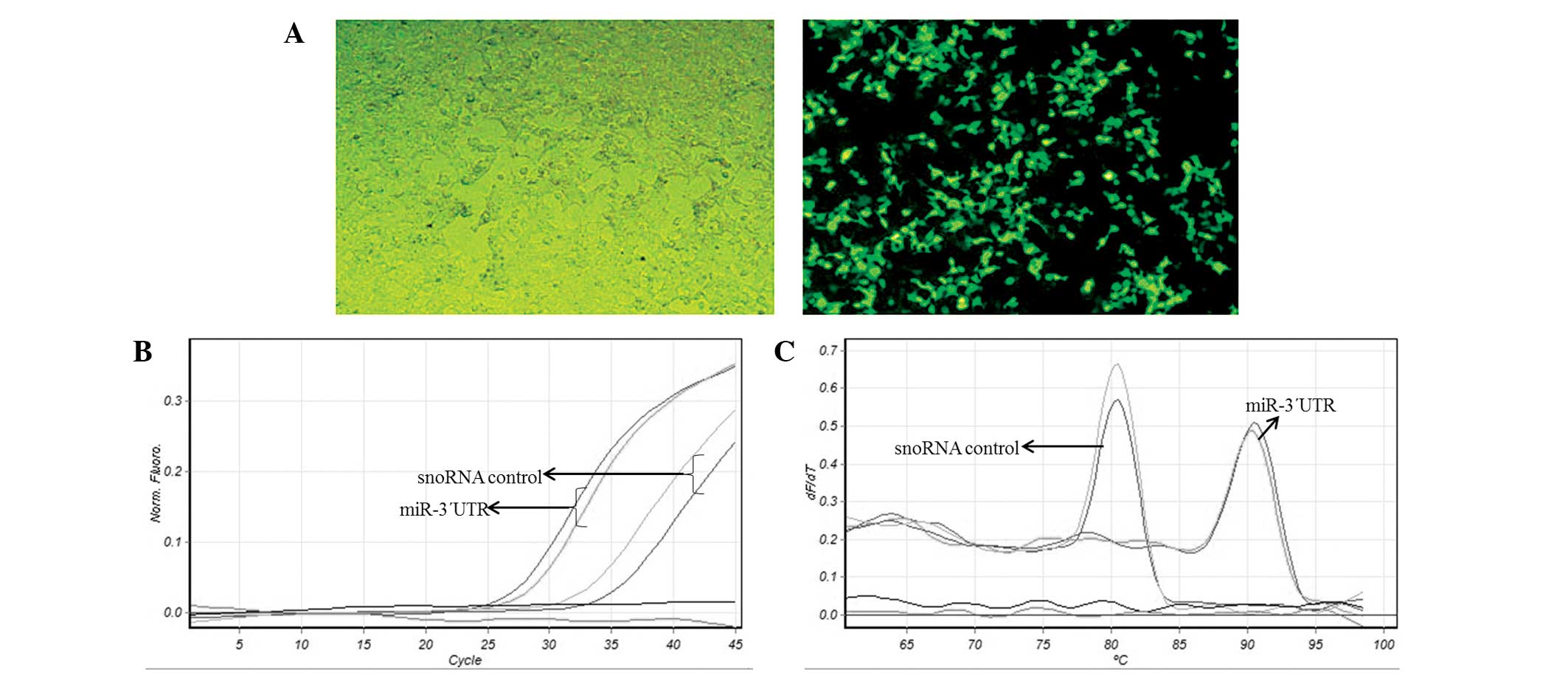
To examine the expression of miR-3-UTR, EGFP levels
were determined by fluorescence microscopy. After 24 h
post-transfection of EGFP-miR-3-UTR into HEK-293T cells, the cells
were analyzed for EGFP expression (Fig.
2A). Stem-loop RT-qPCR was also performed on transfected
HEK-293T cells to detect miR-3-UTR expression levels (Fig. 2B). Melting curve analysis was
performed to check the accuracy of the experiment (Fig. 2C). In addition, the expected size of
the stem-loop RT-qPCR product was determined by agarose gel
electrophoresis, indicating that processing by Drosha-DGCR-8 and
Dicer was performed correctly, to ensure that the anticipated
sequences were successfully incorporated in the RNA-induced
silencing complex (RISC). No miR-3-UTR was observed in cells
transfected with empty EGFP expression plasmids.
Suppression of specific target
sequences by miR-3′-UTR
The knockdown activity levels of miR-3-UTR were
measured using a luciferase activity reporter assay. A 50 nt HIV-1
3′-UTR target sequence was inserted into the 3-UTR of the hRlu gene
in psi-CHECK-2 and co-transfected with artificial miR-3-UTR into
HEK-293T cells.
Dual luciferase assay was used to monitor changes in
the expression levels of Renilla luciferase contained within
the target sequence. The specific suppression activity of miR-3-UTR
was measured by determining the Renilla luciferase
expression, which was normalized to that of Firefly luciferase. As
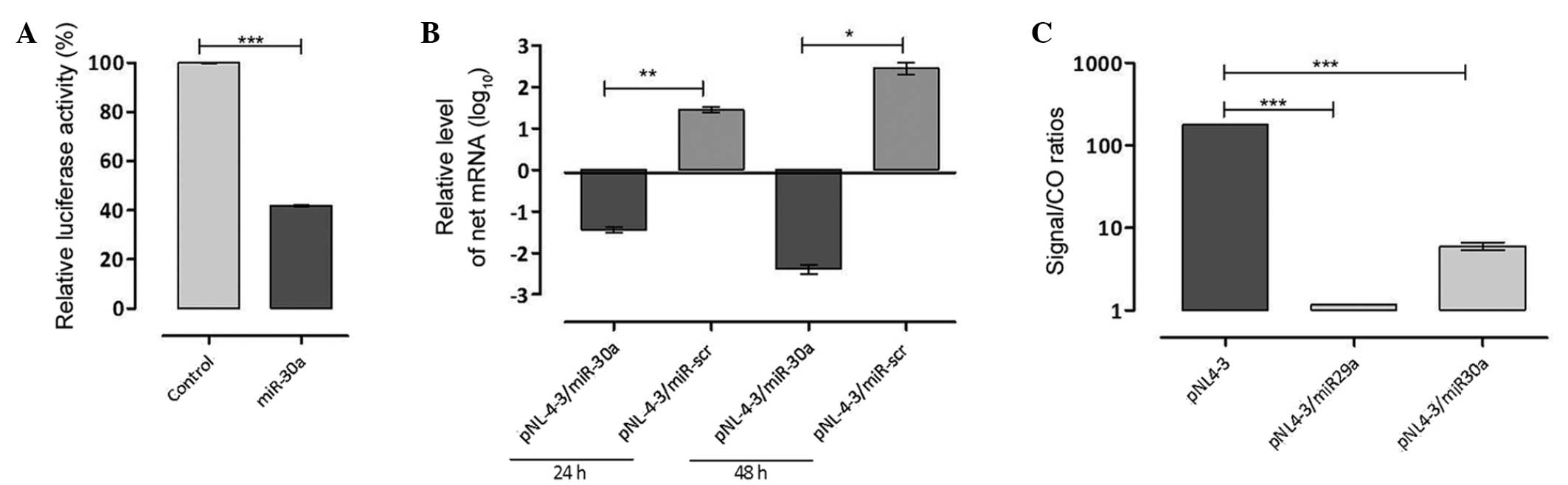
indicated in Fig. 3A, the artificial
miR-3′-UTR was capable of significantly reducing luciferase
activity levels (P<0.0001).
Inhibition of HIV-1 replication by
artificial miR-3-UTR in cell culture
To ascertain whether HIV replication was inhibited
by the miR-3′-UTR construct, the functional inhibition activity of
artificial miR-3′-UTR was analyzed by RT-qPCR. For this purpose,
HEK-293T cells were transfected with artificial miR-3′-UTR
expression plasmid and, simultaneously, challenged with a pNL4-3
strain of HIV-1. The miR-scr expression plasmid was used as a
negative control. 3′-UTR expression levels were normalized to GAPDH
mRNA expression levels (Fig. 3B).
The results obtained from the RT-qPCR analysis indicated that cells
transfected with the pNL4-3 and miR-3′-UTR showed a significant
reduction in 3′-UTR expression levels after 24 and 48 h (P<0.001
and P<0.01, respectively) compared with cells transfected with
the pNL4-3 and miR-scr as a negative control.
The levels of viral capsid p24 in supernatants were
subsequently determined in the transfected cells two days
post-transfection to monitor virus production. To investigate the
inhibitory efficiency, artificial miR-3′-UTR was co-transfected
with pNL4–3. Furthermore, co-transfected hsa-miR-29a and pNL4-3
were used as a positive control, whereas transfected pNL4-3 was
used as a negative control. As indicated in Fig. 3C, inhibitory activity was observed
with artificial miR-3′-UTR as compared with the negative control.
However, the inhibitory activity of artificial miR-3′-UTR was
relatively low when compared with the miR-29a positive control.
Discussion
The advent of HAART to treat HIV-1-positive patients
has improved the lives of patients infected with HIV. Although
postponing the development of AIDS in patients with HIV-1 is the
main advantage of these drugs (20),
there are side effects associated with prolonged use of antiviral
drug therapy, including drug toxicity and emerging drug resistant
variants (21). Several strategies
have been applied to reduce these side effects as well as to
inhibit HIV-1 replication in both in vivo and in
vitro systems. Among these, RNAi has recently been considered
as a promising gene therapy tool (22,23).
Synthetic mature siRNAs were used as first generation anti-HIV-1
RNAi-based gene therapy (24). The
short duration of efficacy in mammalian cells has questioned the
use of siRNAs to treat chronic diseases such as HIV-1 infection
(25). Alternative strategies have
been developed using shRNAs and long hairpin RNAs that are
predominantly expressed by Pol III promoters, which can be
transcribed from expression plasmids (26,27).
High expression levels of transcripts induced by the Pol III
promoter may result in competition with the cellular RNAi and
saturation of the miRNA signaling pathway due to shRNA
overexpression. Thus, induced cell toxicity is an obstacle to this
approach in clinic trials (28).
However, to overcome this problem, decreased cell death has been
observed following the expression with Pol II promoter, as compared
with Pol III promoter (29). To
reduce cell toxicity, an anti-HIV vector under Pol II promoter was
constructed in the present study, and the lowest cell toxicity was
observed using an miR-3′-UTR construct.
Artificial miRNA has also been introduced as a tool
for RNAi-based gene therapy, by incorporating siRNA encoding
sequences into a human pre-miRNA backbone (30,31). In
the present study, an hsa-pre-miR-30a backbone was used to generate
siRNA against HIV-3′-UTR. As artificial miR-3′-UTR has a natural
miRNA backbone, this construct was processed efficiently by nuclear
microprocessor protein complexes and cytosolic Dicer proteins. The
results obtained from the stem-loop RT-qPCR demonstrated that
cellular miRNA backbones were able to cause efficient processing of
guide strand into RISC. The error-prone nature of viral reverse
transcriptase is an obstacle not only to the development of HIV
drug treatments, but also to the effective design of anti-HIV RNAi.
The use of mono anti-HIV siRNA is limited due to the rapid
emergence of viral escape mutants (32,33). To
overcome this limitation, the use of simultaneous multiple shRNAs
against various conserved regions of HIV-1 has been developed.
However, off-target effects are the main limitation of this
approach. Increased off-target risk has been described using
multiple shRNA anti-HIV strategies (34–36). To
reduce the chance of mutant escape, conserved targeting sequences
among different virus strains are important.
In conclusion, the use of a single anti-HIV miRNA
targeting all HIV-1 transcripts can reduce off-target effects,
highlighting the importance of an artificial miR-3′-UTR construct
as an efficient and alternative RNA-based gene therapy. These
results strengthen the evidence in support of controlled clinical
trials targeting viral replication in patients infected with
HIV.
Acknowledgements
The present study was funded and supported by Tehran
University of Medical Sciences (TUMS; grant no. 92-02-27-22878).
This study was also part of a PhD thesis supported by TUMS (grant
no. 240/66). The authors of the present study are grateful to Dr
Keyhan Azadmanesh (Pasteur Institute of Iran) and Dr
Aghaee-Bakhtiari (Stem Cell Technology Research Center, Tehran,
Iran) for their support, Dr Vahid Salimi and Dr Farhad Rezaie for
assisting with fluorescence microscopy, and Ms Maryam Naseri (all
Virology Department, School of Public Health, Tehran University of
Medical Sciences, Tehran, Iran) and Ms Sara Asgharpoor (Stem Cell
Technology Research Center) for their help with practical
experiments.
References
|
1
|
World Health Organization, HIV/AIDS JUNPo
and UNICEF: Global HIV/AIDS response: Epidemic update and health
sector progress towards universal access. Progress Report 2011. WHO
Press. (Geneva). 2011.
|
|
2
|
Ambrosioni J, Calmy A and Hirschel B: HIV
treatment for prevention. J Int AIDS Soc. 14:282011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hao L, Sakurai A, Watanabe T, Sorensen E,
Nidom CA, Newton MA, Ahlquist P and Kawaoka Y: Drosophila RNAi
screen identifies host genes important for influenza virus
replication. Nature. 454:890–893. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kelly EJ, Hadac EM, Greiner S and Russell
SJ: Engineering microRNA responsiveness to decrease virus
pathogenicity. Nat Med. 14:1278–1283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pu C, Wang L, Miao X, Zhang Y, Jiang C,
Liu W, Sun W and Gao Q: Optimized tandem amLRNA mediates stronger
inhibitory effects on hepatitis B virus infection. J Gastrointestin
Liver Dis. 20:271–278. 2011.PubMed/NCBI
|
|
6
|
Ter Brake O, Legrand N, Von Eije KJ,
Centlivre M, Spits H, Weijer K, Blom B and Berkhout B: Evaluation
of safety and efficacy of RNAi against HIV-1 in the human immune
system (Rag-2(−/-)gammac(−/-)) mouse model. Gene Ther. 16:148–153.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Christensen HS, Daher A, Soye KJ, Frankel
LB, Alexander MR, Lainé S, Bannwarth S, Ong CL, Chung SW, Campbell
SM, et al: Small interfering RNAs against the TAR RNA binding
protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency
virus type 1 long terminal repeat expression and viral production.
J Virol. 81:5121–5131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee NS, Dohjima T, Bauer G, Li H, Li MJ,
Ehsani A, Salvaterra P and Rossi J: Expression of small interfering
RNAs targeted against HIV-1 rev transcripts in human cells. Nat
Biotechnol. 20:500–505. 2002.PubMed/NCBI
|
|
9
|
Zhou J, Neff CP, Liu X, Zhang J, Li H,
Smith DD, Swiderski P, Aboellail T, Huang Y, Du Q, et al: Systemic
administration of combinatorial dsiRNAs via nanoparticles
efficiently suppresses HIV-1 infection in humanized mice. Mol Ther.
19:2228–2238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song E, Lee SK, Dykxhoorn DM, Novina C,
Zhang D, Crawford K, Cerny J, Sharp PA, Lieberman J, Manjunath N
and Shankar P: Sustained small interfering RNA-mediated human
immunodeficiency virus type 1 inhibition in primary macrophages. J
Virol. 77:7174–7181. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ter Brake O, Liu YP, Centlivre M, von Eije
KJ, Berkhout B and 't Hooft K: Lentiviral vector design for
multiple shRNA expression and durable HIV-1 inhibition. Mol Ther.
16:557–564. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aagaard LA, Zhang J, von Eije KJ, Li H,
Saetrom P, Amarzguioui M and Rossi JJ: Engineering and optimization
of the miR-106b cluster for ectopic expression of multiplexed
anti-HIV RNAs. Gene Ther. 15:1536–1549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu YP, Haasnoot J, ter Brake O, Berkhout
B and Konstantinova P: Inhibition of HIV-1 by multiple siRNAs
expressed from a single microRNA polycistron. Nucleic Acids Res.
36:2811–2824. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grimm D, Streetz KL, Jopling CL, Storm TA,
Pandey K, Davis CR, Marion P, Salazar F and Kay MA: Fatality in
mice due to oversaturation of cellular microRNA/short hairpin RNA
pathways. Nature. 441:537–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jackson AL, Bartz SR, Schelter J,
Kobayashi SV, Burchard J, Mao M, Li B, Cavet G and Linsley PS:
Expression profiling reveals off-target gene regulation by RNAi.
Nat Biotechnol. 21:635–637. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herrera-Carrillo E and Berkhout B: The
impact of HIV-1 genetic diversity on the efficacy of a
combinatorial RNAi-based gene therapy. Gene Ther. 22:485–495. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scarborough RJ, Adams KL, Daher A and
Gatignol A: Effective inhibition of HIV-1 production by short
hairpin RNAs and small interfering RNAs targeting a highly
conserved Site in HIV-1 Gag RNA is optimized by evaluating
alternative length formats. Antimicrob Agents Chemother.
59:5297–5305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nejati A, Shahmahmoodi S, Shoja Z and
Marashi SM: Efficient inhibition of HIV replication by targeting
3UTR transcripts using new modified miR-30a. 4th World Congress on
Virology (San Antonio, TX, USA). 2014.
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Richman DD, Margolis DM, Delaney M, Greene
WC, Hazuda D and Pomerantz RJ: The challenge of finding a cure for
HIV infection. Science. 323:1304–1307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blankson JN, Persaud D and Siliciano RF:
The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med.
53:557–593. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chung J, Zhang J, Li H, Ouellet DL,
DiGiusto DL and Rossi JJ: Endogenous MCM7 microRNA cluster as a
novel platform to multiplex small interfering and nucleolar RNAs
for combinational HIV-1 gene therapy. Hum Gene Ther. 23:1200–1208.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou J, Neff CP, Swiderski P, Li H, Smith
DD, Aboellail T, Remling-Mulder L, Akkina R and Rossi JJ:
Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a
chemically synthesized aptamer with a sticky bridge. Mol Ther.
21:192–200. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Coburn GA and Cullen BR: Potent and
specific inhibition of human immunodeficiency virus type 1
replication by RNA interference. J Virol. 76:9225–9231. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Novina CD, Murray MF, Dykxhoorn DM,
Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P
and Sharp PA: SiRNA-directed inhibition of HIV-1 infection. Nat
Med. 8:681–686. 2002.PubMed/NCBI
|
|
26
|
Saayman S, Barichievy S, Capovilla A,
Morris KV, Arbuthnot P and Weinberg MS: The efficacy of generating
three independent anti-HIV-1 siRNAs from a single U6 RNA Pol
III-expressed long hairpin RNA. PLoS One. 3:e26022008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
ter Brake O, Konstantinova P, Ceylan M and
Berkhout B: Silencing of HIV-1 with RNA interference: A multiple
shRNA approach. Mol Ther. 14:883–892. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vickers TA, Lima WF, Nichols JG and Crooke
ST: Reduced levels of Ago2 expression result in increased siRNA
competition in mammalian cells. Nucleic Acids Res. 35:6598–6610.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lo HL, Chang T, Yam P, Marcovecchio PM, Li
S, Zaia JA and Yee JK: Inhibition of HIV-1 replication with
designed miRNAs expressed from RNA polymerase II promoters. Gene
Ther. 14:1503–1512. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McManus MT, Petersen CP, Haines BB, Chen J
and Sharp PA: Gene silencing using micro-RNA designed hairpins.
RNA. 8:842–850. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zeng Y, Wagner EJ and Cullen BR: Both
natural and designed micro RNAs can inhibit the expression of
cognate mRNAs when expressed in human cells. Mol Cell. 9:1327–1333.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Das AT, Brummelkamp TR, Westerhout EM,
Vink M, Madiredjo M, Bernards R and Berkhout B: Human
immunodeficiency virus type 1 escapes from RNA
interference-mediated inhibition. J Virol. 78:2601–2605. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
von Eije KJ, ter Brake O and Berkhout B:
Human immunodeficiency virus type 1 escape is restricted when
conserved genome sequences are targeted by RNA interference. J
Virol. 82:2895–2903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Birmingham A, Anderson EM, Reynolds A,
Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E,
Robinson K, Karpilow J, et al: 3′-UTR seed matches, but not overall
identity, are associated with RNAi off-targets. Nat Methods.
3:199–204. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fedorov Y, Anderson EM, Birmingham A,
Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS and
Khvorova A: Off-target effects by siRNA can induce toxic phenotype.
RNA. 12:1188–1196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jackson AL, Burchard J, Schelter J, Chau
BN, Cleary M, Lim L and Linsley PS: Widespread siRNA ‘off-target’
transcript silencing mediated by seed region sequence
complementarity. RNA. 12:1179–1187. 2006. View Article : Google Scholar : PubMed/NCBI
|