Introduction
Infertility is the absence of pregnancy within one
year of unprotected normal intercourse, and remains the leading
reproductive health problem affecting 15% of couples of
reproductive age (1). However, male
infertility (MI), defined as the incapability of a male to conceive
with a fertile female, accounts for 50% of all infertility cases,
and affects 1/20 males worldwide (2,3). Despite
the progress in diagnostic methods to identify MI, such as physical
examination, reproductive history and semen analysis, the
underlying etiology of MI is poorly understood (4,5). In
general, MI is associated with non-genetic risk factors such as
testicular torsion or trauma, seminal tract infections,
hypogonadotropic hypogonadism, cryptorchidism, gonadal dysgenesis,
idiopathic oligozoospermia, reproductive channel obstruction, and
anti-sperm antibodies (6–8). More recently, genetic risk factors in
MI have received significant attention with the identification that
single gene mutations and chromosomal aberrations account for
10–15% of MI cases (9–11).
The H19 gene is one of the first imprinting
genes to have been identified and is expressed from the maternal
allele in humans and mice, with its expression highly restricted to
heart and skeletal muscles in adults (12). The H19 gene is located on the
human 11p15.5 chromosome and on the distal section of mouse
chromosome 7, spanning ~2.5 kb and containing 5 exons and 4
intrinsic factors (13,14). The H19 gene encodes a
non-coding RNA lacking an open reading frame and its expression is
highly regulated by DNA elements (15,16).
H19 has a role in the regulation of body weight and cell
multiplication, and is dysregulated in certain types of cancer
(17,18). Recently, it has been shown that the
H19 gene encodes microRNA (miR), namely miR-675 which is
important in tumorigenesis (19).
H19 expression was present in numerous types of cancer
including hepatocellular carcinoma, choriocarcinoma, breast cancer,
bladder cancer, colorectal cancer, testicular cancer, esophageal
cancer and ovarian cancer (20).
Notably, aberrant methylation in H19 gene alters the onset
of MI and affects individual susceptibility to MI (21,22). The
present study hypothesized that aberrant methylation of the
H19 gene is associated with MI.
Materials and methods
Ethical statement
The study was approved by the academic board and the
ethics committee of the First Affiliated Hospital of South China
University (Hengyang, China). All eligible patients conformed to
the study inclusion criteria. Written-informed consent was obtained
from each eligible patient, and the residual semen following
analysis was used in the present investigation. All experimental
procedures were conducted according to the Declaration of Helsinki
(23).
Subjects and grouping
A total of 15 MI patients (age, 35.5±8.5 years) who
were admitted to the First Affiliated Hospital of South China
University were enrolled in the present study between March 2013
and June 2014 as the experimental group. Semen samples were
collected from the patients, with sperm concentration
≤20×106/ml, sperm (a+b) concentration ≤50%, and
percentage of morphologically normal sperm ≤15%. The inclusion
criteria for the present study were as follows: i) Patients who had
received a routine seminal fluid analysis ≥2 times at the First
Affiliated Hospital of South China University, with consistent
results; ii) seminal plasma fructose and neutral α-glucosidase were
expressed normally; iii) an anti-sperm antibody test was negative;
and iv) white blood cell semen counts wesre
<1×106/ml. Conversely, patients were excluded if they
had a history of hypertension, cardiovascular diseases, metabolic
diseases, acquired immune deficiency syndrome, syphilis, hepatitis
B and other infectious diseases, and a long-term history of heavy
drinking or contact with poison. Furthermore, patients with
varicocele, inflammation of the urinary and reproductive system, or
genital tract infections, including those caused by Chlamydia
trachomatis, Neisseria gonorrhoeae and Ureaplasma
aurealyticum, were excluded. In addition, patients with
infertility caused by diseases such as non-inflammatory sexual
dysfunction of accessory sex gland or infection were excluded from
the investigation.
A total of 15 fertile males (age, 32.5±6.5 years)
with normal semen analyses were enrolled as the control group. The
semen samples were determined to be normal in accordance with the
standards of the World Health Organization (WHO) (24): Normal seminal liquefaction; no sperm
agglutination; volume >2.0 ml; pH>7.2; sperm concentration
>20×106/ml; total sperm number for one ejaculation
≥40×106/ml; sperm progressive motility within 60 min
≥50%, or sperm fast progressive motility≥25%; percentage of
morphologically normal sperm ≥15%; survival rate ≥60%; and white
blood cell concentration <1×106/ml.
Semen samples
The semen samples from MI patients and control
subjects were obtained by masturbation following 2–7 days of sexual
abstinence, collected in disposable semen cups, and immediately
placed at 37°C. Rapidly following seminal liquefaction, the
concentration and motility of the raw semen samples were examined
according to the Fourth Edition of the WHO Laboratory Manual for
the Examination of Human Semen and Sperm-Cervical Mucus Interaction
(24). Staining was performed with a
modified Papanicolaou staining technique recommended by the WHO
(24). Briefly, prepared smears were
fixed in 95% (v/v) ethanol for ~15 min, rehydrated in an ethanol
gradient and stained with hematoxylin (Beyotime Institute of
Biotechnology, Haimen, China) for 4 min. Following washing with
pure-water for 30 sec and 4~8 times immersion in acidic ethanol for
1 sec each, the smears were dehydrated using an ethanol gradient
and stained with Orange G6 (Beijing Leagene Biotech Co., Ltd.,
Beijing, China) for 1 min. Subsequently, the semen samples were
washed three times for 30 sec each with 95% ethanol, followed by
staining with EA-50 Green (Beijing Leagene Biotech Co., Ltd.) for 1
min and washing two times for 30 sec each with 95% ethanol and two
times for 15 sec each with 100% (v/v) ethanol. After air drying and
mounting with neutral balsam (Sigma-Aldrich, St. Louis, MO, USA),
multiple regions were selected on the smears for morphological
evaluation under a BH-2 optical microscope (Olympus Corporation,
Tokyo, Japan), using the domestic WLJY-9000 WeiLi Color Sperm
Analysis System (Beijing Weili New Century Science & Tech.
Deve., Co., Ltd., Beijing, China) for assessing sperm quality.
Following the morphological assessment, the
remaining sperm were separated using Percoll density gradient
centrifugation. Briefly, 2 ml 40% Percoll liquid (Sigma-Aldrich)
was added to the bottom of a conical centrifuge tube. A transfer
pipette was then plunged into the bottom of the tube and 2 ml 80%
Percoll liquid (Sigma-Aldrich) was added below the 40% Percoll
liquid to form a two-layer Percoll density gradient. The semen
samples were added on top of the 40% Percoll, and centrifugal
separation was performed at 400 × g for 20 min at room temperature.
Following discarding of the supernatant, 1 ml Earle's balanced salt
solution (Sigma-Aldrich) was added to the sediment prior to
centrifugation at 1,000 × g for 5 min at room temperature. The
resulting supernatant was discarded and 0.1–0.2 ml sperm suspension
was maintained for the subsequent procedures.
DNA extraction and polymerase chain
reaction (PCR) amplification
Genomic DNA was extracted from the sperm using a
TIANamp Blood DNA kit (cat. no. DP304-02; Tiangen Biotech Co.,
Ltd., Beijing, China) to obtain 30 µl dissolved DNA solution. The
purity of the DNA was assessed by the A260/A280 ratio, and the DNA
concentration was calculated from the absorption readings obtained
from a spectrophotometer (DU 800; Beckman Coulter, Inc., Brea, CA,
USA). Bisulfite modification of the DNA was conducted using an
EpiTect Bisulfite kit (cat. no. 59104; Qiagen China Co., Ltd.,
Shanghai, China) in accordance with the manufacturer's protocol,
and the DNA samples were then preserved at −20°C. The
H19-specific primers used for the reaction were synthesized
by 216 bp fragments of imprinted gene H19 which contained 18
CpG loci (genbank accession no. AFl 25183; nucleotides 7881-809):
H19 forward, 5′-TGGGTATTTTTGGAGGTTTTTTT-3′, and reverse,
5′-ATAAATATCCTATTCCCAAATAA-3′ (Beijing Liuhe Huada Genetic Science
and Technology Co., Ltd., Beijing, China). PCR amplification was
performed using bisulfite-modified genomic DNA as template. The
total volume of the fundamental reaction system was 25 µl, which
consisted of 0.25 µl LA Taq polymerase, 2.5 µl 10X LA Taq
buffer, 4 µl dNTP (all Takara Biotechnology Co., Ltd., Dalian,
China), 3.25 µl deionized water, 5 µl sense primer (2 µM), 5 µl
antisense primer (2 µM), and 5 µl DNA template. The PCR was
conducted on a Mastercycler Gradient PCR thermal cycler (Eppendorf,
Hamburg, Germany), with the following conditions: Denaturation for
15 min at 95°C, 50 cycles of annealing at 95°C for 30 sec, 57°C for
30 sec and 72°C for 30 sec, and a final extension at 72°C for 5
min. The amplification products (5 µl) were analyzed by 2% agarose
gel electrophoresis.
Cloning and sequencing of PCR
products
The PCR products of the H19 gene in each
group were recovered and purified using an EasyPure Quick Gel
Extraction kit (cat. no. EG101; Beijing TransGen Biotech Co., Ltd.,
Beijing, China). All purified PCR products were cloned into pMD18-T
vectors (Takara Biotechnology Co., Ltd.) using a TA ligation system
(10 µl) containing 0.25 µl pMD18-T vector, 1 µl 10X ligase buffer,
1 µl ligase enzyme (both Takara Biotechnology Co., Ltd.), and 7.75
µl DNA fragments. The plasmid was transformed into E. coli DH5α
chemically-competent cells (cat. no. CB101-03; Tiangen Biotech Co.,
Ltd.), which were coated onto a Luria-Bertani agar plate
(Sigma-Aldrich) containing 50 µg/ml ampicillin (Promega
Corporation, Madison, WI, USA) and cultured overnight at 37°C until
white single colonies were observed. The white single colonies were
picked for culturing at 37°C overnight, after which a small
quantity of the plasmids were extracted using the
E.Z.N.A.® Plasmid Mini kit I (cat. no. D6943-01, Omega
Bio-Tek, Inc., Norcross, GA, USA) and verified by enzyme digestion
with RsaI (Fermentas; Thermo Fisher Scientific, Inc.,
Pittsburgh, PA, USA). The products of the enzyme digestion were
separated by agarose gel electrophoresis and positive clones were
identified by staining with ethidium bromide (Omega Bio-Tek, Inc.).
The DL 2000 Marker (Takara Biotechnology Co., Ltd.) served as a
reference. H19-positive bacterial clones were selected from
each group and sent to GENEWIZ, Inc. (Beijing, China) for
sequencing using a universal primer. The sequence analysis was
conducted using a BiQ analyzer 2.0 software for DNA methylation
analysis (http://biq-analyzer.bioinf.mpi-sb.mpg.de).
Statistical analysis
Data were presented as means ± standard deviation.
The statistical comparison of the overall methylation rate of the
H19 gene between groups was analyzed using a t-test.
Statistical analysis was conducted using SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant result.
Results
Baseline characteristics
As shown in Table I,
sperm concentration in the experimental group was ~10-fold lower
compared with that of the control group, a result which was
statistically significant (P<0.01). Sperm (a+b) percentage in
the experimental group was also significantly lower compared with
that in the control group (P<0.05). In addition, statistically
significant differences also existed in the percentage of
morphologically normal sperm between the control and experimental
groups (P<0.05).
 | Table I.Comparison of the sperm parameters in
the semen of the experimental and control groups. |
Table I.
Comparison of the sperm parameters in
the semen of the experimental and control groups.
| Sperm parameter | Control group
(n=15) | Experimental group
(n=15) | P-value |
|---|
| Sperm concentration
(×106/ml) | 113.6±32.1 | 11.8±7.2 | <0.01 |
| Sperm (a+b) (%) | 53.7±4.5 |
18.6±12.1 | <0.01 |
| Morphologically
normal sperm (%) | 19.2±5.3 |
9.2±2.3 | <0.01 |
PCR amplification of the H19
differential methylation region (DMR)
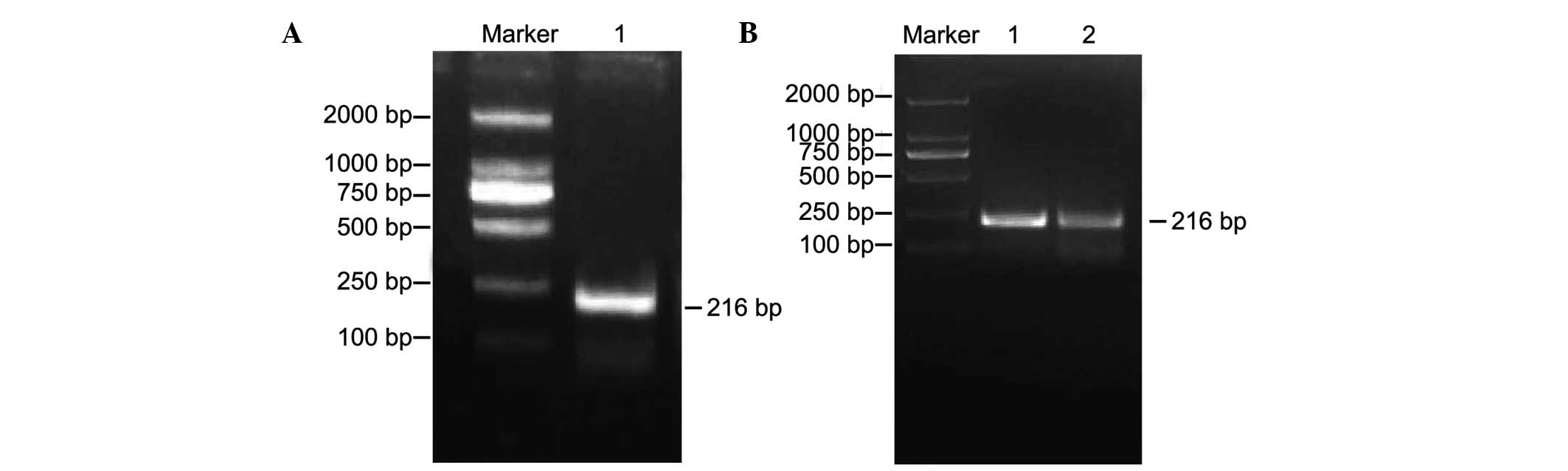
The results of the agarose gel electrophoresis with
a DL2000 marker as a reference revealed a single band of 216 bp PCR
product, as shown in Fig. 1,
representing the amplified DNA fragment of H19 DMR. DNA
amplification products of the H19 DMR in the normal control
and experimental groups are shown in Fig. 1A and B.
Identification of positive H19 DMR
clones
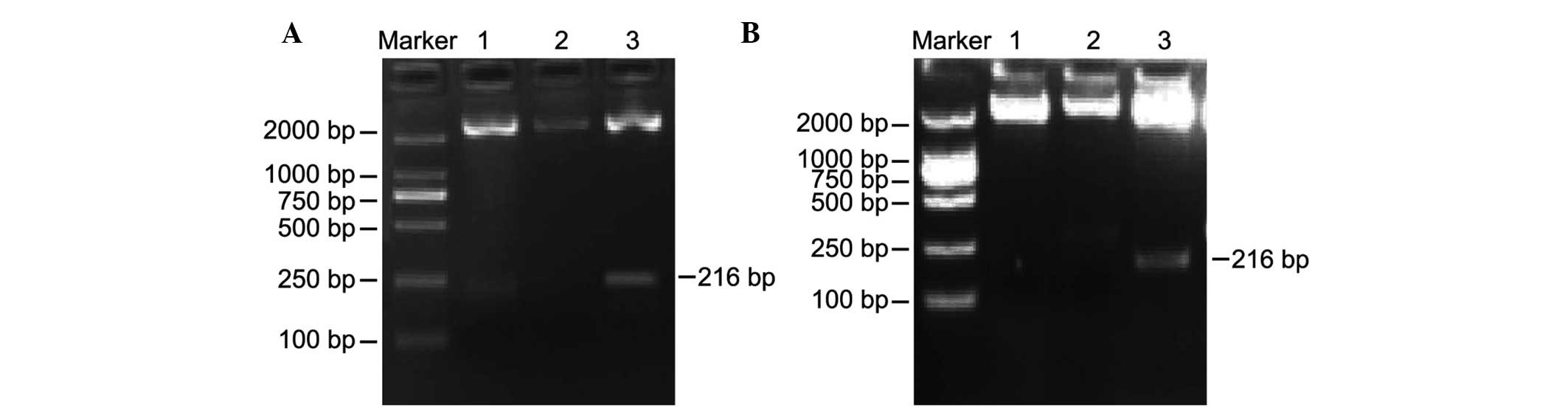
The positive clones underwent electrophoresis
following enzyme digestion with the DL2000 marker as a reference.
As shown in Fig. 2, the results of
the PCR indicated the presence of positive clones of H19 for
the normal control and experimental groups, with one band having a
fragment size of ~2,692 bp, which is consistent with the molecular
size of the pMD18-T vector, and another band having a fragment size
of ~216 bp, which is consistent with the fragment size of
H19 DMR. Fig. 2A shows two
positive clones corresponding to the normal control group following
H19 DMR PCR product cloning, transfer and enzyme-digestion.
Fig. 2B shows one positive clone
corresponding to the experimental group.
Methylation analysis of CpG loci in
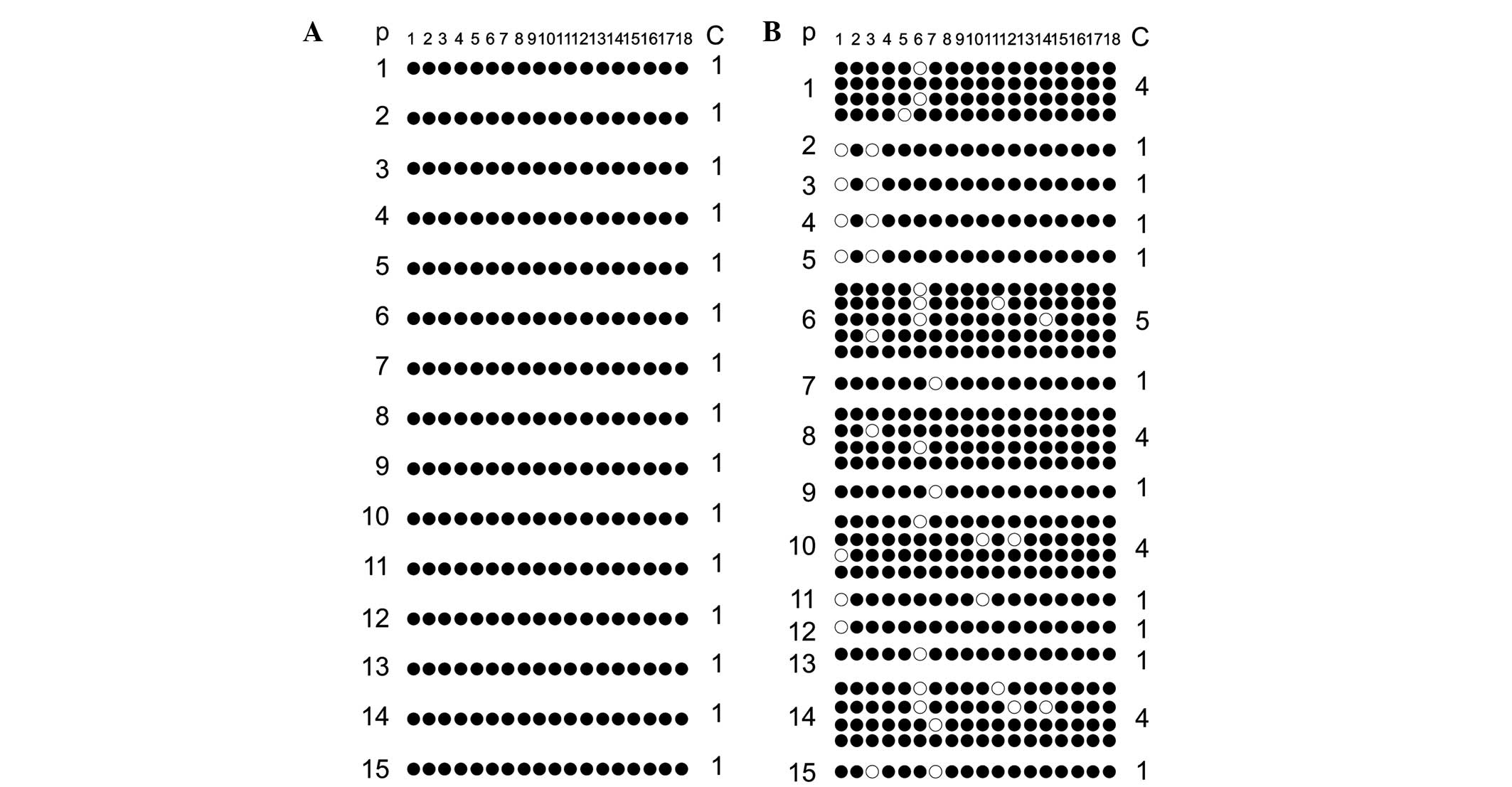
H19 DMR
The methylation scattergram of each locus in the
H19 DMR was obtained using BIQ analyzer software. Fig. 3A shows 18 methylated CpG loci in 15
clones from 15 fertile males in the normal control group and all
sites exhibited 100% methylated status. Similarly, Fig. 3B shows the 18 methylated CpG loci
detected in 15 clones from 15 patients of the experimental group.
It evident that, among the 18 methylated CpG loci, CpG 1, 3, 6, 7,
10, 11, 12 and 14 revealed individual differences in the
methylation pattern, with only 4/31 clones in the experimental
group exhibiting 100% methylation at all 18 sites. As seen in
Fig. 3, comparison of the overall
methylation rate of H19 DMR between the normal control group
and the experimental group was conducted. The methylation rate in
the normal control group was 100% (270/270), whereas that in the
experimental group was 94.1% (525/558), and the comparative
differences were considered statistically significant
(P<0.001).
Comparative analysis of the average
methylation rate of the H19 DMR CpG locus
Following t-test analysis, the average methylation
rate of each H19 CpG in the experimental group was
66.67–100%, whereas that in the normal control group was constant,
namely 100%. In addition, the comparison of the average methylation
rate of each CpG locus between the normal control group and the
experimental group is shown in Table
II. Statistically significant results between the normal
control group and the experimental group were observed to be
present in CpG1, CpG3 and CpG6 (P=0.032, P=0.032, and P=0.014,
respectively).
 | Table II.Comparison of the average methylation
rate (%) in each H19 DMR CpG locus between the normal
control group and the experimental group. |
Table II.
Comparison of the average methylation
rate (%) in each H19 DMR CpG locus between the normal
control group and the experimental group.
| CpG locus | Experimental group
(n=15) | Control group
(n=15) | P-value |
|---|
| CpG1 | 73.33 | 100.00 | 0.03 |
| CpG2 | 100.00 | 100.00 | NA |
| CpG3 | 73.33 | 100.00 | 0.03 |
| CpG4 | 100.00 | 100.00 | NA |
| CpG5 | 100.00 | 100.00 | NA |
| CpG6 | 66.67 | 100.00 | 0.01 |
| CpG7 | 81.82 | 100.00 | 0.07 |
| CpG8 | 100.00 | 100.00 | NA |
| CpG9 | 100.00 | 100.00 | NA |
| CpG10 | 86.67 | 100.00 | 0.14 |
| CpG11 | 93.94 | 100.00 | 0.31 |
| CpG12 | 80.00 | 100.00 | 0.07 |
| CpG13 | 100.00 | 100.00 | NA |
| CpG14 | 93.94 | 100.00 | 0.31 |
| CpG15 | 100.00 | 100.00 | NA |
| CpG16 | 100.00 | 100.00 | NA |
| CpG17 | 100.00 | 100.00 | NA |
| CpG18 | 100.00 | 100.00 | NA |
Discussion
In the present study, the methylation status of the
H19 gene was assessed in both normal males and infertile
males, and the results demonstrated that the levels of DNA
methylation of the H19 gene were lower in the infertile
males compared with the controls. Therefore, aberrant methylation
of the H19 gene may be correlated with the progression of
MI. Genomic imprinting is the allele-specific expression in certain
genes that controls gene expression from both paternal and maternal
genomes and is critical for normal development due to its
significance in placental functions, neurobehavioral processes and
embryonic growth (25,26). In addition, DNA methylation is the
most important silencing mechanism underlying the regulation of
genetic elements, specifically for elements with substantial CpG
dinucleotide content (27). Aberrant
DNA methylation is usually found in human cancers and may account
for chromosomal instability and deregulated gene expression
(28). In fertilization, spermatozoa
have male DNA methylation patterns that contribute to paternal
methylation imprints, and immediately following fertilization the
paternal genome is demethylated, including imprinted genes and
repeat sequences. The present study used bisulfate sequencing and
PCR to measure the degree of methylation of the H19
imprinting control region (ICR), which demonstrated that control
individuals carried 100% methylation of H19 ICR, whereas
infertile males with asthenospermia and oligozoospermia exhibited
H19 hypomethylation (29).
The methylation status of the H19 ICR may serve as an
indicator for aberrant DNA methylation function acting at that
locus, and may contribute to altered gene expression, thus
contributing to MI. Therefore, the decreased methylation of
H19, an important imprinted gene in fertile males, indicates
that aberrant methylation of the H19 genes may be correlated
with the progression of MI, and may serve as a biomarker for
deficiency in human sperm development.
The results of the present study demonstrated that
CpG locus 1, CpG locus 3 and CpG locus 6 in the H19 DMR
displayed aberrant methylation with greater frequency, the reason
for which remains unknown. DNA methylation in the mammalian genome
involves covalent addition of a methyl group at the 5′-end of a
cytosine in the context of CpG. Following t-test analysis, the
methylation status of a total of 18 CpGs located in the H19 DMR of
human sperm in infertile men were compared with the methylation
patterns of normal individuals in order to determine the
specificity and the extent of the loss of DNA methylation
associated with distinct sperm abnormalities. Among the 18 CpGs in
infertile men, CpG1, CpG3, CpG6, CpG7, CpG10, CpG11, CpG12 and
CpG14 exhibited differences in the unmethylated status compared
with the CpGs of healthy men. Analysis of the average methylation
rate of H19 DMR CpG loci and comparison of the degree of
unmethylation at each CpG in the clones analyzed indicated that CpG
locus 1, CpG locus 3 and CpG locus 6 in the H19 DMR are
associated with aberrant methylation.
There were limitations to the present study.
Notably, the study only focused on the analysis of H19 gene
methylation. However, paternal allele expression of IGF is strongly
associated with maternal allele expression of H19, and the
DMR located 2-4-kb upstream of the H19 gene refers to the
ICR of the locus, and its deletion impacts both H19 and IGF2
expression. In addition, the small sample sizes used in the study
may lead to lower statistical significance during the analysis of
H19 gene methylation.
In conclusion, the results of the present study
provide evidence that aberrant methylation of the H19 gene
may be correlated with the progression of MI, and CpG locus 1, CpG
locus 3 and CpG locus 6 in the H19 DMR appear associated
with aberrant methylation.
Acknowledgements
The present study was supported by the Hengyang
Science and Technology Bureau (grant no. 2015KJ44). The authors of
the present study are grateful to the reviewers for their helpful
comments on this manuscript.
References
|
1
|
Kasturi SS, Tannir J and Brannigan RE: The
metabolic syndrome and male infertility. J Androl. 29:251–259.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jungwirth A, Giwercman A, Tournaye H,
Diemer T, Kopa Z, Dohle G and Krausz C: European Association of
Urology Working Group on Male Infertility: European association of
urology guidelines on male infertility: The 2012 update. Eur Urol.
62:324–332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dada R, Kumar M, Jesudasan R, Fernández
JL, Gosálvez J and Agarwal A: Epigenetics and its role in male
infertility. J Assist Reprod Genet. 29:213–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rajender S, Avery K and Agarwal A:
Epigenetics, spermatogenesis and male infertility. Mutat Res.
727:62–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu W, Shen O, Qin Y, Lu J, Niu X, Zhou Z,
Lu C, Xia Y, Wang S and Wang X: Methylenetetrahydrofolate reductase
C677T polymorphism and the risk of male infertility: A
meta-analysis. Int J Androl. 35:18–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferlin A, Vinanzi C, Selice R, Garolla A,
Frigo AC and Foresta C: Toward a pharmacogenetic approach to male
infertility: Polymorphism of follicle-stimulating hormone
beta-subunit promoter. Fertil Steril. 96:1344–1349.e2. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hammoud AO, Meikle AW, Reis LO, Gibson M,
Peterson CM and Carrell DT: Obesity and male infertility: A
practical approach. Semin Reprod Med. 30:486–495. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ross C, Morriss A, Khairy M, Khalaf Y,
Braude P, Coomarasamy A and El-Toukhy T: A systematic review of the
effect of oral antioxidants on male infertility. Reprod Biomed
Online. 20:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ferlin A, Raicu F, Gatta V, Zuccarello D,
Palka G and Foresta C: Male infertility: Role of genetic
background. Reprod Biomed Online. 14:734–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Poongothai J, Gopenath TS and Manonayaki
S: Genetics of human male infertility. Singapore Med J. 50:336–347.
2009.PubMed/NCBI
|
|
11
|
Krausz C and Giachini C: Genetic risk
factors in male infertility. Arch Androl. 53:125–133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gabory A, Ripoche MA, Le Digarcher A,
Watrin F, Ziyyat A, Forné T, Jammes H, Ainscough JF, Surani MA,
Journot L and Dandolo L: H19 acts as a trans regulator of the
imprinted gene network controlling growth in mice. Development.
136:3413–3421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y and Tycko B: Monoallelic
expression of the human H19 gene. Nat Genet. 1:40–44. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reese KJ, Lin S, Verona RI, Schultz RM and
Bartolomei MS: Maintenance of paternal methylation and repression
of the imprinted H19 gene requires MBD3. PLoS Genet. 3:e1372007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cai X and Cullen BR: The imprinted H19
noncoding RNA is a primary microRNA precursor. RNA. 13:313–316.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Steck E, Boeuf S, Gabler J, Werth N,
Schnatzer P, Diederichs S and Richter W: Regulation of H19 and its
encoded microRNA-675 in osteoarthritis and under anabolic and
catabolic in vitro conditions. J Mol Med (Berl). 90:1185–1195.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boissonnas CC, Abdalaoui HE, Haelewyn V,
Fauque P, Dupont JM, Gut I, Vaiman D, Jouannet P, Tost J and Jammes
H: Specific epigenetic alterations of IGF2-H19 locus in spermatozoa
from infertile men. Eur J Hum Genet. 18:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marques CJ, Costa P, Vaz B, Carvalho F,
Fernandes S, Barros A and Sousa M: Abnormal methylation of
imprinted genes in human sperm is associated with oligozoospermia.
Mol Hum Reprod. 14:67–74. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Macklin R: Revising the Declaration of
Helsinki: A work in progress. Indian J Med Ethics. 9:224–226.
2012.PubMed/NCBI
|
|
24
|
World Health Organization: Laboratory
manual of the WHO for the examination of human semen and
sperm-cervical mucus interaction. Ann Ist Super Sanita. 37:I–XII.
2001.(In Italian). PubMed/NCBI
|
|
25
|
Ferguson-Smith AC: Genomic imprinting: The
emergence of an epigenetic paradigm. Nat Rev Genet. 12:565–575.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kobayashi H, Sato A, Otsu E, Hiura H,
Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N and Arima T: Aberrant
DNA methylation of imprinted loci in sperm from oligospermic
patients. Hum Mol Genet. 16:2542–2551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kato Y and Nozaki M: Distinct DNA
methylation dynamics of spermatogenic cell-specific intronless
genes is associated with CpG content. PLoS One. 7:e436582012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dammann RH, Kirsch S, Schagdarsurengin U,
Dansranjavin T, Gradhand E, Schmitt WD and Hauptmann S: Frequent
aberrant methylation of the imprinted IGF2/H19 locus and LINE1
hypomethylation in ovarian carcinoma. Int J Oncol. 36:171–179.
2010.PubMed/NCBI
|
|
29
|
Poplinski A, Tuttelmann F, Kanber D,
Horsthemke B and Gromoll J: Idiopathic male infertility is strongly
associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int
J Androl. 33:642–649. 2010.PubMed/NCBI
|