Introduction
Breast milk, which is commonly recommended for
infants, is of considerable importance to the development of
neonatal gut microflora (1,2). Feeding pre-term infants with breast
milk was shown to reduce the incidence of necrotising enterocolitis
(1). Furthermore, infants gained a
rapid tolerance of enteral nutrition (1). Breast milk protects against infection
and promotes long-term metabolic health, as well as reducing the
occurrence of asthma and other atopic disorders (2). This effect may be the result of the
combined action of breast milk components, including maternal
immunoglobulins, immunocompetent cells and various antimicrobial
compounds (3). As well as its
benefits for an infant's health, breastfeeding has been shown to be
beneficial for the health of the mother, including preventing
complications in the breast such as blocked ducts, mastitis and
breast abscesses, and also reducing the risk of Type II diabetes
and breast and ovarian cancers (4,5).
Enterococcus faecium, which is a commensal
bacterial species in the gastrointestinal tracts of humans and
animals, is a Gram-positive, facultative anaerobic cocci that
occurs singly, in pairs or chains (6). In addition, E. faecium is
commonly found in large numbers on vegetables and plants, and in
soil, surface waters and dairy products (6). E. faecium produces bacteriocins
that inhibit food-borne bacteria and intestinal pathogens (2,7).
Furthermore, various E. faecium strains have been used as
efficient probiotics for humans (1,2).
Enterococci have useful biotechnological and
functional properties, including anti-Listerial activity (7,8). The
present study aimed to determine the antibiotic susceptibility
patterns and antimicrobial activity of E. faecium strains
isolated from human breast milk. Thus, the probiotic potential of
E. faecium strains isolated from breast milk could be
evaluated and the results may influence the development of novel
antibacterials against clinical pathogens.
Materials and methods
Collection of breast milk
The present study analyzed isolates derived from
breast milk samples obtained by a student for their Master of
Science Thesis in 2005. The isolate samples that had been stored in
20% glycerol at −86°C were activated by culturing in M17A broth for
24 h.
Isolation and identification of
bacteria
Breast milk samples were cultured anaerobically on
MRS and M17 agar plates at 37°C for 48 h in order to isolate lactic
acid bacteria. Subsequently, the isolates were examined under a
microscope for cell morphological and Gram-staining analyses. In
addition, the isolates were tested for oxidase and catalase
activities. Sugar fermentation patterns of the isolates were
determined using a API 20 STREP system (bioMérieux, Inc., Durham,
NC, USA), according to the manufacturer's protocol. Isolates were
examined for CO2 production from glucose, and growth at
various temperatures (4, 15 and 45°C), and pH values (pH 3.9–9.6)
were assessed. Ammonia production from arginine was analyzed and
growth under various NaCl concentrations (6, 7.5 and 10%) was
investigated, according to previous studies (9,10).
Isolates were identified using an automated
ribotyping system. Specifically, EcoRI ribotyping was performed
using an automated RiboPrinter® Microbial
Characterisation System (DuPont, Wilmington, DE, USA), according to
the manufacturer's protocol. Ribotype patterns were compared to
patterns stored in the RiboPrinter® database. An isolate
was identified when its ribotype pattern had a similarity of ≥0.86
with a DuPont Identification Library Code (DUP-IDs).
Detection of antibacterial
activity
Antibacterial activity of the isolates was
investigated using a well diffusion assay. Briefly, Mueller-Hinton
Agar was poured into sterile petri dishes, solidified and dried in
a laminar flow cabinet for 30 min at room temperature. Wells of 6
mm diameter were formed in the agar using a cork borer, which were
subsequently filled with 15 µl soft agar. Supernatants from
Enterococcus cultures [108 colony forming unit
(CFU)/ml] were obtained via centrifugation at 2,500 × g for 5 min.
Samples were neutralized by the addition of 5 N NaOH (pH 5.5).
Neutralized supernatants were filter-sterilized
using a 0.22 µm filter membrane. Subsequently, 80 µl neutralized
supernatant was dispensed into the wells and the plates were
overlaid with 8 ml soft agar (0.75% agar) and seeded with 8 µl test
bacteria culture (~107 CFU/ml stationary-phase cells).
The plates were incubated at 37°C for 24 h, and were subsequently
examined for zones of inhibition.
Production of gelatinase and
haemolytic activity
Strains were cultured in M17 broth at 37°C for 18 h
and transferred onto blood agar at a density of 107
CFU/ml. Blood agar plates were incubated at 18–24 h for 37°C, after
which haemolytic activity was recorded. Production of gelatinase
was assessed using trypticase soy agar, supplemented with 1.5%
skimmed milk. Plates were incubated for 18 h at 37°C. Following
incubation, a clear halo surrounding the colonies was considered
positive, as described in a previous study (11).
pH and bile acid resistance. M17 broth containing
200 mM KCl/HCl and 100 mM citric acid/200 mM
Na2HPO4, buffered at pH 1.0–2.0 and pH
3.0–6.5 respectively, was used to determine bacterial growth under
various pH conditions. In order to assess resistance to bile acid,
cultures (108 CFU/ml) were inoculated (1%) into M17
broth with or without Oxgall (0.15 or 0.5%), and incubated at 37°C
for 24 h, after which growth was assessed.
Resistance to phenol and lysozyme
activity
The ability of the isolates to grow on phenol was
investigated by inoculating (2%) cultures into 10 ml M17 broth in
the presence or absence of 0.4% phenol. Growth of the cultures was
then determined following incubation for 24 h at 37°C. In order to
assess the resistance of the isolates to lysozyme activity, E.
faecium strains (1:50) were inoculated into 10 ml M17 broth
with or without lysozyme (100 ppm). Tubes were incubated at 37°C
for 24 h, after which growth was assessed.
Biofilm-formation assays
Biofilm formation on polystyrene microtitre plates
was quantified using a method developed by Heilmann et al
(12). Briefly, 50 µl overnight
cultures were transferred onto the microtitre plates. MRS without
glucose basal medium was supplemented with glucose, fructose,
sucrose and 2% lactose, and 200 µl of this medium was subsequently
transferred to each microplate. Microplates were incubated for 24 h
at 37°C, after which the optical density (OD) of the biofilm was
measured at 570 nm using an Automated Spectrophotometer. Biofim
formation was evaluated as weak, moderate or strong according to OD
measurements, as described previously (13). Biofilm analyses were repeated three
times in triplicate for each strain.
Lactic acid determination
Lactic-acid production was assessed in sterilized
skimmed milk. Briefly, sterilized skimmed milk was inoculated at a
rate of 2/100 ml with active strains of E. faecium and
acidity was assessed by performing a titration. Acidity is
expressed as mg/ml lactic acid, according to a previous study
(14).
Proteolytic activity
Proteolytic activities of the cultures were
determined spectrophotometrically, using a previously described
method (15). This method detects
free tyrosine and tryptophan liberated in the reaction mixture. In
the present study, proteolytic activity was measured in triplicate.
Results were calculated using a calibration curve obtained from
dilutions of tyrosine in distilled water, as described previously
(16), and are expressed as µg/ml
tyrosine.
Hydrogen peroxide
(H2O2)
The level of H2O2 produced by
the isolates was determined spectrophotometrically, according to a
previous study (17). Briefly,
measurements were obtained following a 24 h incubation period in
skimmed milk, and production was monitored at OD350.
H2O2 was quantified using a
H2O2 standard curve, performed with
concentrations ranging from 1–10 µg/ml.
Effect of enzymes, pH and heat on the
antibacterial activity of E. faecium strains
Concentrated supernatants of E. faecium
strains were treated with various enzymes, including catalase (5
µg/ml), α-amylase (1 mg/ml), pepsin (10 U/mg), trypsin (2 mg/ml),
α-chymotrypsin (5 mg/ml), proteinase K (1 mg/ml) and lysozyme (1
mg/ml). Each enzyme was dissolved in sterile 0.05 M sodium
phosphate buffer and added to the E. faecium supernatant to
a final concentration of 1 mg/ml. Following incubation at 37°C for
4 h, the reaction mixtures were heated at 100°C for 10 min to
inactivate the enzymes. In addition, the effect of heat on the
antibacterial activity of the E. faecium strains was
determined. Briefly, the supernatants of the E. faecium
strains were heated at 121°C for 20 min, and the inhibitory
activity against Enterococcus faecalis, L. monocytogenes
1 and Proteus vulgaris was subsequently determined using
the well diffusion method. Experiments were performed in duplicate,
using the untreated supernatant as a control.
Antibiotic susceptibility assay
Susceptibility of the E. faecium strains to
ciprofloxacin (30 µg), gentamicin (120 µg), netilmicin sulfate (10
µg), penicillin G (10 U), vancomycin (30 µg) and cefaclor (30 µg;
all Oxoid, Ltd., Basingstoke, UK) was determined using the
Kirby-Bauer Disk Diffusion method, as previously described
(18). Susceptibility or resistance
of the Enterococcus strains was determined according to the
guidelines outlined by the Clinical and Laboratory Standards
Institute (18).
Results
Identification of breast milk
isolates
A total of 20 isolates were cultured from the breast
milk sample, and identified using various phenotypic and genotypic
tests. The results of cell morphological analyses, and the growth
of the isolates at various temperatures and salinity, are presented
in Table I. All isolates were
Gram-positive, catalase-negative and oxidase-negative. Sugar
fermentation tests were performed using the API ID 32 STREP system.
According to the phenotypic tests, the strains were identified as
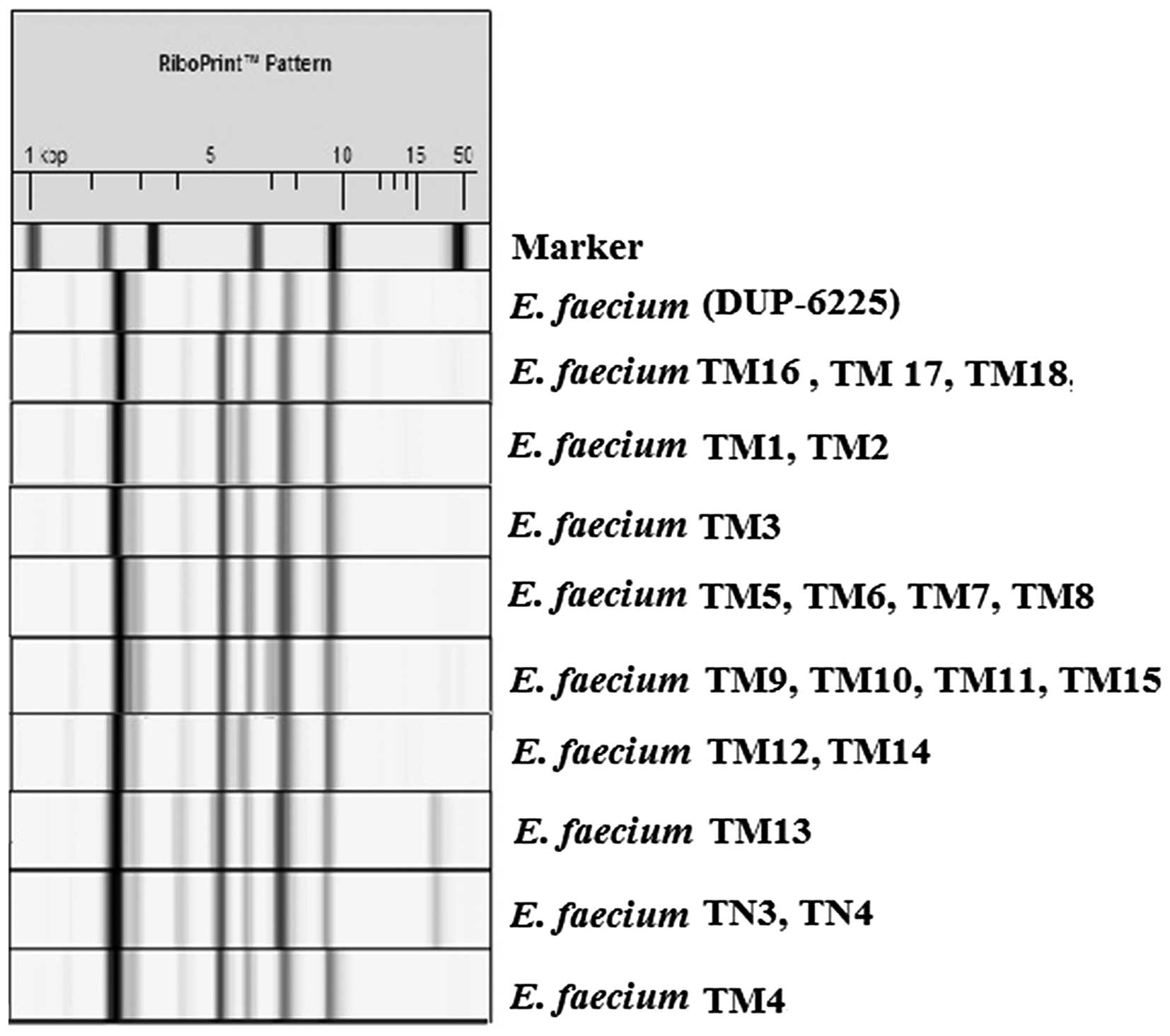
E. faecium, which was confirmed by the automated EcoRI
ribotyping results (Fig. 1). EcoRI
ribotyping differentiated the isolates into two distinct ribotypes,
with similarities ranging from 0.89–0.96. The two distinct
ribotypes belonged to two DUP-IDs: DUP-6225, which was classified
as Lineage I, and DUP-6227, which was classified as Lineage II.
 | Table I.Morphological and physiological
characteristics of the Enterococcus faecium strains isolated
from human breast milk. |
Table I.
Morphological and physiological
characteristics of the Enterococcus faecium strains isolated
from human breast milk.
|
|
|
|
| Temperature
(°C) | NaCl (%) |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Isolate | Gram reaction | Morphology | Catalase | 4 | 15 | 45 | 6 | 7.5 | 10 | pH 3.9–9.6 | H2S production | NH3 production | Biofilm |
|---|
| TM1 | + | c | − | + | + | + | + | − | − | + | − | + | − |
| TM2 | + | c | − | + | + | + | + | − | − | + | − | + | + |
| TM3 | + | c | − | + | + | + | + | + | + | + | − | + | − |
| TM4 | + | c | − | + | + | + | + | + | − | + | − | + | + |
| TM5 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM6 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM7 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM8 | + | c | − | + | + | + | − | − | − | + | − | + | + |
| TM9 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM10 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM11 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM12 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM13 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM14 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM15 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM16 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM17 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TM18 | + | c | − | + | + | + | − | − | − | + | − | + | − |
| TN3 | + | c | − | + | + | + | + | − | − | + | − | + | − |
| TN4 | + | c | − | + | + | + | + | − | − | + | − | + | − |
Antibacterial activity of the
isolates
Antibacterial activities of the isolates against
various test strains are presented in Table II. Bacillus cereus,
Escherichia coli, Klebsiella pneumoniae,
Salmonella enterica, Salmonella typhimurium and
Pseudomonas aeruginosa were not inhibited by E.
faecium. However, the majority of E. faecium isolates
were able to inhibit the growth of P. vulgaris, E.
faecalis, L. monocytogenes 1, Lactococcus lactis,
Lactobacillus plantarum, Leuconostoc
paramesenteroides, Lactobacillus bulgaricus and
Lactobacillus buchneri. A few of the E. faecium
isolates exhibited inhibitory effects against L.
monocytogenes, L. monocytogenes 2 and S. aureus.
Of these, the E. faecium TM13, TM15, TM17, TM18 and TN3
strains exhibited the strongest antibacterial activity against the
tested bacteria (Table II).
Therefore, these strains were used for further analyses.
 | Table II.Inhibitory activity of the
neutralized supernatant of Enterococcus faecium isolates
against various clinical and food-borne pathogens. |
Table II.
Inhibitory activity of the
neutralized supernatant of Enterococcus faecium isolates
against various clinical and food-borne pathogens.
| Isolate | PV | BS | ST | EF* | YE | SA* | LM | LM1* | LM2 | LL | LPl | LPa | LBl | LBc |
|---|
| TM1 | + | − | − | + | − | − | − | ++ | − | + | + | + | + | ++ |
| TM2 | + | − | − | + | − | − | − | ++ | − | + | + | + | + | ++ |
| TM3 | + | − | − | + | − | − | − | ++ | − | + | + | + | + | ++ |
| TM4 | + | − | − | + | − | − | − | ++ | − | + | + | + | + | ++ |
| TM5 | + | − | − | + | − | − | − | ++ | − | + | + | + | + | ++ |
| TM6 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | + | ++ |
| TM7 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | + | ++ |
| TM8 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | + | ++ |
| TM9 | + | − | − | + | − | + | − | + | + | + | + | + | + | + |
| TM10 | + | − | − | + | − | + | + | + | + | + | + | + | + | + |
| TM11 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | + | ++ |
| TM12 | − | − | − | + | − | + | − | + | + | + | + | + | + | + |
| TM13 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | ++ | ++ |
| TM14 | + | − | − | + | − | + | − | − | + | + | + | + | + | + |
| TM15 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | ++ | ++ |
| TM16 | + | − | − | + | + | + | − | + | + | + | + | + | + | + |
| TM17 | + | − | − | + | − | − | − | ++ | − | ++ | + | + | ++ | ++ |
| TM18 | + | − | + | + | − | + | − | ++ | − | ++ | + | + | + | + |
| TN3 | + | ++ | − | + | − | − | + | + | − | + | + | + | + | + |
| TN4 | + | ++ | − | + | − | − | + | + | − | + | + | ++ | + | + |
Production of gelatinase and
haemolytic activity
The E. faecium TM4, TM13, TM15, TM17, TM18
and TN3 strains did not exhibit haemolytic activity. In addition,
no gelatinase activity was detected in these strains (Table III).
 | Table III.Acid and phenol tolerance, resistance
to bile acid, and haemolytic and gelatinase activities of
Enterococcus faecium strains isolated from human breast
milk. |
Table III.
Acid and phenol tolerance, resistance
to bile acid, and haemolytic and gelatinase activities of
Enterococcus faecium strains isolated from human breast
milk.
|
| Acid tolerance |
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Isolate | pH 1 | pH 2 | pH 3 | Phenol
tolerance | Gelatinase
activity | Haemolytic
activity | Resistance to bile
acid | Resistance to
lysozyme |
|---|
| TM4 | − | + | + | − | − | − | + | + |
| TM13 | − | ++ | ++ | − | − | − | + | + |
| TM15 | − | ++ | ++ | − | − | − | + | + |
| TM17 | − | + | + | − | − | − | + | + |
| TM18 | − | ++ | ++ | − | − | − | + | + |
| TN3 | − | ++ | ++ | − | − | − | + | + |
Resistance of isolates to pH, bile
acid, lysozyme activity and phenol and biofilm formation
E. faecium strains exhibited a tolerance to a
pH range of 2.0–9.6 (Table I). None
of the test strains were able to survive at pH 1.0 (Table III). Resistance to bile acid (0.15
and 0.5% Oxgall) was observed in all tested isolates exposed for 24
h. Similarly, resistance to lysozyme activity was detected in all
the isolates tested. Growth of E. faecium strains in the
presence of phenol (0.4%) at 37°C for 24 h was not observed
(Table III). Biofilm formation was
not observed in all tested strains, with the exception of three
strains (Table I). These strains
produced a weak biofilm, according to OD measurements.
Lactic acid and
H2O2 production
In the present study, the E. faecium strains
isolated from human breast milk were slow acid producers; after 24
h of growth, the pH values of the skimmed milk ranged from 4.2–5.5
(data not shown). The amount of lactic acid produced by E.
faecium strains ranged from 12.49–16.59 mg/ml (Table IV). The production of
H2O2 by E. faecium strains is
presented in Table IV. The amount
of H2O2 produced by lactic acid bacteria
ranged from 2.17–1.09 µg/ml. The highest amount of
H2O2 production was observed for E.
faecium TN3.
 | Table IV.Lactic acid and
H2O2 production by, and proteolytic activity
of, Enterococcus faecium strains. |
Table IV.
Lactic acid and
H2O2 production by, and proteolytic activity
of, Enterococcus faecium strains.
| Isolate | Lactic acid
(mg/ml) | Proteolytic
activity (Tyrosine mg/ml) |
H2O2 (µg/ml) |
|---|
| TM13 | 14.33±0.04 | 0.35±0.03 | 1.24±0.02 |
| TM15 | 16.59±0.04 | 0.23±0.00 | 1.31±0.04 |
| TM17 | 13.33±0.00 | 0.67±0.01 | 1.09±0.02 |
| TM18 | 12.49±0.01 | 0.96±0.16 | 1.30±0.00 |
| TN3 | 13.88±0.01 | 0.77±0.03 | 2.17±0.11 |
Proteolytic activity
Proteolytic activities of the isolated strains are
presented in Table IV. The amount
of tyrosine released by these bacteria ranged from 0.23–0.96 mg/ml.
These results suggest that the E. faecium strains exhibit
low proteolytic activity.
Antibacterial activity of TM13, TM15,
TM17, TM18 and TN3 strains
The antibacterial activities of the E.
faecium TM13, TM15, TM17 and TM18 strains were not affected by
treatment with α-amylase, catalase, trypsin, α-chymotrypsin nor
lysozyme; however, proteinase K was able to affect the
antibacterial activity of all strains (Table V). Antibacterial activity of the
E. faecium TN3 supernatant was affected by lysozyme,
catalase and proteinase K treatment. The antibacterial activities
of the supernatants of all E. faecium strains were retained
following heating at 121°C for 5 min (Table V).
 | Table V.Effect of enzymes or heat on the
antibacterial activity of the Enterococcus faecium strains
isolated from breast milk. |
Table V.
Effect of enzymes or heat on the
antibacterial activity of the Enterococcus faecium strains
isolated from breast milk.
| Isolate | Filtrate | Catalase | Trypsin | α-Chymotrypsin | Lysozyme | α-Amylase | Proteinase K | Heating at 121°C
for 20 min |
|---|
| TM13 |
|
|
|
|
|
|
|
|
| EF | + | − | − | − | − | − | − | + |
| PV | + | − | − | − | − | − | − | + |
| LM | + | + | + | + | + | + | − | + |
| TM15 |
|
|
|
|
|
|
|
|
| EF | + | − | − | − | − | − | − | + |
| PV | + | − | − | − | − | − | − | + |
| LM | + | + | + | + | + | + | − | + |
| TM17 |
|
|
|
|
|
|
|
|
| EF | + | − | − | − | − | − | − | + |
| PV | + | − | − | − | − | − | − | + |
| LM | + | + | + | + | + | + | − | + |
| TM18 |
|
|
|
|
|
|
|
|
| EF | + | − | − | − | − | − | − | + |
| PV | + | − | − | − | − | − | − | + |
| LM | + | + | + | + | + | + | − | + |
| TN3 |
|
|
|
|
|
|
|
|
| EF | + | − | − | − | − | − | − | + |
| PV | + | − | − | − | − | − | − | + |
| LM | + | − | + | + | − | + | − | + |
Antibiotic susceptibility
profiles
E. faecium strains were resistant to
ciprofloxacin, netilmicin sulfate and cefaclor; thus a
multiresistant antibiotic profile was observed (Table VI). In addition, the E.
faecium TM13, TM15, TM17, TM18 and TN3 strains were sensitive
to vancomycin (Table VI). Only two
of the 20 strains, TM1 and TM2, were resistant to all tested
antibiotics.
 | Table VI.Antibiotic susceptibility profiles of
Enterococcus faecium strains from human breast milk. |
Table VI.
Antibiotic susceptibility profiles of
Enterococcus faecium strains from human breast milk.
| Isolate | Vancomycin (30
µg) | Ciprofloxacin (30
µg) | Penicillin G (10
U) | Gentamicin (120
µg) | Netilmicin sulfate
(10 µg) | Cefaclor (30
µg) |
|---|
| TM1 | R | R | R | R | R | R |
| TM2 | R | R | R | R | R | R |
| TM3 | R | R | I | R | R | R |
| TM4 | R | R | R | I | R | I |
| TM5 | R | R | S | R | R | R |
| TM6 | S | I | R | S | R | R |
| TM7 | R | R | R | S | R | I |
| TM8 | R | R | R | S | I | R |
| TM9 | S | R | S | I | R | R |
| TM10 | S | R | R | S | R | R |
| TM11 | S | R | I | S | R | R |
| TM12 | S | I | R | S | R | R |
| TM13 | S | R | S | S | I | R |
| TM14 | S | R | R | S | R | R |
| TM15 | S | R | I | S | R | I |
| TM16 | S | R | R | S | R | R |
| TM17 | S | S | S | S | R | S |
| TM18 | S | R | R | S | R | S |
| TN3 | S | R | S | I | I | I |
| TN4 | R | I | R | S | R | R |
Discussion
The present study isolated bacteria from human
breast milk and identified them using conventional tests and an
automated RiboPrinter® Microbial Characterisation
System. According to phenotypic and genotypic characterisation
tests, the isolates from the breast milk were all Gram-positive,
non-spore-forming, catalase- and oxidase-negative, facultative
anaerobic cocci identified as E. faecium. These isolates
were able to grow at pH 9.6. E. faecium TM13, TM15, TM17 and
TM18 strains grew poorly or not at all in the presence of 6.0%
NaCl; this is inconsistent with a previous study in which E.
faecium isolates survived in 6.5% NaCl (19).
Martín et al (20) reported that Lactobacillus
gasseri and E. faecium are the most commonly isolated
microorganisms from human breast milk. In the present study,
ribotyping and phenotypic identification methods suggested that the
isolates were E. faecium. Furthermore, the isolates were
differentiated into two distinct ribotypes using the Automated
RiboPrinter®.
The present study investigated the antibacterial
activity of the supernatants of the E. faecium strains
isolated from human breast milk. The E. faecium strains were
able to inhibit P. vulgaris, E. faecalis, L.
monocytogenes 1, L. lactis, L. plantarum, L.
paramesenteroides, L. bulgaricus and L. buchneri.
In particular, L. monocytogenes 1 was sensitive to all E.
faecium strains, with the exception of TM14. L.
monocytogenes 1 is an ocular surface isolate and so inhibition
of this isolate by E. faecium may have important clinical
applications. L. monocytogenes may cause ocular infections,
including conjunctivitis, keratitis, chorioretinitis and
endophthalmitis, that may lead to blindness. Furthermore, L.
monocytogenes has been shown to cause a serious food-borne
disease with a high mortality rate (21,22).
Antimicrobial activity of E. faecium has
previously been investigated (7,23,24). The
main cause of inhibitory activity may be associated with
antibacterial peptides, namely bacteriocins (23). Kang and Lee (23) reported that the supernatant of E.
faecium strains exhibited inhibitory activity against L.
monocytogenes. Typically, Gram-negative bacteria are not
susceptible to the supernatant of E. faecium; however, in
the present study, P. vulgaris was sensitive to the E.
faecium strains.
Haemolysin is an important enterococcal virulence
factor (19). However, haemolytic
activity was not observed for the E. faecium strains
isolated from breast milk in the present study. In addition, no
gelatinase activity was observed for the E. faecium strains,
which was consistent with previous studies (25,26).
Lactic acid production by the E. faecium
strains was low; whereas the pH of the growth medium was relatively
high. In a previous study, enterococci derived from cheese
exhibited a poor acidifying ability; only a slight decrease in the
pH (<5.0) of milk was observed following incubation for 24 h at
37°C (27). Conversely, an
acidifying potential was demonstrated for E. faecium strains
isolated from bovine milk; the pH of MRS broth was lowered to
~3.85–4.05 following incubation for 48 h (26). In the present study, the Enterococci
isolates were resistant to bile acid following exposure for 24 h,
which is consistent with a previous study (28). This resistance may be due to the
inactivation of various bile components by β-glucuronidase
activity.
In the presence of 0.4% phenol, E. faecium
strains did not grow following incubation for 24 h. In a previous
study, Enterococci strains exhibited a high resistance to phenol
(28). Lysozyme promotes the
hydrolysis of the bacterial cell (29). The E. faecium strains isolated
in the present study exhibited resistance to lysozyme-mediated
hydrolysis, which is consistent with a previous study (28).
Of the 20 E. faecium strains isolated from
breast milk in the present study, only two were biofilm producers,
although these exhibited only weak biofilm-producing abilities.
These results are consistent with the findings for clinical
isolates in a previous study (30).
The strains isolated in the present study may be
considered as slow acid producers. These results are consistent
with those reported by Arizcun et al (31). Furthermore, it has previously been
shown that E. faecium exhibits weak proteolytic activity at
37°C after 72 h (32). Previous
studies demonstrated that the majority of E. faecium
isolates from dairy products exhibited weak proteolytic activities
in milk (27,33).
Enterococci, which are natural inhabitants of the
gastrointestinal tract, are a type of lactic acid bacteria, thus
H2O2 is the primary metabolite that may
contribute to their antagonistic action (34). In the present study, the
antimicrobial effect of the TN3 strain may have been associated
with high H2O2 production. Notably, the
antimicrobial activity of E. faecium TN3 was completely
inhibited following treatment with proteinase K, lysozyme, catalase
and trypsin.
Antimicrobial activity of the E. faecium
TM13, TM15, TM17 and TM18 supernatants was not affected by
treatment with catalase, α-amylase, α-chymotrypsin, lysozyme nor
trypsin. Conversely, the activities of all E. faecium
supernatants were completely inhibited following treatment with
proteinase K, indicating that the antibacterial was proteinaceous
in nature.
Bacteriocins produced by Enterococci are divided
into two classes (35). Class II
bacteriocins are small, cationic, hydrophobic and heat-stable
peptides, and the strains in the present study, with the exception
of the TN3 strain, exhibited properties that resembled the class II
bacteriocins (36,37). These findings are in agreement with a
previous study (23). In addition,
E. faecium has been shown to exhibit high acid resistance at
pH 2.0 and 1.0 (28); however, in
the present study, the E. faecium strains were susceptible
to acid at pH 1.0, which is consistent with a previous study
(28).
The present study demonstrated that the ability of
E. faecium strains to inhibit the growth of E.
faecalis, P. vulgaris and L. monocytogenes was
stable following heating for 20 min at 121°C. This is inconsistent
with a previous study, in which E. faecium isolates lost
their inhibitory activity following heat treatment (26).
Enterococci are known to be resistant to
antibiotics. A high degree of antibiotic resistance is associated
with a combination of over-the-counter antibiotic sales and the
inappropriate use of antibiotics. The most clinically relevant
antibiotics include vancomycin and gentamicin, since these are able
to treat infections caused by multi-drug resistant strains
(7). In the present study, E.
faecium strains were sensitive to vancomycin. However,
antibiotic resistance may not explain the virulence of enterococci.
On the other hand, a multiresistant antibiotic profile was observed
in the present study. Strains of Enterococci resistant to multiple
antibiotics have emerged in the last decade, and have demonstrated
resistance to tetracycline, chloramphenicol and vancomycin
(38). In addition, 85% of E.
faecium clinical isolates were shown to be resistant to
ciprofloxacin in Sweden (39), and a
marked increase in high level resistance to gentamicin and
vancomycin has previously been demonstrated (39).
Antibiotic resistance is a significant problem in
the clinical setting, such that the discovery and development of
novel antibiotics is required. Bacteriotherapy, in which bacteria
are used against pathogenic bacterial strains in a host, has
emerged as a novel area that may be useful in this endeavour
(2). Bacteriocins produced by E.
faecium may have useful clinical applications. The
antibacterial activity of the supernatants of the E. faecium
TM13, TM15, TM17 and TM18 strains were heat stable and sensitive to
proteolysis. E. faecium TM13, TM15, TM17 and TM18 strains
exhibited strong inhibitory activities against L.
monocytogenes. In addition, E. faecium isolated from
breast milk exhibited antibacterial effects against L.
monocytogenes and E. faecalis isolated from human eyes.
These isolates may be considered useful for the development of
novel drugs against bacteria, and may have potential applications
as probiotics and in the food industry.
Acknowledgements
The present study was supported by the Anadolu
University Council of Research Project Fund (grant no. 41020).
References
|
1
|
Lindemann PC, Foshaugen I and Lindemann R:
Characteristics of breast milk and serology of women donating
breast milk to a milk bank. Arc Dis Child Fetal Neonatal Ed.
8:F440–F441. 2004. View Article : Google Scholar
|
|
2
|
Martin R, Langa S, Reviriego C, Jiménez E,
Marín ML, Olivares M, Boza J, Jiménez J, Fernández L, Xuas J and
Rodríguez JM: The commensal microflora of human milk: New
perspectives for food bacteriotherapy and probiotics. Trends Food
Sci Technol. 15:121–127. 2004. View Article : Google Scholar
|
|
3
|
Saavedra JM: Clinical applications of
probiotic agents. Am J Clin Nutr. 73:1147S–1151S. 2001.PubMed/NCBI
|
|
4
|
Ip S, Chung M, Raman G, Chew P, Magula N,
DeVine D, Trikalinos T and Lau J: Breastfeeding and maternal and
infant health outcomes in developed countries. Evid Rep Technol
Assess (Full Rep). 1–186. 2007.
|
|
5
|
Labbok M: Breastfeeding: A woman's
reproductive right. Int J Gynaecol Obstet. 94:277–286. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Giraffa G: Functionality of enterococci in
dairy products. Int J Food Microbiol. 88:215–222. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Foulquié Moreno MR, Sarantinopoulos P,
Tsakalidou EL and De Vuyst L: The role and application of
enterococci in food and health. Int J Food Microbiol. 106:1–24.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cocolin L, Foschino R, Comi G and Fortina
M: Description of the bacteriocins produced by two strains of
Enterococcus faecium isolated from Italian goat milk. Food
Microbiol. 24:752–758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schillinger U and Lucke FK: Identification
of lactobacilli from meat and meat products. Food Microbiol.
4:199–208. 1987. View Article : Google Scholar
|
|
10
|
Stiles ME and Holzapfel WH: Lactic acid
bacteria of foods and their current taxonomy. Int J Food Microbiol.
36:1–29. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Creti R, Imperi M, Bertuccini L, Fabretti
F, Orefici G, Di Rosa R and Baldassarri L: Survey for virulence
determinants among Enterococus faecalis isolated from different
sources. J Med Microbiol. 53:13–20. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stepanovic S, Vukovic D, Dakic I, Savić B
and Svabic-Vlahovic M: A modified microtiter-plate test for
quantification of staphylococcal biofilm formation. J Microbiol
Methods. 40:175–179. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Christensen GD, Simpson WA, Younger JJ,
Baddour LM, Barrett FF, Melton DM and Beachey EH: Adherence of
coagulase-negative staphylococci to plastic tissue culture plates:
A quantitative model for the adherence of staphylococci to medical
devices. J Clin Microbiol. 22:996–1006. 1985.PubMed/NCBI
|
|
14
|
Demirci M and Gunduz H: Dairy Technology
Handbook. Hasad Press; Turkey: pp. 1841994
|
|
15
|
Citti J, Sandine WE and Elliker PR: Some
observations on the Hull method for measurement of proteolysis in
milk. J Dairy Sci. 46:3371963. View Article : Google Scholar
|
|
16
|
Rajagopal SN and Sandine WE: Fissociative
growth and proteolysis of Streptococcus thermophilus and
Lactobacillus bulgaricus in Skim Milk. J Dairy Sci. 73:894–899.
1990. View Article : Google Scholar
|
|
17
|
Patrick WA and Wagner HB: Determination of
hydrogen peroxide in small concentrations. Anal Chem. 21:1279–1280.
1949. View Article : Google Scholar
|
|
18
|
Clinical and Laboratory Standards
Institute, . Performance Standards for Antimimicrobial
Susceptibility TestingTwenty-Third Informational Supplement. CLSI;
Wayne, PA: pp. 1992013
|
|
19
|
Morandi S, Brasca M, Andrighetto C,
Lombardi A and Lodi R: Technological and molecular characterization
of enterococci isolated from north-west Italian dairy products. Int
Dairy J. 16:867–875. 2006. View Article : Google Scholar
|
|
20
|
Martín R, Langa S, Reviriego C, Jimínez E,
Marín ML, Xaus J, Fernández L and Rodríguez JM: Human milk is a
source of lactic acid bacteria for the infant gut. J Pediatr.
143:754–758. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shoughy SS and Tabbara KF: Listeria
monocytogenes endophthalmitis following keratoconjunctivitis. Clin
Ophthalmol. 8:301–304. 2014.PubMed/NCBI
|
|
22
|
Gandhi M and Chikindas ML: Listeria: A
foodborne pathogen that knows how to survive. Int J Food Microbiol.
113:1–15. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kang JH and Lee MS: Characterization of a
bacteriocin produced by Enterococcus faecium GM-1 isolated from an
infant. J Appl Microbiol. 98:1169–1176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohmomo S, Murata S, Katayama N,
Nitisinprasart S, Kobayashi M, Nakajima T, Yajima M and Nakanishi
K: Purification and some characteristics of enterocin ON-157, a
bacteriocin produced by Enterococcus faecium NIAI 157. J Appl
Microbiol. 88:81–89. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mannu L, Paba A, Daga E, Comunian R,
Zanetti S, Duprè I and Sechi LA: Comparison of the incidence of
virulence determinants and antibiotic resistance between
Enterococcus faecium strains of dairy, animal and clinical origin.
Int J Food Microbiol. 88:291–304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Banwo K, Sanni A and Tan H: Technological
properties and probiotic potential of Enterococcus faecium strains
isolated from cow milk. J Appl Microbiol. 114:229–241. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Andrighetto C, Knijff E, Lombardi A,
Torriani S, Vancanneyt M, Kersters K, Swings J and Dellaglio F:
Phenotypic and genetic diversity of enterococci isolated from
Italian cheeses. J Dairy Res. 68:303–316. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vassos D, Bezırtzoglou EA, Voıdarou C,
Alexopoulos A and Maıpa V: Biochemical and antimicrobial profile of
Enterococcus faecium and E. faecalis isolated from traditional
dairy products and infant intestinal microbiota. Microb Ecol Health
Dis. 21:241–250. 2009. View Article : Google Scholar
|
|
29
|
Bottazzi V, Battistotti B, Bosi F,
Corradini C and Dell'Acqua E: Effects of lysozyme on the
thermophilous lactic ferments. 20th International Dairy Congress.
Paris. pp. 535–536. 1978; (In French).
|
|
30
|
Di Rosa R, Creti R, Venditti M, D'Amelio
R, Arciola CR, Montanaro L and Baldassarri L: Relationship between
biofilm formation, the enterococcal surface protein (Esp) and
gelatinase in clinical isolates of Enterococcus faecalis and
Enterococcus faecium. FEMS Microbiol Lett. 256:145–150. 2006.
View Article : Google Scholar
|
|
31
|
Arizcun C, Barcina Y and Torre P:
Identification and characterization of proteolytic activity of
Enterococcus spp. isolated from milk and Roncal and Idiazábal
cheese. Int J Food Microbiol. 38:17–24. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Suzzi G, Caruso M, Gardini F, Lombardi A,
Vannini L, Guerzoni ME, Andrighetto C and Lanorte MT: A survey of
the enterococci isolated from an artisanal Italian goat's cheese
(semicotto caprino). J Appl Microbiol. 89:267–274. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sarantinopoulos P, Kalantzopoulos G and
Tsakalidou E: Effect of Enterococcus faecium on microbiological,
physicochemical and sensory characteristics of Greek Feta cheese.
Int J Food Microbiol. 76:93–105. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Juven BJ, Weisslowicz H and Harel S:
Detection of hydrogen peroxide produced by meat lactic starter
cultures. J Appl Bacteriol. 65:357–360. 1988. View Article : Google Scholar
|
|
35
|
Cotter PD, Hill C and Ross RP:
Bacteriocins: Developing innate immunity for food. Nat Rev
Microbiol. 3:777–788. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perez R H, Zendo T and Sonomoto K: Novel
bacteriocins from lactic acid bacteria (LAB): Various structures
and applications. Microb Cell Fact. 13(Suppl 1): S32014. View Article : Google Scholar
|
|
37
|
Klaenhammer TR: Genetics of bacteriocins
produced by lactic acid bacteria. FEMS Microbiol Rev. 12:39–85.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yousif NM, Dawyndt P, Abriouel H, Wijaya
A, Schillinger U, Vancanneyt M, Swings J, Dirar HA, Holzapfel WH
and Franz CM: Molecular characterization, technological properties
and safety aspects of enterococci from ‘Hussuwa’, an African
fermented sorghum product. J Appl Microbiol. 98:216–228. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Billström H, Top J, Edlund C and Lund B:
Frequent occurrence of multidrug-resistant CC17 Enterococcus
faecium among clinical isolates in Sweden. J Appl Microbiol.
108:1810–1816. 2010. View Article : Google Scholar : PubMed/NCBI
|