Introduction
Leucine-rich repeats and immunoglobulin-like domain
protein 1 (LRIG1) is located at chromosome 3p14, which is
recurrently deleted in human cancers (1,2). A
previous study demonstrated that LRIG1 was a negative regulating
factor of numerous oncogenic receptor tyrosine kinases (RTKs)
(3). In two independent studies,
LRIG1 was shown to downregulate the expression of four members of
the epidermal growth factor (EGF) family of receptor tyrosine
kinases (ERBB receptors), including the EGF receptor (EGFR), human
epidermal growth factor receptor 2 (HER2), HER3 and HER4 (3,4). In
addition, LRIG1 was shown to reduce the levels of EGFR by
accelerating receptor ubiquitylation and lysosomal degradation
(4). Furthermore, LRIG1 has been
reported to regulate ERBB receptor degradation and act as a tumor
suppressor (5). However, at present,
no correlation has been demonstrated between LRIG1 and
EGFR expression in colorectal cancer (6). Thus, the various functions of
LRIG1 in cancer remain uncertain.
Previous studies reported that LRIG1
overexpression suppressed the growth of the PC3 prostate cancer
cell line, and that this effect was overcome by androgen in
patients with increased LRIG1 and androgen activities (7,8). These
findings suggested that LRIG1 may be unable to elicit sufficient
tumor suppressive activity in a high-androgen environment. The
prognostic significance of the LRIG1 protein in cervical cancer has
been associated with tumor suppressors, oncogenes and numerous
factors, including cancer subtype and stage (9). LRIG1 expression has been shown
to be downregulated in a number of cancer types, although it was
reported to be overexpressed in prostate and colorectal tumors
(7,8). The expression level and subcellular
location of LRIG proteins have a prognostic value in brain tumors
(10). Due to this heterogeneity in
results, whether the function of LRIG1 suppresses or promotes tumor
growth remains unclear.
Phosphatase and tensin homolog (PTEN) is a type I
protein tyrosine phosphatase containing five domains. The
phosphatase and tensin domains of PTEN are recurrently
deleted from chromosome 10q23 in oncogenic events (11). PTEN targets a wide range of
molecules, thereby regulating tumorigenic functions such as
apoptosis, the cell cycle, cell adhesion and cell migration
(12). The function of LRIG1 and its
regulatory association with the PTEN gene in esophageal
squamous cell carcinoma (ESCC) remains unclear. Therefore, the
present study aimed to investigate the association between
LRIG1 and PTEN in ESCC cell lines and patient
samples.
Materials and methods
Reagents
The pEGFP-N1-LRIG1 plasmid was provided by Professor
Guo Dongsheng (Huazhong University of Science & Technology,
Wuhan, China). The empty pEGFP-N1 vector, used as a control, was
provided by Xinjiang Medical University (Urumqi, China). Inhibitors
of phosphoinositide 3-kinase (PI3K; LY294002), mitogen-activated
protein kinase (MAPK) kinase 1 (MEK; PD98059, U0126), p38-MAPK2
(SB203580), janus kinase 2 (JAK2; AG-490),
Ca2+/calmodulin-dependent protein kinase II (KN-62) and
calcineurin (FK-506) were purchased from Sigma-Aldrich (St. Louis,
MO, USA). The protein kinase C (PKC) inhibitor, bisindolylmaleimide
I,was obtained from Cell Signaling Technology, Inc. (Danvers, MA,
USA). Anti-LRIG1 antibody was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Anti-PTEN antibody was
purchased from Wuhan Boster Biological Technology, Ltd. (Wuhan,
China).
Cell culture
Eca-109 human esophageal cancer cells were purchased
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). KYSE-450 human esophageal cancer cells were
purchased from the Beijing Institute for Cancer Research (Beijing
Cancer Hospital, Beijing, China). All cells were maintained
according to the manufacturer's protocols. Briefly, the cells were
cultured at a density of 4×104 in Roswell Park Memorial
Institute-1640 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 5% fetal bovine serum (Hangzhou
Sijiqing Biological Engineering Materials Co., Ltd., Hangzhou,
China) and 100 µ/ml penicillin/streptomycin in six-well plates in a
5% CO2 humidified incubator at 37°C. After achieving 80%
confluence, the Eca-109, and KYSE-450 cells were pretreated with
LY294002 (10 µM), PD98059 (50 µM), U0126 (10 µM),
bisindolylmaleimide I (4 µM), SB203580 (10 µM), AG-490 (10 µM),
KN-62 (10 µM) and FK-506 (10 µM) for 1 h at 37°C, followed by
transfection with pEGFP-N1-LRIG1 or pEGFP-N1 (control) plasmids
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) for 48 h.
Patients
A total of 48 pairs of ESCC and nearby normal
esophageal tissue samples were obtained from Hazak patients at the
Affiliated Tumor Hospital of Xinjiang Medical University between
January 2008 and June 2009. Informed consent was obtained from all
patients, and the present study was approved by the research
ethical committee of the Affiliated Tumor Hospital of Xinjiang
Medical University. All patients underwent a radical esophagectomy
without preoperative chemotherapy or radiotherapy, and the
pathological diagnosis of ESCC was confirmed by histopathological
analyses. Tumors were classified as stage I (n=1), IIa
(n=13), IIb (n=5) or III (n=29), according to
the criteria of the Unio Internationale Contra Cancrum
tumor-node-metastasis classification system (13) of malignant tumors. Tumors and coupled
normal esophageal tissues from the same patient were harvested and
analyzed by senior pathologists, followed by freezing in liquid
nitrogen within 30 min postoperatively and storing at −80°C until
experimental use.
Immunohistochemical analysis
Cells (4×104) were cultured in RPMI-1640
supplemented with 5% fetal bovine serum and 100 µ/ml
penicillin/streptomycin in six-well plates. The plates were then
placed in a 5% CO2 humidified incubator at 37°C.
Following 24 h of growth, cells attached to the glass slide were
fixed with 4% poly formaldehyde and counted by immunohistochemical
staining. Further steps were conducted in a hydrated chamber at
room temperature. Endogenous peroxidase activity was inhibited
using a peroxidase block (Wuhan Boster Biological Technology, Ltd.)
for 5 min, after which the slides were incubated with 20% normal
goat serum in 50 mM Tris-HCl (pH 7.4). The cells attached to the
glass slide were incubated with goat anti-human LRIG1 monoclonal
antibody (1:100) overnight at 4°C. Subsequently, the cells were
incubated with rabbit horseradish peroxidase (HRP)-conjugated
anti-goat IgG secondary antibody (1:6,000; cat. no. BA1060; Wuhan
Boster Biological Technology, Ltd.) at 37°C for 30 min, following
washing with phosphate-buffered saline (PBS), and stained with
3,3′-diaminobenzidine. The slides were visualized by fluorescent
inverted microscopy.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tissue samples and
ESCC cell lines using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. cDNA
was synthesized from 1 µg RNA in a 20 µl reaction mixture using the
Reverse Transcription System (cat. no. A3500; Promega Corporation,
Madison, WI, USA). cDNA was amplified using the following primers:
GAPDH forward, 5′-GACCTGACCTGCCGCCTA-3′ and reverse,
5′-AGGAGTGGGTGTCGCTGT-3′; LRIG1 forward,
5′-ACCTGTGAACCTTGGAGGAAT-3′ and reverse,
5′-TCATCGCACACGAAACTCTC-3′; and PTEN forward,
5′-AAAGGGACGAACTGTGGTGTAATG-3′ and reverse,
5′-TGGTCCTTACTTCCCCATAGAA-3′. qPCR was performed using the SYBR
Green PCR Master Mix (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. PCR was performed in a
reaction volume of 20 µl containing 10 µl 2X PCR Master, 2 µl cDNA,
7 µl water, 0.5 µl forward primer and 0.5 µl reverse primer. The
thermal cycling conditions were as follows: 5 min at 95°C, followed
by 40 cycles (94°C, 30 sec; 60°C, 30 sec; 72°C, 30 sec). Relative
mRNA expression levels were determined by normalization to GAPDH
using the 2−ΔΔCq method (14).
Western blot analysis
Cells and tissues (following tissue homogenization)
were lysed using cell lysis buffer (5 mM Tris-HCl, 1 mM EGTA, 1 mM
EDTA, 150 mM NaCl, 1% NP-40, 0.1% SDS and 1 mM PMSF; pH 7.5)
containing 1% Triton X-100 with protease inhibitors at 4°C for 30
min. Protein lysates were purified by centrifugation (15,000 × g;
4°C; 10 min), after which equal quantities of protein (60 µg) were
separated from various cells and tissue samples and boiled for 5
min. The proteins were then separated by 10% SDS-PAGE and
transferred onto nitrocellulose membranes. The membranes were
blocked overnight at 4°C using blocking solution containing 1%
bovine serum albumin (Sangon Biotech Co., Ltd., Shanghai, China),
5% skimmed milk and 0.05% Tween-20. Subsequently, the membranes
were incubated overnight at 4°C with polyclonal goat anti-human
LRIG1 (1:250; cat. no. SC-134435; Santa Cruz Biotechnology, Inc.),
monoclonal rabbit anti-human PTEN (1:100; cat. no. BA1377; Wuhan
Boster Biotechnology, Co., Ltd.), and monoclonal rabbit anti-human
GAPDH (1:400; cat. no. BM1623; Wuhan Boster Biotechnology, Co.,
Ltd.) antibodies, followed by washing three times with
Tris-buffered saline supplemented with Tween-20 and incubation
overnight at 4°C with monoclonal HRP-conjugated goat anti-rabbit
(cat. no. BA1055; Wuhan Boster Biotechnology Co., Ltd.) and rabbit
anti-goat (cat. no. BA1060; Wuhan Boster Biotechnology Co., Ltd.)
secondary antibodies (1:6,000). The protein complexes were
evaluated using an enhanced chemiluminescence system (Wuhan Boster
Biological Technology, Ltd.), and the blots were visualized using
3,3′-diaminobenzidine (Beijing Zhongshan Golden Bridge
Biotechnology, Co., Ltd.). Western blot bands were quantified using
Quantity One software version 4.6.2 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), which measured the band intensity of each
group. Band intensities were normalized to GAPDH as an internal
control.
Apoptosis analysis
The level of apoptosis was evaluated by Annexin
V-fluorescein isothiocyanate (FITC) staining and flow cytometry.
Following pretreatment with various inhibitors and transfection
with pEGFP-N1-LRIG1 plasmids, the cells were harvested, washed with
pre-chilled PBS and resuspended in 1X binding buffer at a density
of ~1×106 cells/ml. Subsequently, ~100 µl solution was
mixed with 5 µl Annexin V-FITC and 5 µl propidium iodide (PI) for
15 min, followed by addition of 400 µl 1X binding buffer. Apoptosis
was assessed within 1 h using the BD FACSCalibur (BD Biosciences,
San Jose, CA, USA). The percentage of apoptotic cells was assessed
using CXP cytometer software version 2.0 (Beckman Coulter, Inc.,
Brea, CA, USA).
Cell cycle distribution analysis
Cells were cultured in six-well plates until
reaching ~60–70% confluence, followed by transfection with
pEGFP-N1-LRIG1 plasmid. After 48 h, the cells were harvested using
0.25% trypsin, washed with 1X PBS and centrifuged (1,500 × g; 5
min) at 4°C. The resulting cell pellets were resuspended in Eca-109
and KYSE-450 cells and fixed with 75% ethanol for 1 h at 4°C,
followed by washing with PBS, resuspending in 0.05 mg/ml PI
solution containing RNase A and incubation in the dark at 4°C for
30 min. Subsequently, the cell cycle distribution was analyzed
using a flow cytometer.
Statistical analysis
Standard two-tailed t-tests were performed on two
data sets. Representative data from three experiments are
presented, in which bars represent the mean ± standard deviation.
Summary statistics of patient data were calculated to demonstrate
patient characteristics. Fisher's exact test was used to evaluate
the association between two categorical variables. SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA) was used to perform
Fisher's exact test and t-tests to analyze the differences in gene
expression between the ESCC and non-cancerous mucosa. P≤0.05 was
considered to indicate a statistically significant difference.
Results
LRIG1 downregulates PTEN expression in
esophageal cancer cell lines
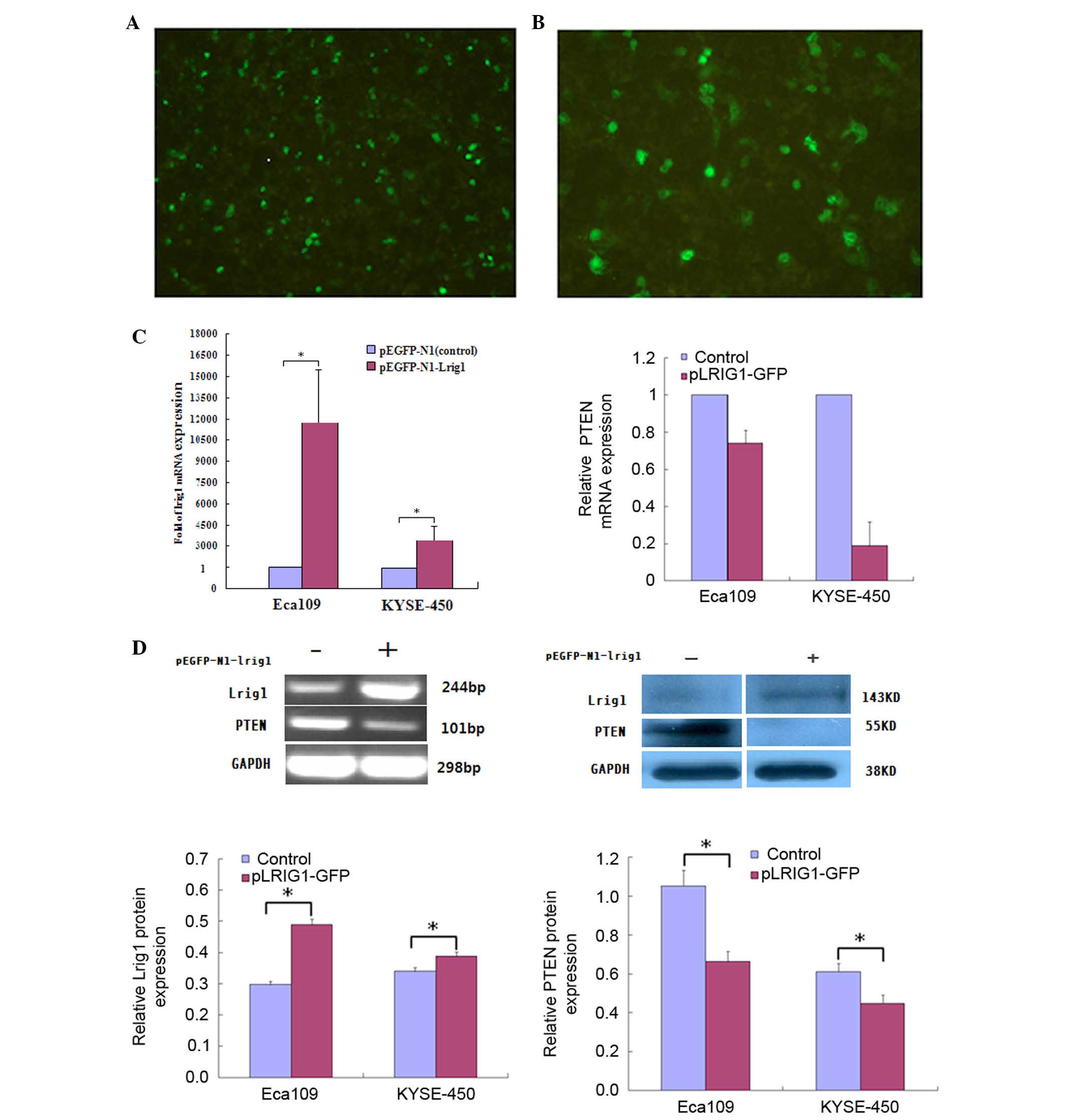
The pEGFP-N1-LRIG1 plasmid was transfected into
Eca-109 and KYSE-450 cell lines, in which LRIG1 expression
levels were intrinsically low. LRIG1 expression was
inversely correlated with PTEN expression in Eca-109 and
KYSE-450 cell lines following transfection of the cells. At 48 h
after transfection, fluorescence microscopy detected green
fluorescence in the transfected cells (Fig. 1A and B). The mRNA expression levels
of LRIG1 were examined by RT-qPCR at 48 h following
transfection, and demonstrated a significant increase (magnitude,
>212) in the expression levels of LRIG1 mRNA
in the transfected cells, as compared with the negative controls
(Fig. 1C). In addition, western
blotting demonstrated that the protein expression levels of LRIG1
were significantly increased by ~10.1% in KYSE-450 cells and 50.5%
in Eca-109 cells transfected with pEGFP-N1-LRIG1 plasmid, as
compared with the negative controls (P<0.05; Fig. 1D).
RT-qPCR and western blotting demonstrated a
correlation between PTEN and LRIG1 expression levels
in Eca-109 and KYSE-450 cell lines following transfection with the
pLRIG1-GFP-N1 plasmid (Fig. 1C and
D). Upregulation of LRIG1 expression markedly decreased
the mRNA and significantly decreased the protein expression levels
(P<0.05) of PTEN in the transfected cells (Fig. 1C and D), as compared with the
negative controls.
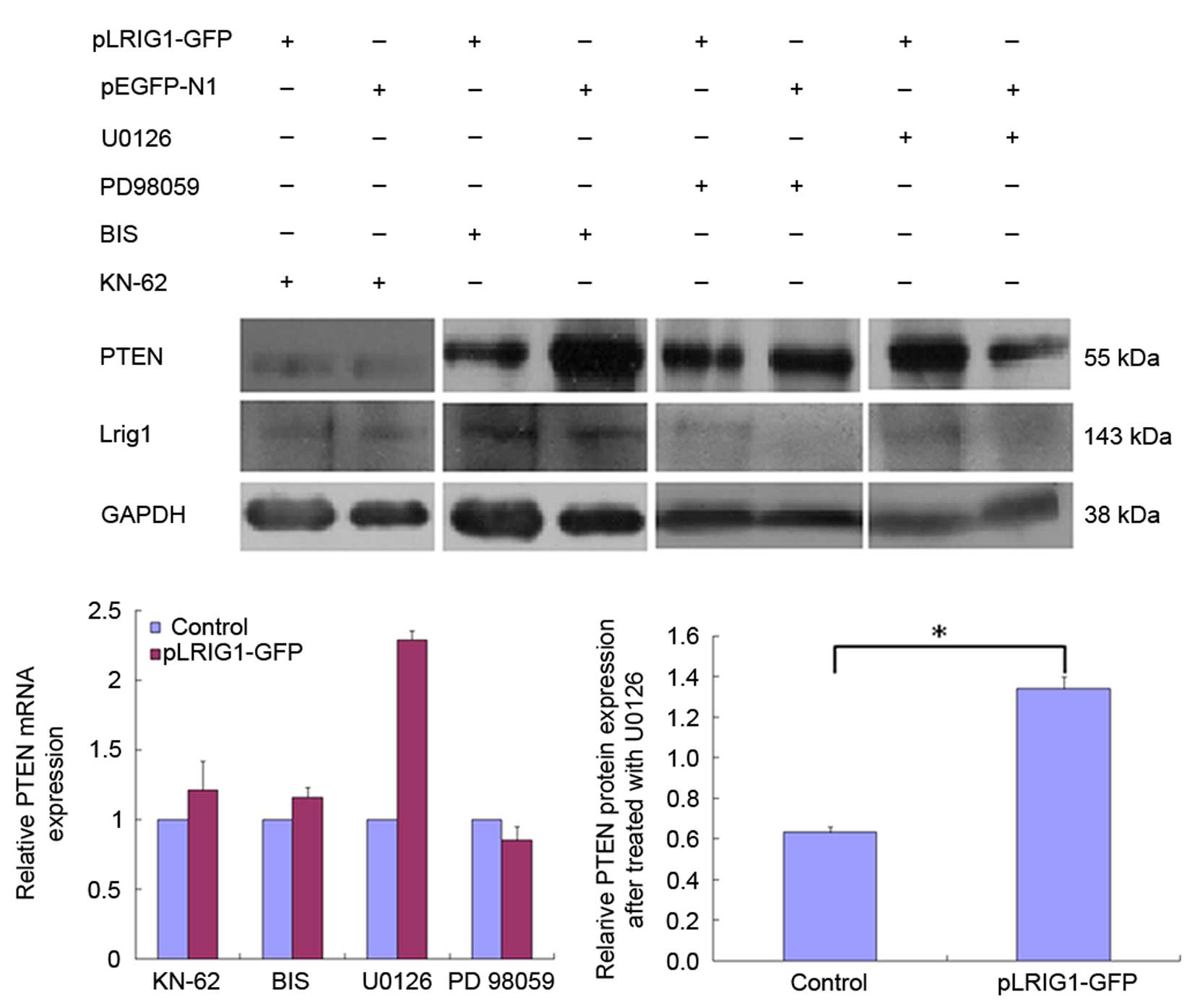
The cells were pretreated with various inhibitors to
elucidate the signal transduction pathways involved in the
LRIG1-mediated inhibition of PTEN expression. The ability of
LRIG1 to downregulate PTEN expression was diminished by
pretreatment of Eca-109 and KYSE-450 cells with U0126 and KN-62
inhibitors (Fig. 2). These results
suggest that LRIG1-induced downregulation of PTEN in
esophageal cancer cells involves the MEK signaling pathway.
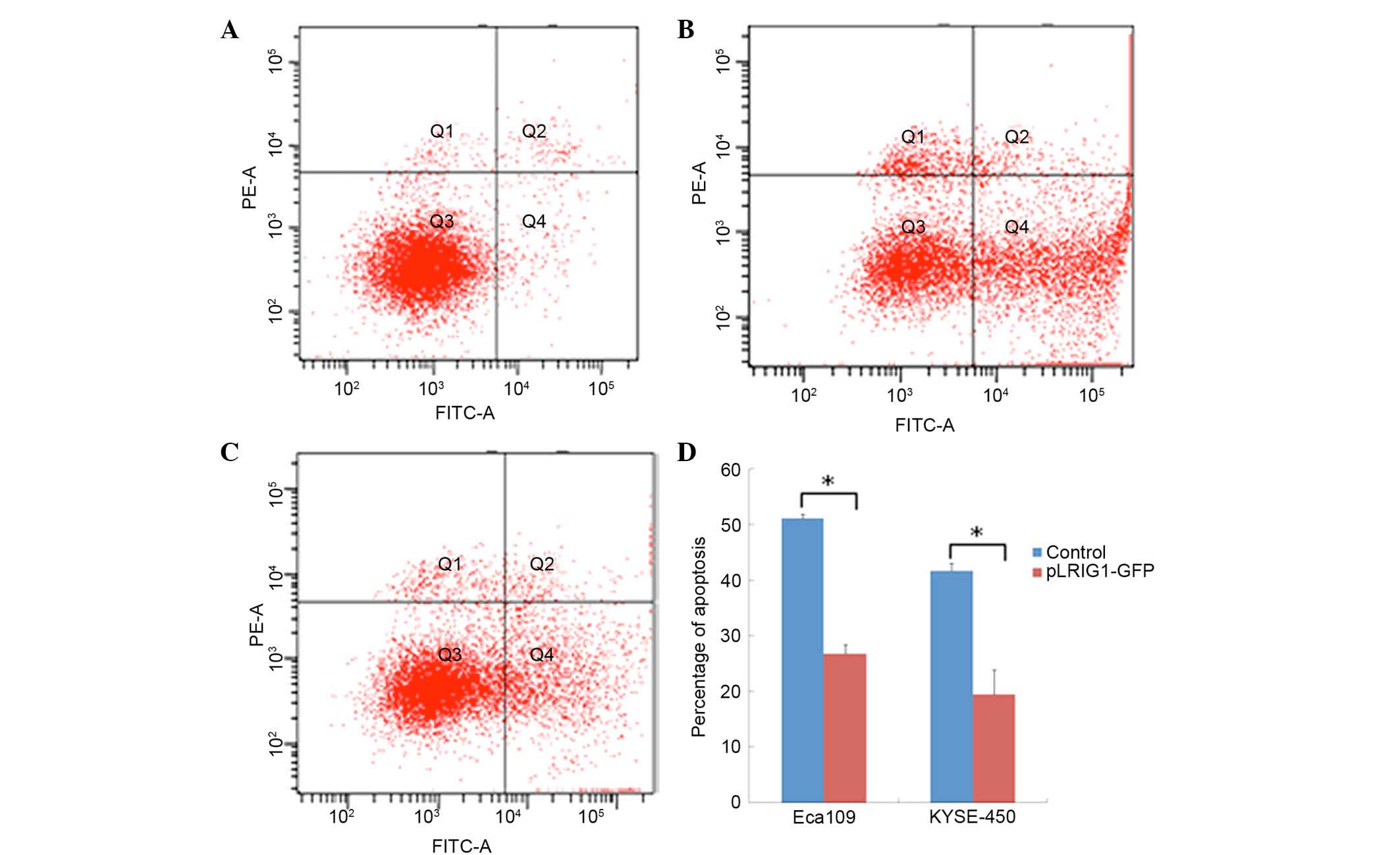
Overexpression of LRIG1 reduces
apoptosis in Eca-109 and KYSE-450 cells
The effect of LRIG1 overexpression on the
apoptosis of Eca-109 and KYSE-450 cells transfected with the
pEGFP-N1-LRIG1 plasmid was assessed by flow cytometry. The
percentage of apoptotic Eca-109 cells was significantly decreased
from 51.10% in control cells transfected with pEGFP-N1 plasmid to
26.76% in Eca-109 cells transfected with pEGFP-N1-LRIG1 plasmid
(P<0.05; Fig. 3). Similarly, the
percentage of apoptotic KYSE-450 cells was significantly decreased
from 41.70% in control cells to 19.47% in LRIG1
overexpressing cells (P<0.05; Fig.
3).
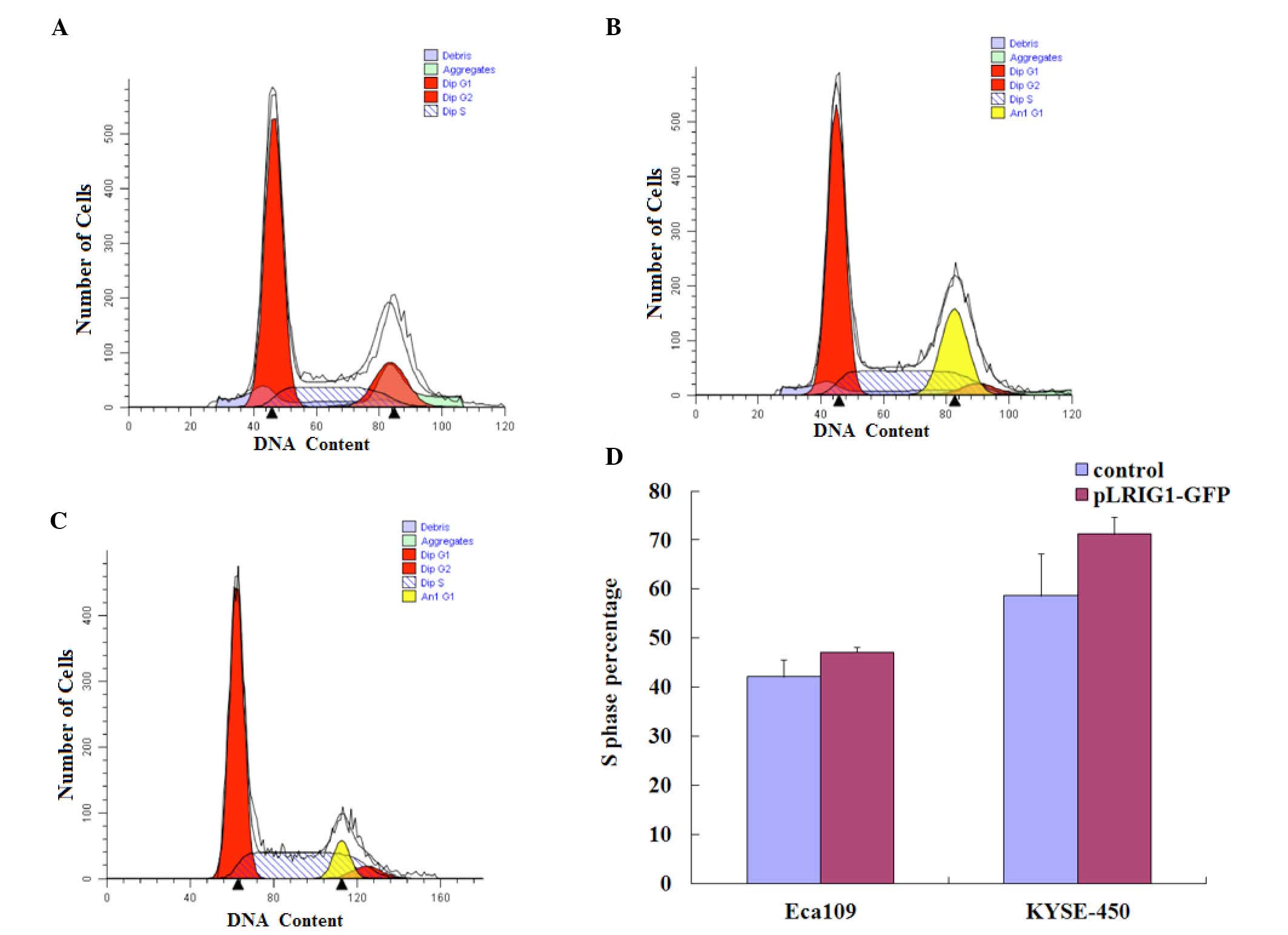
LRIG1 overexpression arrests the cell
cycle in the S-phase
The effects of LRIG1 overexpression on the
cell cycle distribution of Eca-109 and KYSE-450 cells was assessed
by flow cytometry (Fig. 4A-C). As
compared with the control cells, LRIG1 overexpression
increased the percentage of Eca-109 and KYSE-450 cells in the
S-phase of the cell cycle. The percentage of Eca-109 cells in the
S-phase was increased from 42.11% of the control cells to 47.19% of
the LRIG1 overexpressing cells (Fig. 4D). Similarly, the percentage of
KYSE-450 cells in the S-phase was increased from 58.64% of the
control cells to 71.24% of the LRIG1 overexpression cells
(Fig. 4D).
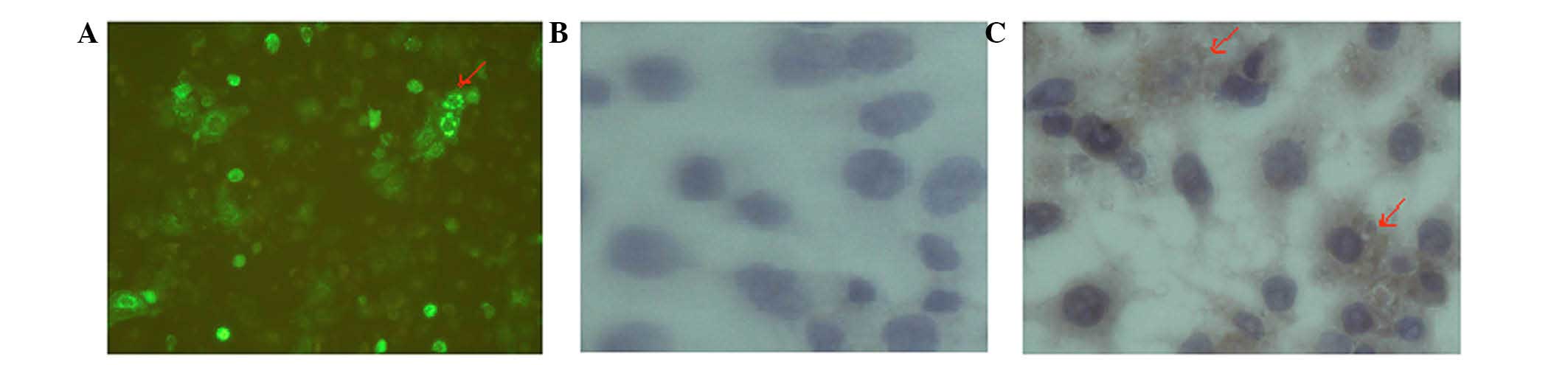
LRIG1 protein localization in Eca-109
and KYSE-450 cells
The localization of LRIG1 in Eca-109 and KYSE-450
cells was determined by immunohistochemical analyses. The
perinuclear staining of LRIG1 was observed by microscopy in Eca-109
cells following transfection of the cells with the pLRIG1-GFP
plasmid. In addition, the LRIG1 protein was primarily observed in
the cytoplasm of KYSE-450 cells (Fig.
5).
PTEN mRNA expression negatively
correlated with LRIG1 in cancerous tissue
Total RNA was isolated from ESCC tissues and
corresponding normal distant tissues from the same patient, and the
mRNA expression levels of LRIG1 and PTEN were
determined by RT-qPCR. The mRNA expression levels of PTEN
were inversely correlated to the expression levels of LRIG1
in the cancerous tissue (R=0.478; P=0.002; Table I). No significant correlation was
observed between clinical pathological factors and the expression
levels of LRIG1 or PTEN.
 | Table I.Association between LRIG1 and
PTEN mRNA expression in esophageal squamous cell carcinoma
cancer tissues and matched normal tissues (n=48). |
Table I.
Association between LRIG1 and
PTEN mRNA expression in esophageal squamous cell carcinoma
cancer tissues and matched normal tissues (n=48).
|
| LRIG1
(tumor) |
|
| LRIG1
(normal) |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Gene | + | − | R | P-value | + | − | R | P-value |
|---|
| PTEN |
|
| 0.478 | 0.002 |
|
| 0.181 | 0.183 |
| + | 29 | 9 |
|
| 18 | 19 |
|
|
| − | 2 | 8 |
|
| 3 | 8 |
|
|
Discussion
LRIG1, a membrane-associated protein, has been shown
to inhibit growth factor signal transduction from oncogenic RTKs,
including EGFR, and MET and RET proto-oncogenes (15–17). The
downregulation of LRIG1 expression results in spontaneous
tumor formation, which suggests that LRIG1 may serve as a
tumor suppressor in certain types of cancer (2). However, LRIG1 has been shown to
be overexpressed in numerous cancers, including leukemia,
astrocytoma and prostate cancers, thus suggesting that LRIG1
may perform general downregulation in all types of human tumors
(7,18), and that the role of LRIG1 may
be masked by other factors. Therefore, the expression and function
of LRIG1 should be carefully evaluated. In a previous study
performed by the authors, it was demonstrated that LRIG1
expression was not associated with EGFR expression, although it was
correlated with HER2 expression in ESCC (19), which was consistent with the
demonstrated correlation between increased gene copy numbers of
HER2 and LRIG1 in a previous study (20).
The progression of esophageal cancer has been
associated with the loss of tumor suppressor genes, including
PTEN (21). PTEN may be an
important biological marker in the treatment of human esophageal
cancer (21). The present study
demonstrated a significant correlation between LRIG1 and
PTEN mRNA expression levels in ESCC cell lines, and in tumor
samples from patients with ESCC. Notably, LRIG1
overexpression downregulated PTEN expression in Eca-109 and
KYSE-450 cell lines, indicating that LRIG1 may have an oncogenic
role. The expression and subcellular localization of LRIG1 may be
associated with specific clinicopathological features of ependymoma
tumors, and thus may be of great importance in carcinogenesis
(22). In human oligodendroglioma,
the cytoplasmic and perinuclear localizations of LRIG1 were
associated with the expression of various genes; thus suggesting
that LRIG1 may perform various functions. In the present study,
immunohistochemical analysis demonstrated that LRIG1 was
predominantly expressed in the cytoplasm in ESCC cell lines.
Notably, a number of perinuclear LRIG1 were observed following
transfection with LRIG1 plasmid.
PTEN selectively inhibits the activation of
extracellular signal-regulated kinase (ERK) in the MAPK signaling
pathway (23,24). The absence of PTEN in cancer cells
has typically been associated with the increased activation of the
PI3K/Akt signaling pathway, leading to malignant cancer
transformation and progression (25,26). In
the present study, LRIG1-mediated downregulation of PTEN
mRNA expression was diminished by pretreatment of ESCC cell lines
with bisindolylmaleimide I (PKC inhibitor), PD98059 (ERK 1/2
inhibitor) and U0126 (MEK 1/2 inhibitor). These results suggest
that LRIG1 regulates PTEN expression via the MAPK/MEK/ERK signaling
pathway.
The apoptosis of Eca-109 and KYSE-450 cells was
significantly inhibited following transfection of the cells with
the pLRIG1-EGF plasmid, compared with the control cells.
Furthermore, LRIG1 overexpression increased the percentage of
Eca-109 and KYSE-450 cells in the S-phase of the cell cycle. These
results suggested that LRIG1 inhibited cell apoptosis and
promoted the growth of cancer cells, serving as an oncogene in
ESCC. In addition, these results indicated that LRIG1 may
function as an oncogene by regulating tumor-suppressor genes in a
manner that is dependent on the subcellular localization of LRIG1.
The overexpression of cytoplasmic LRIG1 in ESCC cell lines may have
promoted cell growth by suppressing apoptosis and accelerating the
cell cycle.
In conclusion, the present study demonstrated that
LRIG1 overexpression downregulated PTEN expression in
ESCC cell lines by activating the MAPK/MEK signaling pathway. In
addition, LRIG1 was shown to function as a proto-oncogene by
promoting tumor cell proliferation and simultaneously decreasing
cellular apoptosis. LRIG1 followed a non-canonical mechanism of
interaction with various signal transduction pathways to elicit
novel functions in the cytoplasm of tumor cells.
Acknowledgements
The present study was supported by the Nature
Science Foundation of China (grant nos. 30950012 and 81360304) and
the Xinjiang Endemic Molecular Biology Laboratory, China (grant no.
xjdx0208-2011-02).
References
|
1
|
Nilsson J, Vallbo C, Guo D, Golovleva I,
Hallberg B, Henriksson R and Hedman H: Cloning, characterization
and expression of human LIG1. Biochem Biophys Res Commun.
284:31–37. 2001. View Article : Google Scholar
|
|
2
|
Hedman H, Nilsson J, Guo D and Henriksson
R: Is LRIG1 a tumour suIs LRIG1 a tumour suppressor gene at
chromosome 3p14.3? Acta Oncol. 41:352–354. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laederich MB, Funes-Duran M, Yen L,
Ingalla E, Wu X, Carraway KL III and Sweeney C: The leucine-rich
repeat protein LRIG1 Is a negative regulator of ErbB family
receptor tyrosine kinases. J Biol Chem. 279:47050–47056. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gur G, Rubin C, Katz M, Amit I, Citri A,
Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al:
LRIG1 restricts growth factor signaling by enhancing receptor
ubiquitylation and degradation. EMBO J. 23:3270–3281. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Poulin EJ and Coffey RJ: LRIG1 is
a triple threat: ERBB negative regulator, intestinal stem cell
marker and tumour suppressor. Br J Cancer. 108:1765–1770. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ljuslinder I, Golovleva I, Palmqvist R,
Oberg A, Stenling R, Jonsson Y, Hedman H, Henriksson R and Malmer
B: LRIG1 expression in colorectal cancer. Acta Oncol. 46:1118–112.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomasson M, Wang B, Hammarsten P, Dahlman
A, Persson JL, Josefsson A, Stattin P, Granfors T, Egevad L,
Henriksson R, et al: LRIG1 and the liar paradox in prostate cancer:
A study of the expression and clinical significance of LRIG1 in
prostate cancer. Int J Cancer. 128:2843–2852. 2010. View Article : Google Scholar
|
|
8
|
Thomasson M, Hedman H, Ljungberg B and
Henriksson R: Gene expression pattern of the epidermal growth
factor receptor family and LRIG1 in renal cell carcinoma. BMC Res
Notes. 5:2162012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lindström AK, Ekman K, Stendahl U, Tot T,
Henriksson R, Hedman H and Hellberg D: LRIG1 and squamous
epithelial uterine cervical cancer: Correlation to prognosis, other
tumor markers, sex steroid hormones and smoking nternational. Int J
Glynecol Cancer. 18:312–317. 2008. View Article : Google Scholar
|
|
10
|
Guo D, Nilsson J, Haapasalo H, Raheem O,
Bergenheim T, Hedman H and Henriksson R: Perinuclear leucine-rich
repeats and immunoglobulin-like domain proteins (LRIG1-3) as
prognostic indicators in astrocytic tumors. Acta Neuropathol.
111:238–246. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mak LH and Woscholski R: Targeting PTEN
using small molecule inhibitors. Methods 77–78. 63–68. 2015.
View Article : Google Scholar
|
|
12
|
Zhang H, Sun Z and Kong Y: Expression of
PTEN, Her-2 and Glut-1 proteins in endometrial intraepithelial
neoplasia and endometrioid. Xin Jiang Yi Ke Da Xue Xue Bao.
34:142–146. 2011.
|
|
13
|
Edge SB, Byrd DR and Compton CC: AJCC
Cancer Staging Manual. 7th. New York: Springer; pp. 103–111.
2009
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JM, Kim B, Lee SB, Jeong Y, Oh YM,
Song YJ, Jung S, Choi J, Lee S, Cheong KH, et al: Cbl-independent
degradation of Met: Ways to avoid agonism of bivalent Met-targeting
antibody. Oncogene. 33:34–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goldoni S, Iozzo RA, Kay P, Campbell S,
McQuillan A, Agnew C, Zhu JX, Keene DR, Reed CC and Iozzo RV: A
soluble ectodomain of LRIG1 inhibits cancer cell growth by
attenuating basal and ligand-dependent EGFR activity. Oncogene.
26:368–381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ledda F, Bieraugel O, Fard SS, Vilar M and
Paratcha G: Lrig1 Is an endogenous inhibitor of Ret receptor
tyrosine kinase activation, downstream signaling and biological
responses to GDNF. J Neurosci. 28:39–49. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thomasson M, Wang B, Hammarsten P, Dahlman
A, Persson JL, Josefsson A, Stattin P, Granfors T, Egevad L,
Henriksson R, et al: LRIG1 and the liar paradox in prostate cancer:
A study of the expression and clinical significance of LRIG1 in
prostate cancer. Int J Cancer. 128:2843–2852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang XF, Li H, Wang HJ, Li XM, Chen Y,
Pang ZL, Chen HM and Li HW: Research of Lrig1 in esophageal
squamous cell carcinoma. Chin J Dig. 33:123–124. 2013.
|
|
20
|
Ljuslinder I, Golovleva I, Henriksson R,
Grankvist K, Malmer B and Hedman H: Co-incidental increase in gene
copy number of ERBB2 and LRIG1 in breast cancer. Breast Cancer Res.
11:4032009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hou G, Lu Z, Liu M, Liu H and Xue L:
Mutational analysis of the pten gene and its effects in esophageal
squamous cell carcinoma. Dig Dis Sci. 56:1315–1322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yi W, Haapasalo H, Holmlund C, Järvelä S,
Raheem O, Bergenheim AT, Hedman H and Henriksson R: Expression of
leucine-rich repeats and immunoglobulin-like domains (Lrig)
proteins in human ependymoma relates to tumor location, who grade
and patient age. Clin Neuropathol. 28:21–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chappell WH, Steelman LS, Long JM, Kempf
RC, Abrams SL, Franklin RA, Bäsecke J, Stivala F, Donia M, Fagone
P, et al: Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors:
Rationale and importance to inhibiting these pathways in human
health. Oncotarget. 2:135–164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chetram MA and Hinton CV: PTEN regulation
of ERK1/2 signaling in cancer. J Recept Signal Transduct Res.
32:190–195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maehama T and Dixon JE: The tumor
suppressor, PTEN/MMAC1, dephosphorylates the lipid second
messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem.
273:13375–13378. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chetram MA, Don-Salu-Hewage AS and Hinton
CV: ROS enhances CXCR4-mediated functions through inactivation of
PTEN in prostate cancer cells. Biochem Biophys Res Commun.
410:195–200. 2011. View Article : Google Scholar : PubMed/NCBI
|