Introduction
The Gram-negative bacteria Brucella causes
Brucellosis, a zoonotic disease that is widely disseminated
throughout the world (1). In humans,
Brucellosis causes undulant fever, arthritis and myocarditis
(2). Brucella spp. are able
to survive and multiply inside the placenta and fetus of pregnant
mammals, causing abortion during pregnancy (3). The interactions between Brucella
and their hosts are extremely complex, as these facultative
intracellular parasites are able to adapt to the harsh environment
of host cells, which include oxidative damage, nitrosative damage,
acidic pH, antimicrobial peptides and nutrient deprivation
(4,5). Brucella spp. are able to achieve
this by regulating gene expression differently when growing in
vitro or in vivo. However, the mechanisms underlying
this survival and multiplication within host cells require further
characterization.
Brucella spp. have a tropism for cells
containing erythritol, such as embryo-trophoblasts found in the
placenta (6), and erythritol has a
growth-promoting effect on some Brucella strains. Therefore,
Brucella spp. are able to colonize and reproduce in embryo
trophoblast cells, which can cause placentitis and result in
abortion (7). Furthermore, the
virulence of Brucella spp. is correlated with erythritol
metabolism (8), and the
Brucella-encoded catabolic erythritol pathways are required
for intracellular survival (9).
Erythritol usage relies on the ery operon, which consists of
the genes eryA, eryB, eryC and eryD
(10). The eryA gene encodes
a 519 amino acids (AA) putative erythritol kinase (10). The eryB gene encodes an
erythritol phosphate dehydrogenase (10). The eryC gene product has been
assigned as a D-erythrulose-1-phosphate dehydrogenase, and the
eryD gene encodes a regulator of ery operon
expression (10–12). Although ery operon expression
is correlated with erythritol metabolism, growth conditions can
regulate gene expression (10). To
understand ery operon regulation in Brucella
melitensis during infection, we examined gene expression at
several timepoints following growth in HPT-8 trophoblast cells. The
results help to characterize the mechanisms required for
Brucella spp. pathogenesis.
Materials and methods
Bacterial strains, plasmids and growth
conditions
Table I lists the
strains and constructed plasmids used in this study. Brucella
abortus 2308 was obtained from the Chinese Center of Disease
Prevention and Control (CDC; Beijing, China). B. melitensis
027 strain was isolated from Xinjiang, China, and was identified by
the CDC. Brucella strains were cultured in tryptic soy agar
(TSA) or tryptic soy broth (TSB; Sigma-Aldrich, St. Louis, MO,
USA). Plates were incubated at a temperature of 37°C in an
atmosphere enriched with 5% CO2. Escherichia coli
strain JM109 (Promega Corporation, Madison, WI, USA) was grown in
Luria-Bertani (LB) media. The culture media were supplemented with
50 µg/ml ampicillin (Invitrogen; Thermo Fisher Scientific, Inc.,
Carlsbad, CA, USA). The plasmid pMD18-T Simple Vector was purchased
from Takara Bio, Inc. (Otsu, Japan). The standard curves were
constructed using pMD18-T Simple Vector.
 | Table I.Bacterial strains and plasmids used
in this study. |
Table I.
Bacterial strains and plasmids used
in this study.
| Name | Description | Source |
|---|
| Bacteria
strain |
|
|
|
Brucella abortus
2308 | Wild-type, virulent
strain | China CDC |
|
Brucella melitensis
027 | Biotype 3, virulent
strain (China), identified by China CDC | Present study |
|
2308Δery | Δery
promoter mutant of strain 2308 | Present study |
|
027Δery | Δery
promoter mutant of strain 027 | Present study |
|
Escherichia coli
JM109 | endA1,
recA1, gyrA96, thi, hsdR17
(rk-, mk+), relA1, |
|
|
| supE44,
Δ(lac-proAB), [F', traD36, proAB,
laqIqZΔM15] | Promega |
| Plasmid |
|
|
| pMD18-T
simple vector | Broad-host range
vector; Ampr | Takara |
|
pMD18-eryA | pMD18-T containing
87 bp fragment amplified with | Present study |
|
| eryA-RT-S
and eryA-RT-A including a fraction of eryA |
|
|
pMD18-eryB | pMD18-T containing
104 bp fragment amplified with | Present study |
|
| eryB-RT-S
and eryB-RT-A including a fraction of eryB |
|
|
pMD18-eryC | pMD18-T containing
118 bp fragment amplified with | Present study |
|
| eryC-RT-S
and eryC-RT-A including a fraction of eryC |
|
|
pMD18-eryD | pMD18-T containing
120 bp fragment amplified with | Present study |
|
| eryD-RT-S
and eryD-RT-A including a fraction of eryD |
|
|
pMD18-16S rRNA | pMD18-T containing
88 bp fragment amplified with | Present study |
|
| 16s-RNA-S and
16s-RNA-A including a fraction of 16S rRNA |
|
Cells
Murine macrophages (RAW 264.7) and human
trophoblasts (HPT-8) were used in this study. HPT-8 cells and RAW
264.7 murine macrophage were purchased from the Cell Resource
Center, IBMS, CAMS/PUMC (Beijing, China).
Construction of 2308Δery and
027Δery
Deletion of the ery operon in B.
abortus 2308 and B. melitensis 027 (2308Δery and
027Δery) was performed as previously described (13).
Growth curve of 2308Δery and
027Δery
To monitor the growth of B. abortus 2308,
B. melitensis 027, 2308Δery and 027Δery, cells
were cultured in TSB to an optical density at 600 nm (OD600) of
0.6, then diluted with TSB to an OD600 of 0.05 and cultured in
rotary shaker (100.62 × g) at 37°C for 48 h. Aliquots of cultures
were collected at 4-h intervals, and bacterial growth was measured
at OD600.
Erythritol sensitivity of 2308Δery and
027Δery
To detect erythritol sensitivity in 2308Δery
and 027Δery, stationary phase pre-cultures of B.
abortus 2308, B. melitensis 027, 2308Δery and
027Δery were diluted in TSB containing 20 mM erythritol
(Sigma-Aldrich), and grown for 48 h. The bacterial growth was
measured at an OD600.
Evaluation of 2308Δery and 027Δery
attenuation in RAW 264.7 murine macrophages
RAW 264.7 murine macrophages were used to assess the
intracellular survival of B. abortus 2308, B.
melitensis 027, 2308Δery and 027Δery. RAW 264.7
cells were infected as previously described (14). Briefly, 5×105 cells/well
were cultured in 24-well plates for 16 h at 37°C and infected with
Brucella at a multiplicity of infection (MOI) of 100.
Culture plates were centrifuged for 5 min at 350 × g at room
temperature and placed in an incubator at 37°C with 5%
CO2 atmosphere. At 45 min post-infection, the cells were
washed twice with media and then incubated with Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Rockville, MD, USA) containing 50 µg/ml gentamicin (Invitrogen;
Thermo Fisher Scientific, Inc.) for 1 h to kill extracellular
bacteria. The media was then replaced with DMEM containing 25 µg/ml
gentamicin (incubation point 0 min). At 4, 12, 24 and 48 h
post-infection, the number of colony-forming units (CFU) was
obtained by plating serial dilutions of the lysates on TSA plates.
All assays were performed in triplicate and repeated at least three
times.
HPT-8 cells invasion assay
HPT-8 cells were infected by B. abortus 2308
and B. melitensis 027 in media with or without erythritol
(20 mM). HPT-8 cells were grown at 37°C in a 5% CO2
atmosphere in DMEM containing 20% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.). Cells were seeded (1×106) in
12-well culture dishes 24 h prior to each infection assay. HPT-8
cells were infected at a MOI of 100 bacteria per cell as previously
described (14,15). Culture plates were centrifuged for 5
min at 350 × g at room temperature. Post-infection, cells were
grown in the presence of erythritol at a concentration of 1% as a
nutritional supplement or at 20 mM for induction of the ery
operon and placed in an incubator at 37°C with 5% CO2
atmosphere. Cells were washed three times with phosphate-buffered
saline (PBS) and monolayers of cells were further incubated with
culture media supplemented with 50 µg/ml gentamicin for 1 h to kill
extracellular bacteria. The cells were washed with DMEM containing
10% fetal bovine serum to remove gentamicin, then the cells were
lysed with TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.).
RNA isolation and reverse
transcription
Total RNA (1 µg) from HPT-8 cells at 0 min
(bacterial culture), 20 min, 1 h, 2 h, 3 h, 4 h and 12 h
post-infection was isolated (Qiagen RNeasy Mini-kits; Qiagen,
Hilden, Germany) and cDNA was generated using random hexamer
primers and MMLV-RT according to the manufacturer's recommendations
(Gibco; Thermo Fisher Scientific, Inc.). Genomic DNA was removed
using a DNase RT kit (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. The 10 µl reaction
mixture system containing 1 µg RNA, 1U DNase, 1 µl DNase buffer and
ddH2O was added to 10 µl. The mixture system was mixed and allowed
to rest for 10 min at room temperature. A total of 1 µl 25 mM EDTA
was added and incubated for 10 min at 65°C. The DNA polymerase was
obtained from Invitrogen (Thermo Fisher Scientific, Inc.). cDNA was
stored at −80°C and used as a template for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Oligonucleotide primers
TaqMan primers for 16S rRNA (housekeeping gene),
eryA, eryB, eryC and eryD genes were
designed using Primer Express 5.0 software (Applied Biosystems,
Palo Alto, CA, USA) according to sequences in GenBank (http://www.ncbi.nlm.nih.gov/genbank/)
(Table II).
 | Table II.Primer sequences used in this
study. |
Table II.
Primer sequences used in this
study.
| Primer | Sequence |
|---|
| 16S
rRNA-RT-sense |
GCGGCTCACTGGTCCATTAC |
| 16S
rRNA-RT-anti-sense |
CGTTTACGGCGTGGACTACC |
|
eryA-RT-sense |
CGCACACGCCAGTATGATGA |
|
eryA-RT-anti-sense |
CGACCCGTCGATGATTTCAG |
|
eryB-RT-sense |
GAGATTGCCAATGCCGATTA |
|
eryB-RT-anti-sense |
GCACCATAGAGCCGTCCATA |
|
eryC-RT-sense |
GCTTTCGCTCAACACCAATC |
|
eryC-RT-anti-sense |
CATGGGTAAGCTGGAGGTCA |
|
eryD-RT-sense |
CGTGGAAAACGCCGATATGA |
|
eryD-RT-anti-sense |
GTCCGTTCGTCGGTGATGAG |
Construction of recombinant
plasmids
Ery operon (eryA, eryB,
eryC and eryD) and 16S rRNA open reading frames were
amplified by PCR with specific primers (see Table II; Premier Biosoft, Palo Alto, CA,
USA) from the B. abortus 2308 genome. The amplified DNA
fragments and pMD18-T simple vectors were ligated overnight at 16°C
using T4 DNA ligase (Takara Bio, Inc.). The ligation reaction was
transformed into E. coli JM109, and insert-containing
plasmids were identified by restriction analysis or PCR. Positive
recombinant plasmids were sequenced to confirm the correct
construction.
Transcriptional analysis of ery operon
genes by RT-qPCR
The concentration and purity of recombinant plasmids
(dilution, ×100) were measured using a Nanodrop 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.) and used to
compute target gene copy numbers. A three-step, 45-cycle RT-qPCR
method was conducted using a LightCycler® 480 System
(Roche Diagnostics, Basel, Switzerland). A linear standard curve
was created with the LightCycler® 480 System Software
and serial dilutions of recombinant plasmids containing the genes
encoding 16S rRNA, eryA, eryB, eryC and
eryD. RT-qPCR was conducted with the following reaction
conditions: 5 min at 95°C, followed by 45 cycles at 58°C for 30 sec
and 72°C for 30 sec. A negative control (no cDNA) and RT control
(no reverse transcription) were used in the experiments. All assays
were performed in triplicate and repeated at least three times. The
expression levels of the target genes were calculated by comparison
with cycle threshold (Ct) and determined at 0 min, 20 min, 1 h, 2
h, 3 h, 4 h and 12 h post-infection. 16S rRNA expression was used
as a reference value to compare the relative expression levels at
the various time points. Target genes were amplified in triplicate
using the LightCycler® 480 System, and the data were
presented as the mean of each triplicate with standard deviations
(SDs). All assays were repeated a minimum of three times.
Statistical analysis
The data were analyzed using Student's t-test and
expressed as the mean value ± SD. The differences between groups
were analyzed by analysis of variance using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
ery operon was successfully deleted in
B. abortus 2308 and B. melitensis 027
The ery operon deletion was confirmed by PCR
in the 2308Δery and 027Δery clones (Fig. 1A). Bacteriological analysis and
typing of the mutant showed that deletion of the ery gene
was stable after passage in culture media (Fig. 1B).
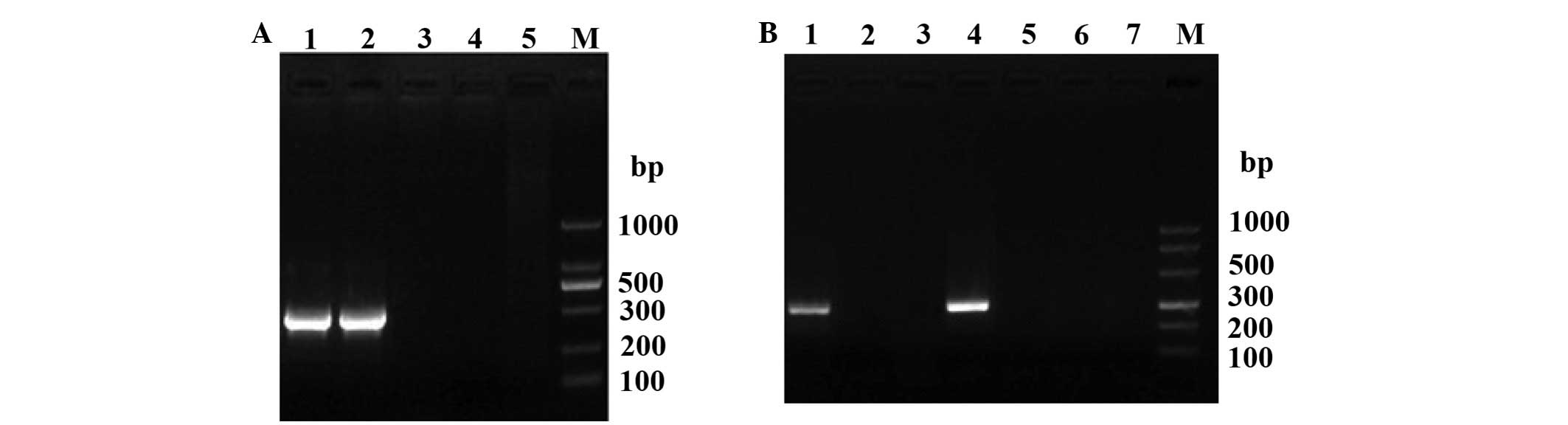 | Figure 1.Identification of construction of the
2308Δery and 027Δery. (A) Polymerase chain reaction
identification of 2308Δery and 027Δery. Lanes: 1, the
strain 2308; 2, the strain 027; 3, 2308Δery mutant strain;
4, 027Δery mutant strain; 5, negative control; M, DNA
marker. (B) Genetic stability detection of 2308Δery and
027Δery. Lanes: 1, the strain 2308; 2 and 3, 2308Δery
mutant strain; 4, the strain 027; 5 and 6, 027Δery mutant
strain; 7, negative control; M, DNA marker. |
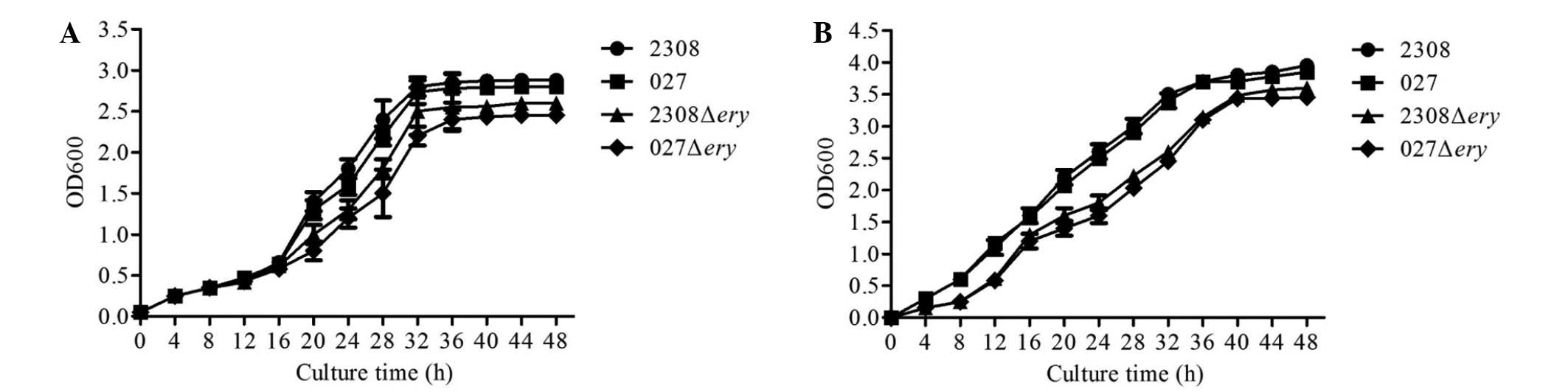
Growth curve of 2308Δery and
027Δery
To test whether deletion of the ery operon
affected the growth of 2308 or 027, we measured the bacterial
growth in nutrient-replete (TSB 7.0) media. When cultured in normal
TSB media, 2308Δery and 027Δery displayed a similar
lag phase and reached the stationary phase at a similar optical
density compared with 2308 and 027 (Fig.
2A).
Erythritol growth response in 2308Δery
and 027Δery
To test whether the 2308Δery and
027Δery strains were sensitive to erythritol when grown in
broth containing erythritol (20 mM), we measured the bacterial
growth. The results showed that the virulent strains 2308 and 027
grew well in broth containing erythritol; however, the
2308Δery and 027Δery mutants grew at a slower rate
(Fig. 2B). These results indicated
that 2308Δery and 027Δery do not respond to
erythritol, and that the virulent strains 2308 and 027 may utilize
erythritol for growth.
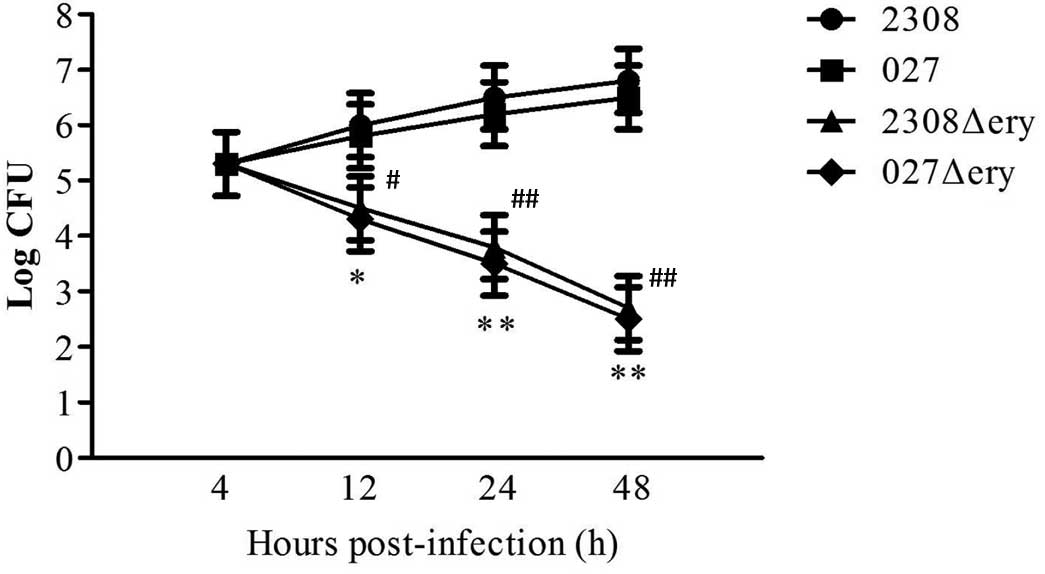
2308Δery and 027Δery are attenuated
and experience reduced survival in macrophages
To assess whether the ery operon influences
virulence, RAW 264.7 macrophages were infected with
2308Δery, 027Δery, 2308 and 027. The surviving
bacteria were enumerated, and there was no difference in the number
of surviving bacteria 4 h post-infection (Fig. 3; P>0.05). This indicated that
deletion of the ery operon does not affect macrophage
invasion. However, at 12 h post-infection, there was a 1.50-log
decrease (P<0.05) in the number of 2308Δery compared to
2308, and there was a 1.55-log decrease (P<0.05) in the number
of 027Δery compared to 027 (Fig.
3). At 24 h post-infection there was a 2.70-log decrease in the
number of 2308Δery compared to 2308 (Fig. 3; P<0.01); and there was a 2.70-log
decrease in the number of 027Δery compared to 027 (Fig. 3; P<0.01). At 48 h post-infection,
there was a 4.10-log decrease in the number of 2308Δery
compared to 2308 (Fig. 3;
P<0.01); and there was a 4.00-log decrease in the number of
027Δery compared to 027 (Fig.
3; P<0.01). Therefore, 2308Δery and 027Δery
mutants had a replication defect in RAW264.7 macrophages.
Preparation of standard curves of
different genes by RT-qPCR assay
The equations for the linear regression line for the
standard curves of different genes generated by RT-qPCR assay and
the corresponding R2 value are as follows: 16S
rRNA, y=0.238x+11.041, R2=0.986; eryA,
y=−0.267x+12.217, R2=0.986; eryB,
y=−0.266x+11.081, R2=0.983; eryC,
y=−0.216x+10.165, R2=0.977; and eryD,
y=−0.297x+11.414, R2=0.995. Based on the slope
rates of these regression lines, the levels of amplification
efficiency (E=10−a-1) were 72.98% for 16S rRNA, 84.93%
for eryA, 84.50% for eryB, 64.44% for eryC,
98.15% for eryD. Melt curve (unpublished) analysis of
amplification products indicated that there was a single peak with
a Tm of 86.8°C for 16S rRNA, 86.2°C for eryA, 84.6°C for
eryB, 90.06°C for eryC and 91.75°C for
eryD.
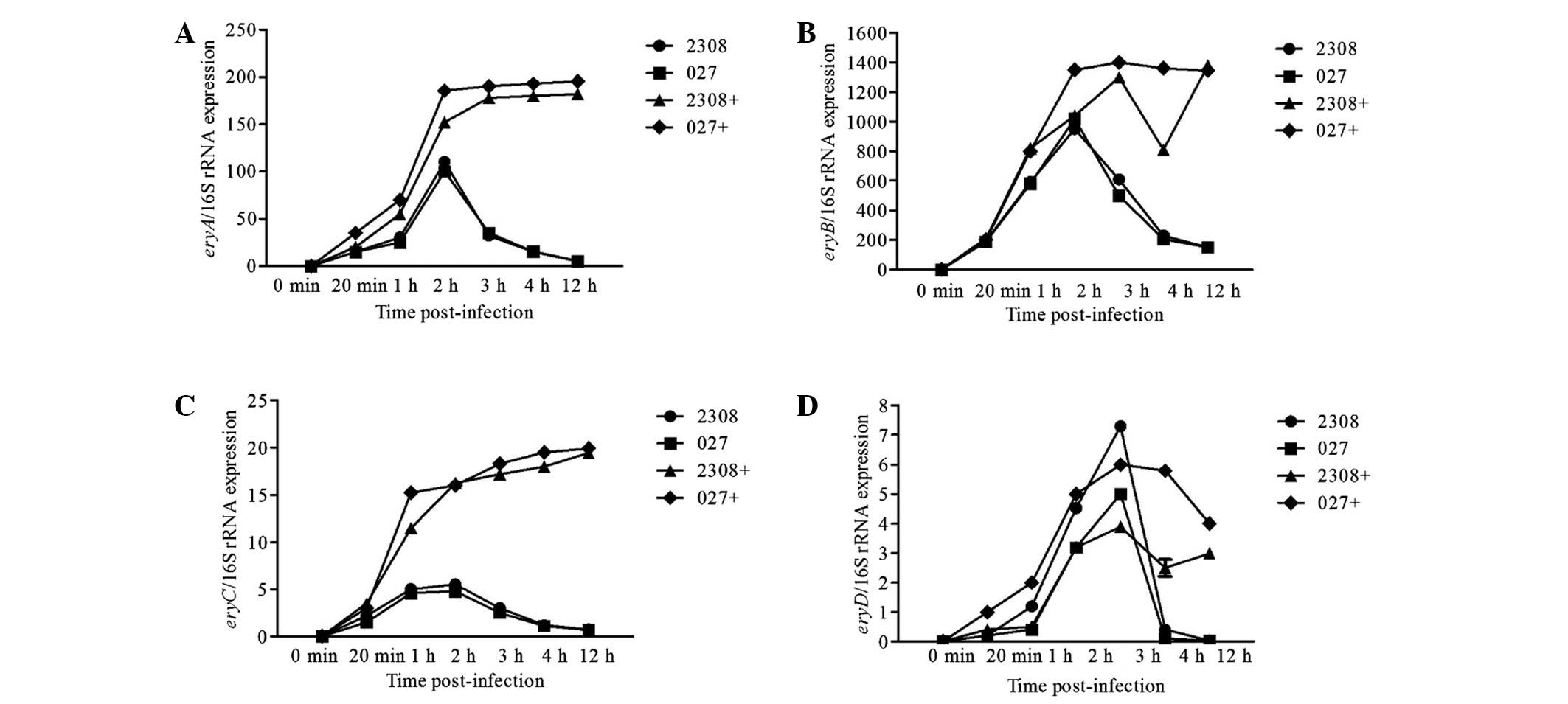
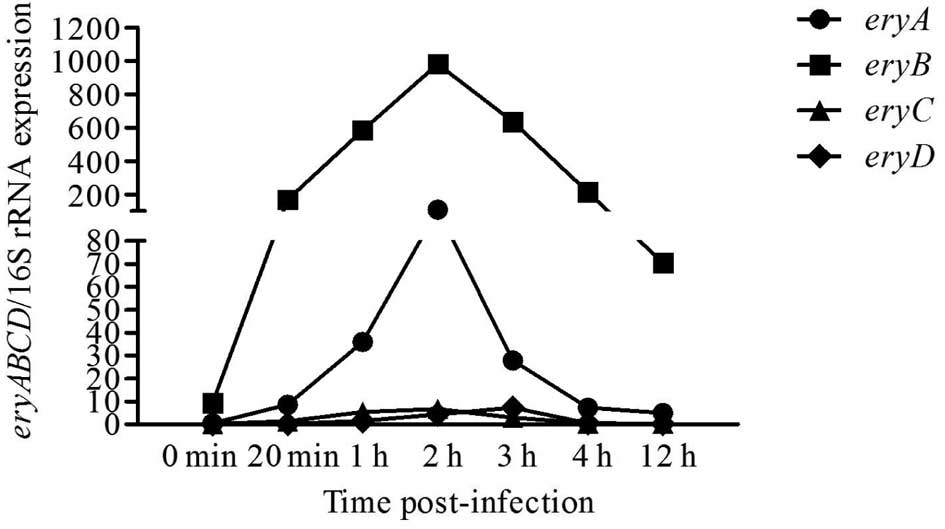
Ery operon gene expression following
infection
We analyzed gene expression at 0 min, 20 min, 1 h, 2
h, 3 h, 4 h and 12 h post-infection. The relative expression levels
of eryA, eryB, eryC and eryD were
significantly higher in Brucella grown with medium
containing erythritol. Furthermore, the relative expression levels
of eryA, eryB and eryC were highest
(P<0.01) at 2 h post-infection (Fig.
4A-C), but were highest (P<0.01) at 3 h post-infection for
eryD (P<0.05) (Fig. 4D).
Gene expression levels were calculated based on the comparison to
the expression of 16S rRNA. Based on these calculations,
eryB was the most highly expressed gene followed by
eryA, eryC and eryD (Fig. 5).
Discussion
The majority of Brucella spp. utilize
erythritol to promote growth (except B. abortus S19).
Metabolism and usage of erythritol are regulated by the 7.7-kb
ery operon, which consists of four genes eryA,
eryB, eryC and eryD (EryA, 519 AA; EryB, 502
AA; EryC, 309 AA and EryD, 316 AA) (16–18). The
functions of these four proteases are similar to xylulose kinase
(E. coli xylB), glycerol-3-phosphate dehydrogenase (E.
coli glpD), hydrogenase (Alcaligenes hydrogenophilus
hupL), operon regulators (Rhodobacter sphaeroides smoC
and Klebsiella pneumoniae dalR) (10). In addition, the promoter of the
ery operon also includes an integration host factor binding
site. This study demonstrates that the expression of the ery
operon in HPT-8 cells is higher than in culture medium, and
suggests that the B. melitensis 027 ery operon is
induced by erythritol within HPT-8 cells. Furthermore, the
expression of the eryD gene may repress the expression of
the other genes in the operon. This is the first demonstration of
the expression and regulation of the ery operon of B.
melitensis in human trophoblast cells.
Brucella is detected in non-professional
phagocytes 30 min post-infection (19). Therefore, the infection time used in
this study was 20 min, at which time Brucella were
detectable within host cells. Once entry has been established a
brucellosome is formed, a process that lasts 2–3 h. To study the
expression of the ery operon during the initial stage of
infection, we measured ery operon expression at 20 min, 1 h,
2 h, 3 h and 4 h post-infection in HPT-8 cells. In addition, the
expression of the ery operon was measured at 12 h
post-infection and was markedly decreased, which may be due to
changes in the intracellular environment induced by
Brucella. Notably, HPT-8 cells easily detached from the
culture flask after infection. In general, longer infection times
correlated with higher levels of detachment. In this study, we used
expression of the 16S rRNA gene as a reference to eliminate
determine genes expression levels at 20 min, 1 h, 2 h, 3 h, 4 h and
12 h post-infection.
Numerous genes from pathogen bacteria, such as
Brucella and Mycobacterium tuberculosis, are
regulated by environment signals in vitro (20). Mariani et al (17) investigated the expression of 14 genes
in Mycobacterium tuberculosis H37Rv in both medium and
macrophages. Five of these genes were expressed in media and
macrophages, four genes were expressed in media, and the remaining
five genes were only expressed in macrophages. In this study, we
characterized the expression of the ery operon in media and
at different timepoints following infection.
All four genes of the ery operon are
essential for optimal Brucella virulence. The function of
the genes in the ery operon control erythritol catabolism,
which has been postulated to increase virulence in the host
environment (21). Cells infected
with Brucella were grown in liquid media with or without
erythritol at a concentration of 20 mM. During the early phase of
infection without erythritol, the expression levels of
eryA-C are highest at 2 h post-infection; eryD is
highest at 3 h post-infection. Therefore, as eryD expression
increased the expression of eryA-C declined (Fig. 4). When erythritol was present in the
media, expression was similar to the above-mentioned results.
However, the expression levels of eryA-C peaked at 2 h then
remained constant, whereas eryD peaked at 3 h and then
declined (Fig. 5). Two explanations
may account for this phenomenon. First, EryD protein expression may
inhibit transcription of the ery operon (7). A second explanation may involve
acidification of the brucellosome at 3 h post-infection, as
multiplication in the acidified-brucellosome may represent a
starvation condition due to decreased ery operon expression
induced by erythritol.
Trophoblastic cells contain high levels of
erythritols and are targeted for infection by Brucella. It
has been previously reported that Brucella preferentially
utilizes the carbon source erythritol. This study demonstrates that
the expression of the ery operon is significantly lower in
media compared with trophoplastic cells.
Therefore, the present results indicate that the
expression of the ery operon of B. melitensis 027
differs when grown in media or HPT-8 cells, and that this
expression is regulated by environmental signals. Specifically, the
ery operon is induced by erythritol after entry into HPT-8
cells. However, a number of questions remain to be answered before
we can understand the role of the Ery system in
Brucella virulence.
Acknowledgements
The present study was supported by grants from the
International Science and Technology Cooperation Project of China
(grant nos. 2013BC005 and 2015DFR31110), the Outstanding Youth
Project of Shihezi University (grant no. 2012ZRKXJQ02), the
National Natural Science Foundation of China (grant nos. 31460650
and 31260596), and the University Key Research Project of Henan
Province (grant no. 16A230013).
References
|
1
|
Bercovich Z: The use of skin delayed-type
hypersensitivity as an adjunct test to diagnose brucellosis in
cattle: A review. Vet Q. 22:123–130. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pappas G, Papadimitriou P, Akritidis N,
Christou L and Tsianos EV: The new global map of human brucellosis.
Lancet Infect Dis. 6:91–99. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boschiroli ML, Foulongne V and O'Callaghan
D: Brucellosis: A worldwide zoonosis. Curr Opin Microbiol. 4:58–64.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Köhler S, Michaux-Charachon S, Porte F,
Ramuz M and Liautard JP: What is the nature of the replicative
niche of a stealthy bug named Brucella? Trends Microbiol.
11:215–219. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roop RM II, Gee JM, Robertson GT,
Richardson JM, Ng WL and Winkler ME: Brucella stationary-phase gene
expression and virulence. Ann Rev Microbiol. 57:57–76. 2003.
View Article : Google Scholar
|
|
6
|
Anderson JD and Smith H: The metabolism of
erythritol by Brucella abortus. J Gen Microbiol. 38:109–124. 1965.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meyer ME: Metabolic characterization of
the genus Brucella VI. Growth stimulation by i-erythritol compared
with strain virulence for guinea pigs. J Bacteriol. 93:996–1000.
1967.
|
|
8
|
Keppie J, Williams AE, Witt K and Smith H:
The role of erythritol in the tissue localization of the Brucellae.
Br J Exp Pathol. 46:104–108. 1965.PubMed/NCBI
|
|
9
|
Delrue RM, Lestrate P, Tibor A, Letesson
JJ and De Bolle X: Brucella pathogenesis, genes identified from
random large-scale screens. FEMS Microbiol Lett. 231:1–12. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sangari FJ, Agüero J and García-Lobo JM:
The genes for erythritol catabolism are organized as an inducible
operon in Brucella abortus. Microbiology. 146:487–495. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burkhardt S, de Bagüés MP Jiménez,
Liautard JP and Köhler S: Analysis of the behavior of eryC mutants
of Brucella suis attenuated in macrophages. Infect Immun.
73:6782–6790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eoh H, Jeon BY, Kim Z, Kim SC and Cho SN:
Expression and validation of D-erythrulose 1-phosphate
dehydrogenase from Brucella abortus: A diagnostic reagent for
bovine brucellosis. J Vet Diagn Invest. 22:524–530. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Yin S, Guo F, Meng R, Chen C,
Zhang H, Li Z, Fu Q, Shi H, Hu S, et al: A potent Brucella abortus
2308 Δery live vaccine allows for the differentiation between
natural and vaccinated infection. J Microbiol. 52:681–688. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hernández-Castro R, Verdugo-Rodríguez A,
Puente JL and Suárez-Güemes F: The BMEI0216 gene of Brucella
melitensis is required for internalization in HeLa cells. Microb
Pathog. 44:28–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pizarro-Cerdá J, Méresse S, Parton RG, van
der Goot G, Sola-Landa A, Lopez-Goñi I, Moreno E and Gorvel JP:
Brucella abortus transits through the autophagic pathway and
replicates in the endoplasmic reticulum of nonprofessional
phagocytes. Infect Immun. 66:5711–5724. 1998.PubMed/NCBI
|
|
16
|
Lillo AM, Tetzlaff CN, Sangari FJ and Cane
DE: Functional expression and characterization of EryA, the
erythritol kinase of Brucella abortus and enzymatic synthesis of
l-Erythritol-4-phosphate. Bioorg Med Chem Lett. 13:737–739. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mariani F, Cappelli G, Riccardi G and
Colizzi V: Mycobacterium tuberculosis H37Rv comparative
gene-expression analysis in synthetic medium and human macrophage.
Gene. 253:281–291. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smith H, Williams AE, Pearce JH, Keppie J,
Harris-smith PW, Fitz-George RB and Witt K: Foetal erythritol: A
cause of the localization of Brucella abortus in bovine contagious
abortion. Nature. 193:47–49. 1962. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Watanabe K, Tachibana M, Tanaka S, Furuoka
H, Horiuchi M, Suzuki H and Watarai M: Heat shock cognate protein
70 contributes to Brucella invasion into trophoblast giant cells
that cause infectious abortion. BMC Microbiol. 8:2122008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Chen Z, Qiao F, Zhong Z, Xu J,
Wang Z, Du X, Qu Q, Yuan J, Jia L, et al: The type IV secretion
system affects the expression of Omp25/Omp31 and the outer membrane
properties of Brucella melitensis. FEMS Microbiol Lett. 303:92–100.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dizon-Townson DS, Lu J, Morgan TK and Ward
KJ: Genetic expression by fetal chorionic villi during the first
trimester of human gestation. Am J Obstet Gynecol. 183:706–711.
2000. View Article : Google Scholar : PubMed/NCBI
|