Introduction
Osteosarcoma is the most prevalent malignant primary
sarcoma of the bone in children and adolescents, and is a leading
cause of cancer-associated mortality in young adults, accounting
for 3–4% of all malignancies and ~30% of malignant bone tumors in
adolescents (1). The development of
effective chemotherapeutic treatment has improved five-year
survival rates of patients with localized osteosarcoma in the US,
from 20 to between 65 and 70% and the success of limb-sparing
surgery over the past several decades (1); however, individuals with metastatic
osteosarcoma continue to exhibit poor prognoses, with long-term
survival rates only observed in <20% of patients. Despite the
advancement of chemotherapeutic treatment, negative side effects,
metastasis, disease recurrence and drug resistance have perpetuated
poor outcomes for patients. Therefore, exploiting novel treatment
strategies is crucial for osteosarcoma therapy.
The autophagy pathway has a critical role in
maintaining cellular homeostasis by delivering macromolecules and
organelles from the cytoplasm to lysosomes for degradation
(2,3). Various stress conditions, including
energy deprivation, nutrient starvation and oxidative stress, may
rapidly induce autophagy, which has a critical role in maintaining
cell homeostasis and survival (4).
However, consistent and prolonged activity of autophagy may induce
autophagic cell death. Dysregulation of autophagy has been observed
in various disease states, including infectious disease, cancer and
neurodegenerative diseases (4,5).
The role of autophagy in cancer is complicated, and
a previous study has referred to it as a ‘double-edged sword’
(2). In particular, it has been
demonstrated that autophagy suppresses mammary tumorigenesis driven
by WNT1 activation (6); however,
there is also evidence to suggest that autophagy is an important
mechanism for cancer cell survival (7). Therefore, whether autophagy inhibits or
propagates cancer may depend on the type of cancer, therapeutic
strategy, or both. A comprehensive understanding of all the
autophagy factors is required to predict whether autophagy will
protect cancer cells or kill them.
Drug resistance is a primary cause of cancer
treatment failure. Many strategies to overcome drug resistance in
cancer have been studied. For example, Wang et al (8) have previously reported that
HMGB1-mediated autophagy promotes neuroblastoma cell
chemoresistance; protective autophagy has been demonstrated to
promote lapatinib resistance in HER2-positive breast cancer cells
(9); Giuliano et al (10) have demonstrated that inhibition of
autophagy leads to sunitinib resistance in renal clear cell
carcinoma; and in a study conducted by Crystal et al
(11), MEK activation was revealed
to promote ceritinib resistance and MEK inhibitor treatment was
able to reverse resistance to ceritinib. A recent study indicated
that caveolin-1 (CAV-1) was highly expressed in cancer stem cells
and decreased cells' chemosensitivity (12). CAV-1 inhibition has previously been
shown to be associated with autophagic induction in human breast
cancer cells (13). Furthermore, it
has been demonstrated that CAV-1 deletion increases basal
autophagy, due to an increase in the complex of autophagy-related
proteins 5 and 12 (Atg5-Atg12), and that a CAV-1 binding motif
mutation broke this complex and accelerated autophagy (14). In addition, previous studies have
reported that CAV-1 deficiency was an independent factor for the
poor prognosis of colorectal cancer, demonstrating that loss of
CAV-1 may increase drug resistance and cancer metastasis (15,16).
In present study, Saos-2 and U-2 OS cells were
cultured with gradually increasing concentrations of Taxol, in
order to establish drug-resistant cell lines. The findings of the
present study suggest that further investigation into the
association between CAV-1 and Taxol resistance is warranted.
Materials and methods
Cell culture and lentivirus
infection
Human osteosarcoma cell lines were purchased from
American Type Culture Collection (Manassas, VA, USA). Saos-2/Taxol
and U-2 OS/Taxol cells were established via gradually increasing
the concentration of Taxol, every fortnight (5, 10, 20, 50, 100,
150, 200, 250, and 300 ng/ml). DNA oligonucleotides carrying small
hairpin (sh) RNA (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) were constructed into pLKO.1 plasmids (Addgene;
Cambridge, MA, USA). Packaging (psPAX2) and envelope (pMD2.G)
plasmids (Addgene, Inc) were transfected into HEK293T cells with
recombinant plasmids. The supernatant containing lentiviruses were
collected after 36 h. The short hairpin (sh)RNA used to assess
caveolin-1 (CAV-1) and autophagy related protein 5 (Atg5) were as
follows: i) shCAV-1#1, CATCTACAAGCCCAACAAC; ii) shCAV-1#2,
AGACGAGCTGAGCGAGAAG; iii) shAtg5#1, ATTGGCTCAATTCCATGAA; iv)
shAtg5#2, GCTACTCTGGATGGGATTG; and v) control shRNA,
CACACCGTTTCGTGGCTTT. The following inhibitory compounds (all 10 µM
in culture medium) were used to treat cells in the present study:
i) Bafilomycin A1 (autophagy inhibitor) for 4 h (Baf A1; cat. no.
ALX-380–063-M001; Enzo Life Sciences, Inc., Farmingdale, NY, USA);
ii) MK-2206 (Akt inhibitor) for 1 h (cat. no. 1888–500; BioVision,
Inc., Milpitas, CA, USA); iii) SP600125 (JNK inhibitor) for 1 h
(cat. no. S5567; Sigma-Aldrich, St. Louis, MO, USA); and iv)
LY294002 (PI3K inhibitor) for 1 h (cat. no. L9908;
Sigma-Aldrich).
Cell viability assay
Cell viability was analyzed via MTT assay using a
Roche Cell Proliferation Kit I (Roche Diagnostics, Basel,
Switzerland; cat. no. 11465007001) according to the manufacturer's
protocol. All experiments were performed in triplicate. Results
were plotted using Prism5 software (GraphPad Software, Inc., La
Jolla, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA from osteosarcoma cells was extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and 4 µg was used for reverse transcription (RT). RT was performed
using a first-strand cDNA synthesis kit (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. qPCR analysis was
carried out using a SYBR Green kit (TransGen Biotech Co. Ltd.,
Beijing, China) on an ABI 7900 system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR reactions (20 µl total volume)
comprised the following: SYBR, 10 µl; cDNA, 1 µl; forward primer,
0.25 µl; reverse primer, 0.25 µl; ROX reference dye, 0.1 µl; and
double-distilled H2O, 8.4 µl. Primers were designed as
follows: CAV-1, forward 5′-AACACGTAGCTGCCCTTCAG-3′ and reverse
5′-GGATGGGAACGGTGTAGAGAT-3′; and ACTB, forward
5′-TGTTTGAGACCTTCAACACCC-3′ and reverse 5′-AGCACTGTGTTGGCGTACAG-3′.
The amplification conditions were as follows: Pre-denaturation at
94°C for 5 min, 40 cycles of denaturation at 94°C for 30 sec,
annealing at 59°C for 30 sec, extension at 72°C for 25 sec and a
final extension at 72°C for 10 min. Cq values were measured during
the exponential amplification phase. Relative gene expression was
calculated using the 2−ΔΔCq method (17), with ACTB as the internal control
gene. For each gene RT-qPCR was performed in triplicate.
Western blot analysis
Cells were lysed in RIPA buffer (Sigma-Aldrich). The
concentration of the protein samples was determined by the Bradford
method (18). Lysates were denatured
at 100°C for 10 min and subsequently cooled on ice. A total of 40
µg protein was separated by 8–15% SDS-PAGE and transferred onto a
polyvinylidene difluoride membrane (GE Healthcare Life Sciences,
Chalfont, UK). The primary antibodies used in the present study
were: i) β-actin (cat. no. A1978; Sigma-Aldrich; mouse; monoclonal;
1:5,000 in 5% w/v milk); ii) microtubule associated protein 1 light
chain 3 [(LC3; cat. no. 2775; rabbit polyclonal; 1:5,000 in 5% w/v
bovine serum albumin (BSA; Sangon Biotech Co., Ltd., Shanghai,
China); iii) Atg5 (cat. no. 2630; rabbit polyclonal, 1:2,000 in 5%
w/v BSA); iv) phosphorylated janus kinase (p-JNK; T183/Y185; cat.
no. 4668; rabbit; polyclonal; 1:1,000 in 5% w/v BSA); v) JNK (cat.
no. 9252; rabbit; polyclonal; 1:2,000 in 5% w/v BSA); vi) CAV-1
(cat. no. 3238; rabbit; polyclonal; 1:2,000 in 5% w/v BSA); vii)
Akt (cat. no. 9272; rabbit; polyclonal; 1:2,000 in 5% w/v BSA);
viii) p-Akt (S473; cat. no. 4060; rabbit; polyclonal; 1:2,000 in 5%
w/v BSA); ix) Atg7 (cat. no. 2631; rabbit; polyclonal; 1:2,000 in
5% w/v BSA); and x) Beclin1 (cat. no. 3495; rabbit; polyclonal;
1:2,000 in 5% w/v BSA) (all Cell Signaling Technologies, Inc.,
Danvers, MA, USA).
Laser scanning confocal
microscopy
Cells stably expressing ref fluorescent protein
(RFP)-LC3 were cultured in live cell imaging culture dishes
(Livefocus, Jiangsu, China; cat. no. C-L-8) and subjected to serum
starvation in Hank's balanced salt solution (Thermo Fisher
Scientific, Inc.) or lentivirus. Living cells were visualized using
a Zeiss LSM510 meta-confocal system (Zeiss AG, Oberkochen,
Germany). A total of 200 cells were detected at each condition, and
significant differences were analyzed by Student's
t–test.
Statistical analysis
Statistical analyses were conducted using GraphPad
Prism5 (GraphPad Software, Inc.). Significant differences were
analyzed by performing a Student's t–test. P<0.05 was
considered to indicate a statistically significant difference.
Results
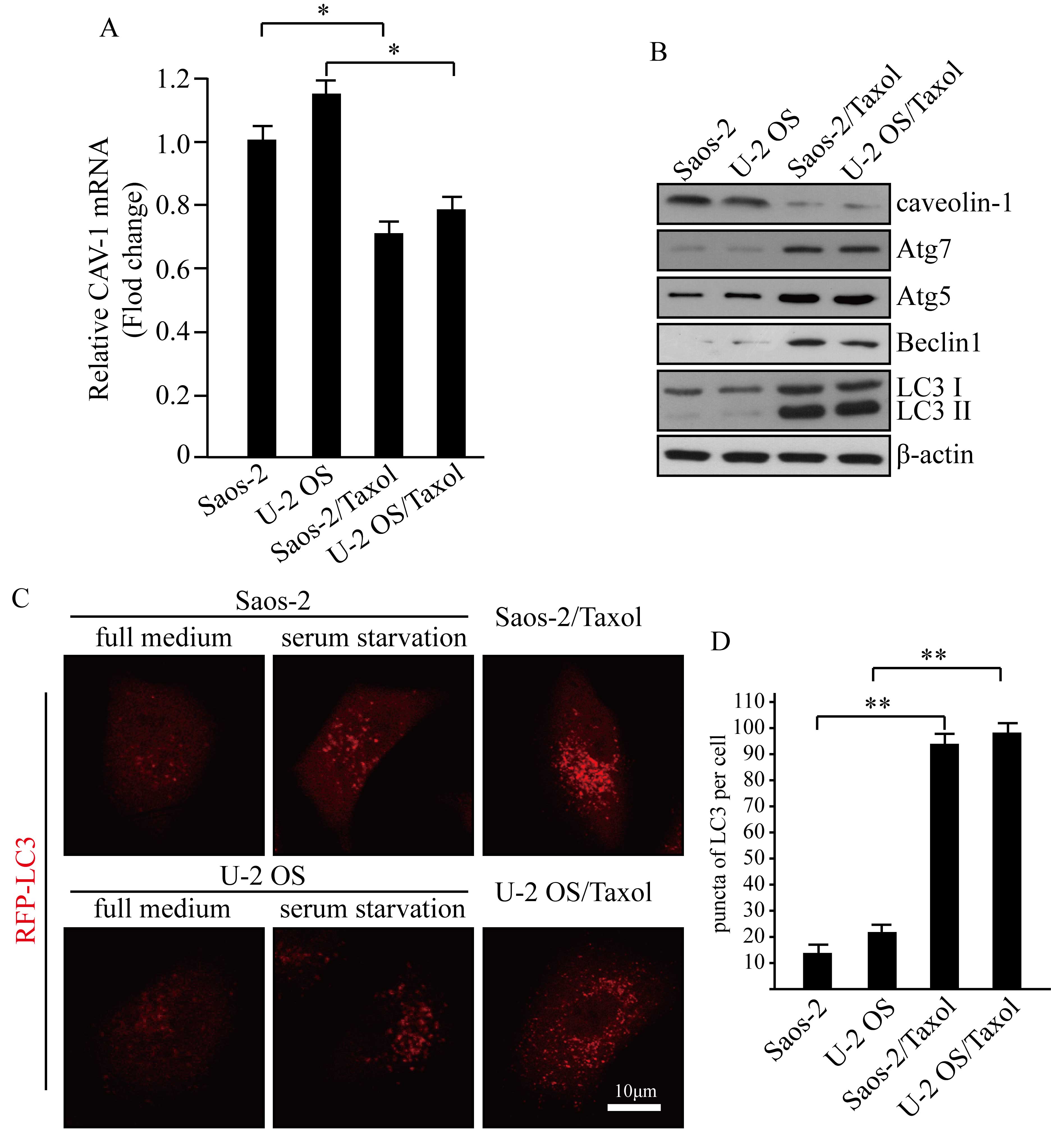
CAV-1 expression is reduced in
Taxol-resistant human osteosarcoma cells
A recent study indicated that CAV-1 was highly
expressed in cancer stem cells and decreased cells'
chemosensitivity (12). To explore
the expression levels of caveolin-1 in Taxol-resistant osteosarcoma
cells, a qPCR assay was performed to detect CAV-1 mRNA expression
levels (Fig. 1A). The results
indicated that CAV-1 expression levels were significantly decreased
in Taxol-resistant human osteosarcoma cells, compared with their
non-resistant counterparts (P<0.05; Fig. 1A). Western blot analysis was
performed to examine the protein expression levels of CAV-1 in
Saos-2/Taxol, U-2 OS/Taxol and their drug-sensitive parent cells
(Fig. 1B). These findings were
consistent with the results of qPCR analysis. Caveolin-1 expression
was markedly reduced in Taxol-resistant human osteosarcoma cells,
as compared with the Soas-2 and U-2 OS controls.
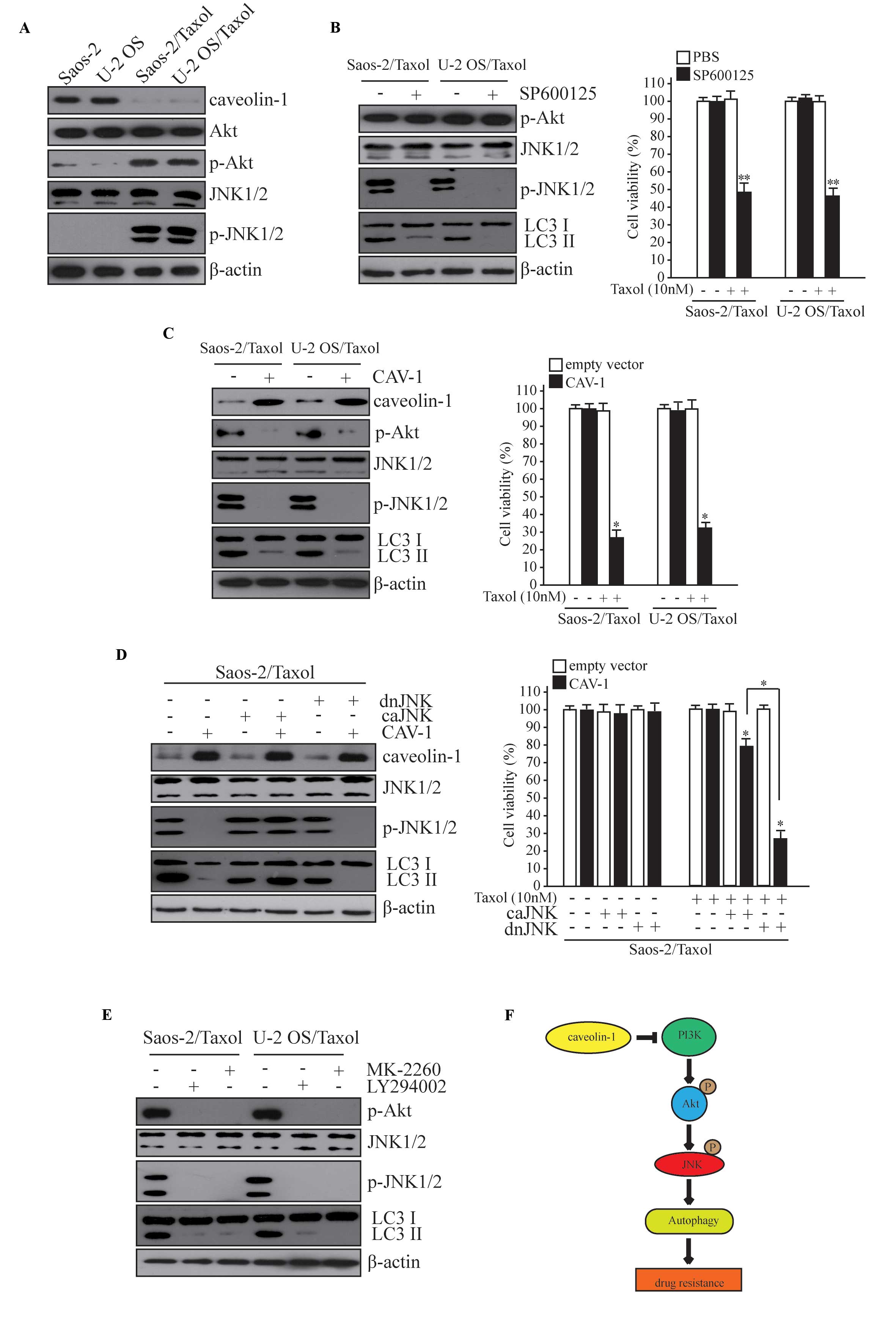
Basal autophagy is higher in
Saos-2/Taxol and U-2 OS/Taxol cells, compared with Saos-2 and U-2
OS cells
CAV-1 inhibition has previously been shown to be
correlated with autophagic induction in human breast cancer cells
(13). Autophagy is a regulatory
mechanism that protects cells from stress; however, prolonged
consistent autophagy may cause type II programmed cell death
(4). To detect autophagy in the
present study, canonical markers of autophagy in Saos-2/Taxol, U-2
OS/Taxol and their drug sensitive parents cells were analyzed
(Fig. 1B). Atg5, Atg7, LC3 II and
Beclin1 were demonstrated to be highly expressed in Saos-2/Taxol
and U-2 OS/Taxol cells. LC3I to LC3II conversion indicates a more
pronounced formation of autophagosomes, and the increase of Atg5,
Atg7 and Beclin1 is typically suggestive of autophagy onset
(3). CAV-1 levels were demonstrated
to be reversely correlated with basal autophagy onset in human
osteosarcoma cancer cells in the context of Taxol resistance. For
the observation of autophagosome formation, it was established that
cells stably expressing RFP-LC3 mediated via a
lentivirus-associated mechanism. Significantly increased basal
autophagy was exhibited by RFP-LC3 puncta in Taxol-resistant cells,
with serum starvation used as a positive control for autophagy
(Fig. 1C and D; P<0.01).
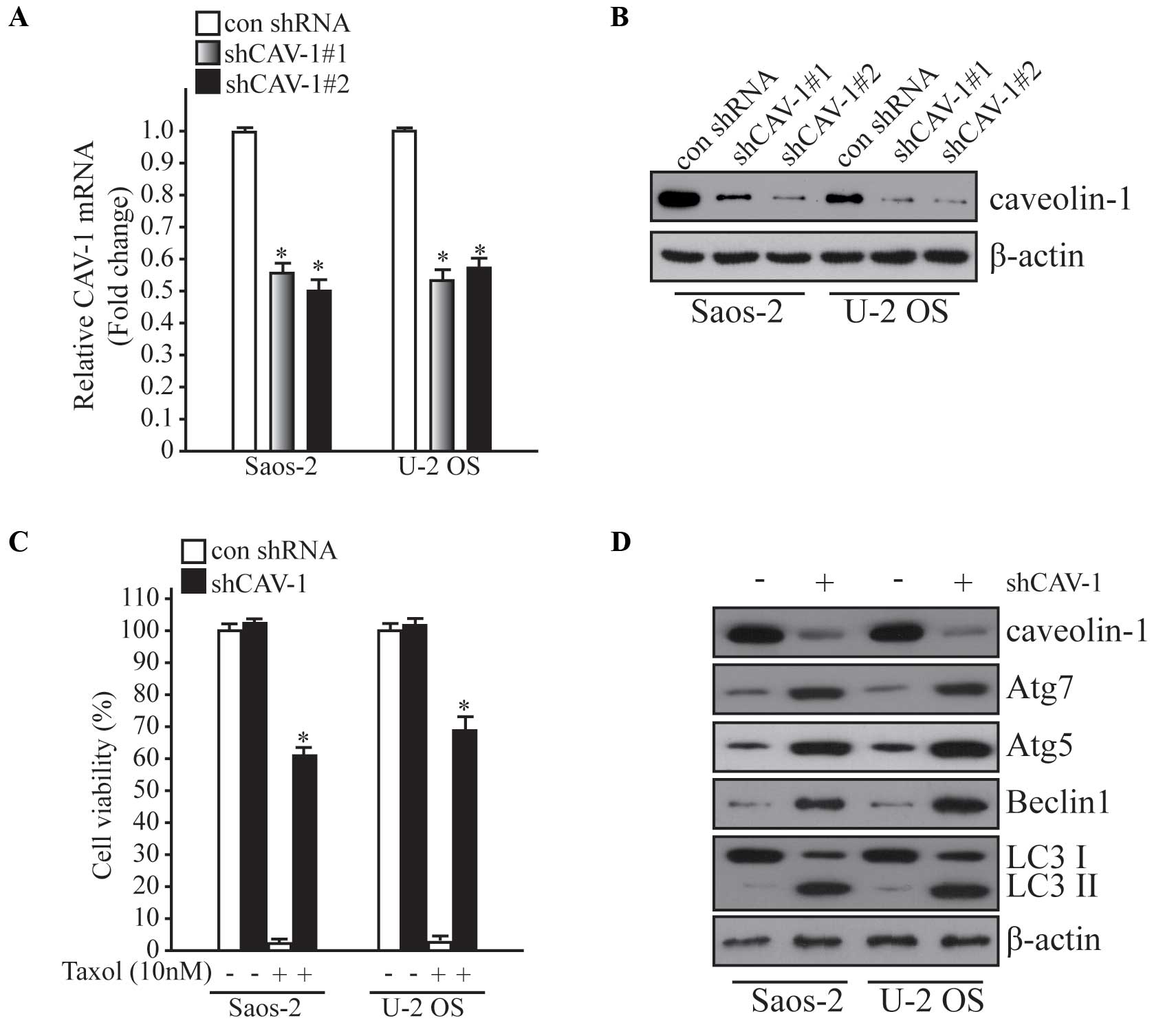
CAV-1 knockdown induces Taxol
resistance and autophagy in Saos-2 and U-2 OS cells
To explore the effect of CAV-1 in Taxol resistance
and autophagy, a CAV-1 knockdown system was established using
lentiviral transduction. To investigate the silencing efficiency of
two different shRNA targeting CAV-1, qPCR and western blot analysis
were performed in Saos-2 and U-2 OS cells (Fig. 2A and B). Following knockdown of
CAV-1, cells were treated with Taxol (10 nM) and cell viability was
examined using an MTT assay kit. As shown in Fig. 2C, knockdown of CAV-1 was induced
resistance to Taxol, and when autophagy-related markers were
detected, autophagy was significantly enhanced after CAV-1
knockdown (P<0.05). These results indicate that deficiency of
CAV-1 may induce autophagy and Taxol resistance in human
osteosarcoma cells.
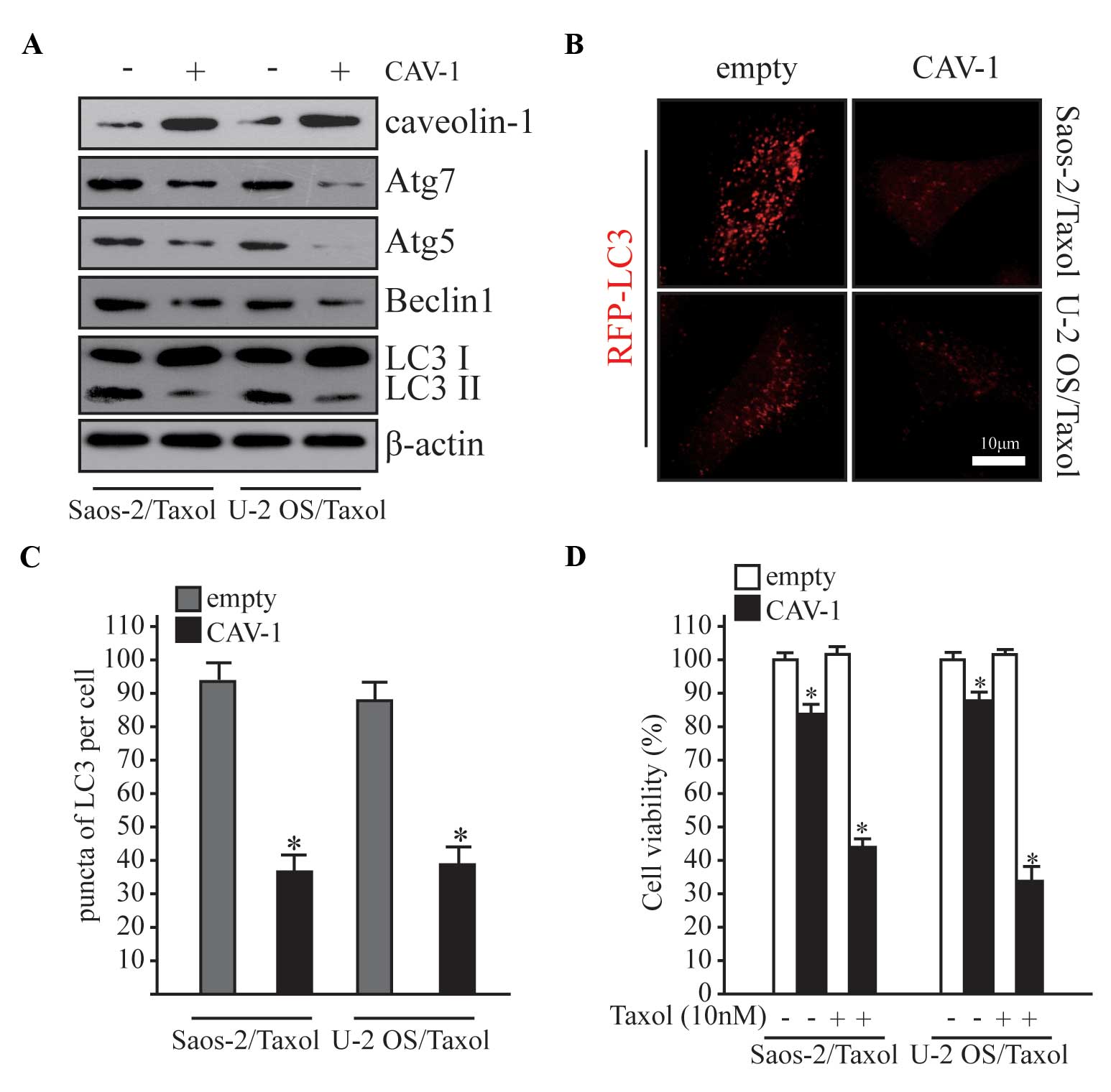
Overexpression of CAV-1 prevents
autophagy and Taxol resistance in Saos-2/Taxol and U-2 OS/Taxol
cells
As shown in Fig. 1B,
CAV-1 expression was significantly reduced in Taxol-resistant human
osteosarcoma cells. To further elucidate the function of CAV-1 in
autophagy and Taxol resistance, CAV-1 was overexpressed in
Saos-2/Taxol and U-2 OS/Taxol cells (Fig. 3A), and alterations in autophagy and
drug resistance were subsequently examined. Autophagy was
significantly inhibited following CAV-1 overexpression in Taxol
resistance cells (Fig. 3B and C;
P<0.05). MTT assay was performed to evaluate cell viability, and
the viability of Taxol resistant cells was significantly decreased
after CAV-1 was overexpressed (Fig.
3D; P<0.05). Notably, cell viability was significantly
reduced in the overexpression group even without Taxol treatment
(Fig. 3D; P<0.05).
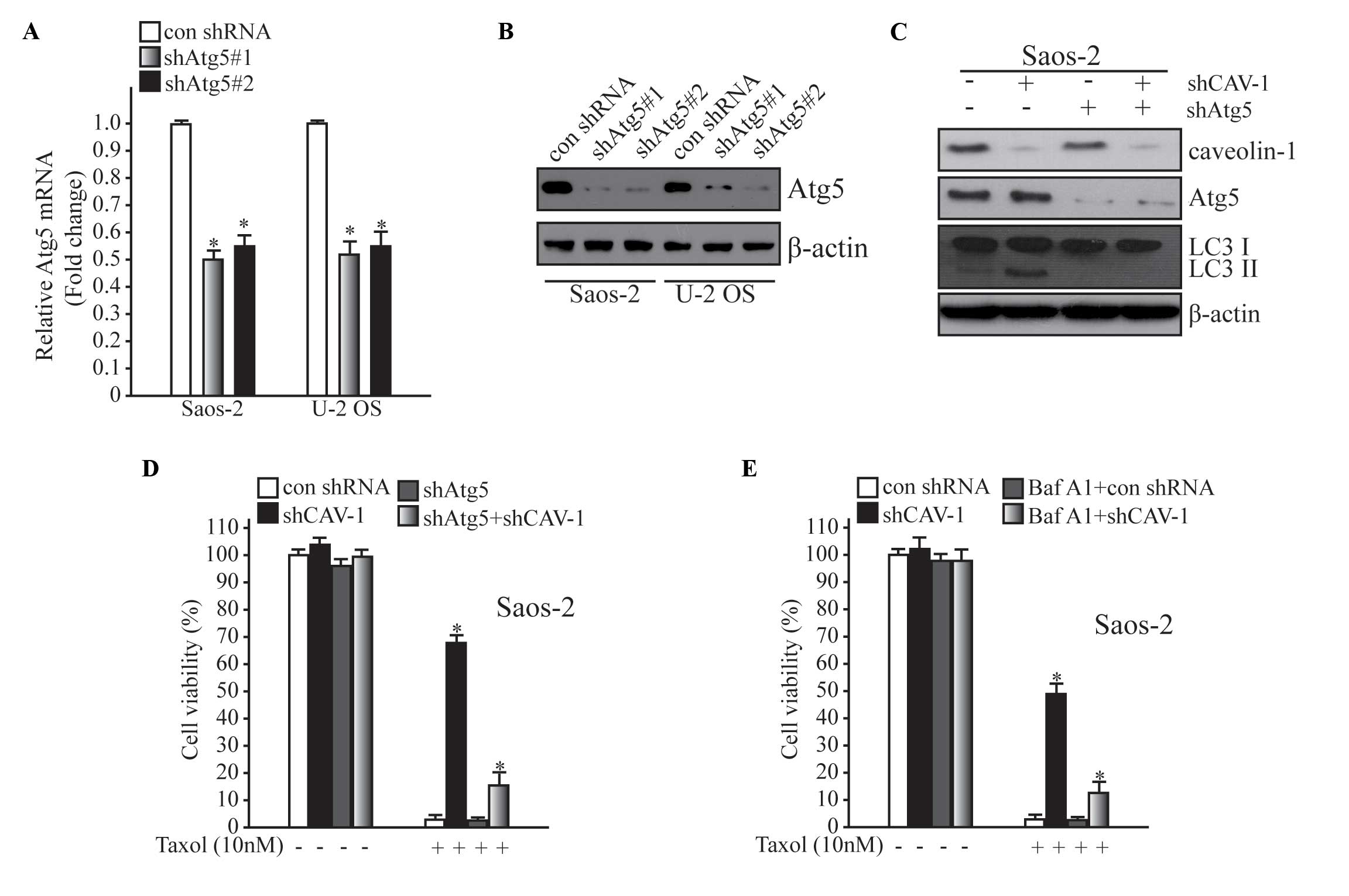
Inhibition of autophagy weakens Taxol
resistance mediated by CAV-1 deficiency
It has previously been reported that various
clinical anti-cancer agents, including tamoxifen, cetuximab and
imatinib, are able to induce autophagy in cell culture and animal
models (5). The present study
revealed that autophagy was increased in drug-resistant cells. To
assess the role of autophagy in drug resistance, autophagy was
inhibited using two different methods; including inhibition of
autophagy at the genetic level and administration of an autophagy
inhibitor, Baf A1 (10 µM). In the present study, Atg5 knockdown was
stimulated to hinder autophagy activation (Fig. 4A and B). As shown in Fig. 4C, basal autophagy and autophagy
induced by CAV-1 deficiency were blocked by downregulating Atg5.
Subsequently, it was demonstrated that Taxol resistance mediated by
the loss of CAV-1 in Fig. 2C was
markedly decreased after the inhibition of autophagy by
downregulating Atg5 (Fig. 4C). When
cells were treated with Baf A1 (10 µM), drug resistance was
declined following autophagy inhibition (Fig. 4D). These results suggest that
CAV-1-associated Taxol resistance involves the autophagy
pathway.
CAV-1 reduces Taxol resistance and
autophagy by JNK signaling
To investigate how CAV-1 reduces Taxol resistance
and autophagy, the role of JNKs in this process was analyzed as
JNKs have previously been reported to mediate Taxol resistance in
ovarian carcinoma cells (19), and
JNK1-mediated phosphorylation of B cell lymphoma-2 has an important
role in the regulation of autophagy (20). To compare Taxol-resistant cells and
their parent cells, the activation of the phosphorylation of JNK
and upstream Akt was increased, suggesting an increase in JNK
activity (Fig. 5A). The effect of
JNK on autophagy and Taxol resistance of osteosarcoma cells was
subsequently investigated. Inhibition of JNK activity was achieved
by treating cells with a JNK inhibitor (SP600125), which
significantly decreased the level of autophagy and the Taxol
resistance of Saos-2/Taxol and U-2 OS cells (Fig. 5B; P<0.05). However, overexpression
of CAV-1 abolished the activity of JNK and Akt, and decreased the
level of autophagy and Taxol resistance of Saos-2/Taxol and U-2 OS
cells (Fig. 5C; P<0.05). The
expression of a constitutively active JNK rescued the inhibiting
effect of CAV-1 on autophagy and Taxol resistance (Fig. 5D). These results indicate that CAV-1
inhibits autophagy and Taxol resistance via JNK signaling. To
identify the pathway by which CAV-1 inhibits the activity of JNK,
the role of PI3K and Akt in this process was examined. Inhibition
of either PI3K or Akt activity using a PI3K inhibitor (LY294002) or
Akt inhibitor (MK-2206) decreased JNK activation (Fig. 5E). These results indicate that CAV-1
may inhibit JNK activity via a decrease in PI3K-Akt activity and
suggests that CAV-1 may inhibit autophagy and Taxol resistance via
the PI3K-Akt-JNK pathway.
Discussion
Osteosarcoma is the most prevalent malignant primary
sarcoma of bone in children and adolescents, and is a leading cause
of cancer-associated mortality in young adults, accounting for 3–4%
of all malignancies in adolescents and ~30% of malignant bone
tumors (1). Standard therapy is
typically multimodal, including neoadjuvant chemotherapy and
subsequent amputation or limb-sparing reconstructive surgeries,
with adjuvant chemotherapy (1).
Chemotherapy plays a significant role in improving patient survival
and decreasing mortality in cancer patients (21,22);
however, drug resistance, whether acquired or otherwise, threatens
the clinical outcomes and prognoses of patients with cancer.
Genetic alterations are thought to be key factors in
the majority of solid tumors. There are numerous molecules
correlated with drug resistance, which have not been demonstrated
to be causative factors; therefore, these molecular markers are not
ideal targets for developing pharmacological agents (5). However, in recent decades, genetic
diagnosis and gene therapy have become fast-growing areas of
research. Progression in molecular technologies has promoted the
elucidation of the mechanisms underlying carcinogenesis, Autophagy
mechanisms have captured increasing attention, and
autophagy-related genes have become viable prospects for novel
potential targets in cancer treatment (23,24).
Autophagy is characterized by the bulk degradation
of damaged organelles and misfolding proteins, and is a critical
process for cellular homeostasis, differentiation, viability and
mammalian development (25).
Autophagy has also been demonstrated to cause cell death in certain
conditions, working as a tumor suppressing mechanism during the
initiation stage of cancer progression (5). Conversely, once the tumor has formed,
autophagy assists in the prevention of cell death induced by
anticancer therapeutic agents (26).
Therefore, autophagy has two inverse effects in different
contexts.
Autophagy has been identified in various tissues,
and has been demonstrated to correlate significantly with cancer,
cardiomyopathies, neurodegenerative diseases and bacterial
infections (4,5). Various proteins are associated with the
detection of autophagic activity, including Atg5, Atg7, Beclin1 and
the microtubule-associated protein LC3. LC3-I and LC3-II are two
cellular forms of LC3 protein; LC3-I is the cytoplasmic form,
whereas LC3-II is located in the autophagosomal membrane (7). Therefore, an increase in the conversion
of LC3-I to LC3-II is correlated with the extent of autophagosome
formation.
To date, the function of CAV-1 in autophagy remains
unclear. In the present study, CAV-1 loss was observed in
drug-resistant osteosarcoma cells and induced the activation of
autophagy. In a previous study, loss of CAV-1 was also reported in
various types of malignancies including colon cancer, breast cancer
and drug-resistant ovarian carcinoma (27). Furthermore, it has been demonstrated
that CAV-1 deletion increased basal autophagy due to an increase in
Atg5-Atg12, whereas CAV-1 binding motif mutation broke the
association between Atg5 and Atg12 and accelerated autophagy
(14). The present study also
provided evidence that CAV-1 loss contributed to autophagy and the
Taxol resistance of Saos-2 and U-2 OS cells via the PI3K-Akt-JNK
pathway; whereas the overexpression of CAV-1 inhibited autophagy
and declined Saos-2/Taxol and U-2 OS/Taxol resistance to Taxol.
Interrupting autophagy genetically (via the knockdown of Atg5) or
with an autophagy inhibitor (Baf A1) impaired the drug resistance
induced by CAV-1 deficiency. Previous studies have similarly
reported that CAV-1 deficiency was an independent factor for the
poor prognosis of colorectal cancer individuals, demonstrating that
loss of CAV-1 may assist drug resistance and cancer metastasis
(15,16).
In conclusion, the results of the present study
demonstrated a novel function of CAV-1 in human osteosarcoma cells,
indicating that attenuating autophagy may be a promising potential
therapeutic approach. In the future, detection of CAV-1 may be a
good indicator with which to evaluate the treatment and prognosis
of osteosarcoma.
Acknowledgements
This work was supported by Science Computing and
Intelligent Information processing of GuangXi Higher Education Key
Laboratory (grant no. GXSCIIP201502).
References
|
1
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Galluzzi L, Pietrocola F, Levine B and
Kroemer G: Metabolic control of autophagy. Cell. 159:1263–1276.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi AM, Ryter SW and Levine B: Autophagy
in human health and disease. N Engl J Med. 368:651–662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cicchini M, Chakrabarti R, Kongara S,
Price S, Nahar R, Lozy F, Zhong H, Vazquez A, Kang Y and Karantza
V: Autophagy regulator BECN1 suppresses mammary tumorigenesis
driven by WNT1 activation and following parity. Autophagy.
10:2036–2052. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang F, Kumano M, Beraldi E, Fazli L, Du
C, Moore S, Sorensen P, Zoubeidi A and Gleave ME: Clusterin
facilitates stress-induced lipidation of LC3 and autophagosome
biogenesis to enhance cancer cell survival. Nat Commun. 5:57752014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Zhang H, Sun M, Yin Z and Qian J:
High mobility group box 1-mediated autophagy promotes neuroblastoma
cell chemoresistance. Oncol Rep. 34:2969–2976. 2015.PubMed/NCBI
|
|
9
|
Chen S, Zhu X, Qiao H, Ye M, Lai X, Yu S,
Ding L, Wen A and Zhang J: Protective autophagy promotes the
resistance of HER2-positive breast cancer cells to lapatinib.
Tumour Biol. 37:2321–2331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giuliano S, Cormerais Y, Dufies M, Grépin
R, Colosetti P, Belaid A, Parola J, Martin A, Lacas-Gervais S,
Mazure NM, et al: Resistance to sunitinib in renal clear cell
carcinoma results from sequestration in lysosomes and inhibition of
the autophagic flux. Autophagy. 11:1891–1904. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Crystal AS, Shaw AT, Sequist LV, Friboulet
L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A,
Greninger P, et al: Patient-derived models of acquired resistance
can identify effective drug combinations for cancer. Science.
346:1480–1486. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J and Chen J: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi Y, Tan SH, Ng S, Zhou J, Yang ND, Koo
GB, McMahon KA, Parton RG, Hill MM, Del Pozo MA, et al: Critical
role of CAV1/caveolin-1 in cell stress responses in human breast
cancer cells via modulation of lysosomal function and autophagy.
Autophagy. 11:769–784. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shiroto T, Romero N, Sugiyama T,
Sartoretto JL, Kalwa H, Yan Z, Shimokawa H and Michel T: Caveolin-1
is a critical determinant of autophagy, metabolic switching, and
oxidative stress in vascular endothelium. PLoS One. 9:e878712014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu KN, Queenan M, Brody JR, Potoczek M,
Sotgia F, Lisanti MP and Witkiewicz AK: Loss of stromal caveolin-1
expression in malignant melanoma metastases predicts poor survival.
Cell Cycle. 10:4250–4255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Z, Han FH, Yang SB, Hua LX, Wu JH and
Zhan WH: Loss of stromal caveolin-1 expression in colorectal cancer
predicts poor survival. World J Gastroenterol. 21:1140–1147. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pu J, Schindler C, Jia R, Jarnik M,
Backlund P and Bonifacino JS: BORC, a multisubunit complex that
regulates lysosome positioning. Dev Cell. 33:176–188. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun NK, Huang SL, Lu HP, Chang TC and Chao
CC: Integrative transcriptomics-based identification of cryptic
drivers of taxol-resistance genes in ovarian carcinoma cells:
Analysis of the androgen receptor. Oncotarget. 6:27065–27082. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei Y, Pattingre S, Sinha S, Bassik M and
Levine B: JNK1-mediated phosphorylation of Bcl-2 regulates
starvation-induced autophagy. Mol Cell. 30:678–688. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Dang TA, Shen J, Hicks J,
Chintagumpala M, Lau CC and Man TK: Plasma proteome predicts
chemotherapy response in osteosarcoma patients. Oncol Rep.
25:303–314. 2011.PubMed/NCBI
|
|
23
|
Shibutani ST, Saitoh T, Nowag H, Münz C
and Yoshimori T: Autophagy and autophagy-related proteins in the
immune system. Nat Immunol. 16:1014–1024. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao XH, Naomoto Y, Hao HF, Watanabe N,
Sakurama K, Noma K, Motoki T, Tomono Y, Fukazawa T, Shirakawa Y, et
al: Autophagy: Can it become a potential therapeutic target? Int J
Mol Med. 25:493–503. 2010.PubMed/NCBI
|
|
25
|
Axe EL, Walker SA, Manifava M, Chandra P,
Roderick HL, Habermann A, Griffiths G and Ktistakis NT:
Autophagosome formation from membrane compartments enriched in
phosphatidylinositol 3-phosphate and dynamically connected to the
endoplasmic reticulum. J Cell Biol. 182:685–701. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levine B: Cell biology: Autophagy and
cancer. Nature. 446:745–747. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Williams TM and Lisanti MP: Caveolin-1 in
oncogenic transformation, cancer, and metastasis. Am J Physiol Cell
Physiol. 288:C494–C506. 2005. View Article : Google Scholar : PubMed/NCBI
|