Introduction
Asthma is a chronic airway disease characterized by
the infiltration of various inflammatory cells. The most common
form, allergic asthma, is thought to be associated with abnormal
CD4+ T helper (Th) 2 immunity (1). A classical Th2 cell response may lead
to IgE production, eosinophils recruitment, goblet cell
hyperplasia, and mucus overproduction by producing Th2 cytokines,
such as interleukin (IL)-4, IL-5, and IL-13 (2). IL-9, which is a pleiotropic cytokine,
was initially considered to be a Th2-specific cytokine. More
recently, a distinct subset of CD4+ Th9 cells was found
to secrete IL-9 (3,4). It has been reported that Th9 cells are
involved in human and murine atopic disease and immunity to
intestinal parasites via IL-9 (5,6).
Moreover, IL-9-producing CD4+ T (Th9) cells are able to
enhance IgE production and regulate mast cell accumulation in the
lungs during allergic inflammation (7).
Similar to CD4+ Th cells, CD8+
cytotoxic T lymphocytes (Tc) may also have an important role in
allergic asthma pathology (8,9).
Previous studies have found that Tc2 cells aggravate asthma by
secreting type 2 cytokines (10,11). On
the other hand, Tc1 cells have been demonstrated to be beneficial
for airway inflammation by secreting interferon (IFN)-γ (11). Under Th9-polarizing conditions, naïve
CD8+ T cells regulated by the transcription factors
signal transducer and activator of transcription 6 (STAT6) and
interferon regulatory factor 4 (IRF4) are able to differentiate
into IL-9-producing CD8+ T (Tc9) cells, a unique
CD8+ T cell subset (12).
It has been reported that tumor-specific Tc9 cells elicit great
antitumor responses depending on IL-9 production after adoptive
transfer (13). Tc9 cells are also
increased in mice and humans with atopic dermatitis and are able to
promote Th2-mediated airway inflammation in mice (12). However, whether Tc9 cells are
abnormal in asthmatic patients remains unknown, and it is also
unclear whether Tc9 cells have a role in allergic asthma.
The present study investigated the frequency of Tc9
cells and IL-9 expression levels in asthmatic patients and analyzed
their association with disease clinical features.
Materials and methods
Subjects
The study protocol was approved by the Ethics
Committee of Zhongnan Hospital of Wuhan University (Wuhan, China),
and all subjects signed informed consent. A total of 28 allergic
asthmatic patients (13 male, 15 female; mean age, 32.71±5.23),
recruited between January 2015 and March 2015 from the Outpatient
Department of Zhongnan Hospital of Wuhan University, and 20 healthy
controls (9 male, 11 female; mean age, 30.35±4.55) were recruited
in this study. Diagnosis of asthma was established according to the
Global Initiative for Asthma (14):
i) clinical history of current symptoms; ii) reversible airway
obstruction; iii) positive skin prick tests (≥3 mm) to at least one
of 10 common allergens (including Dermatophagoides
pteronyssinus, Dermatophagoides farinae, mixed grass
pollens, mixed tree pollens, dog hair, cat fur, fluffed cotton,
feather, cockroach, and Alternaria) (15); iv) no a history of smoking; v) no
other allergic diseases, autoimmune or neoplastic diseases; and vi)
no history of using systematic steroids in the past month. Asthma
severity was determined according to international European
Respiratory Society/American Thoracic Society guidelines (16). Clinical characteristics of subjects
are presented in Table I. Peripheral
blood samples from all subjects were collected into sterile
vacutainers with ethylenediaminetetraacetic acid. Blood eosinophil
count was measured using automatic hematology analyzer. Prior to
the lung function test, fractioned exhaled nitric oxide was
measured by a portable nitric oxide analyzer at an exhalation flow
rate of 50 ml/sec. FeNO measurements were performed according to
the American Thoracic Society and European Respiratory Society 2005
guidelines methods (17). Normal
FeNO values were set as 5–35 ppb for healthy adults.
 | Table I.Clinical characteristics of
subjects. |
Table I.
Clinical characteristics of
subjects.
| Characteristic | Healthy controls
(n=20) | Allergic asthmatics
(n=28) |
|---|
| Age (years) | 30.35±4.55 | 32.71±5.23 |
| Sex
(male/female) | 9/11 | 13/15 |
| FEV1 (% of
predicted) | 112.65±7.74 |
75.68±16.99a |
| Serum IgE
(ng/ml) | 239.68±126.76 |
1553.60±653.11a |
| Blood eosinophils
(109/l) | 0.17±0.09 |
0.42±0.19a |
| FeNO (ppb) | 19.8±6.30 |
43.75±13.85a |
Cell isolation and culture
Heparinized peripheral blood samples from all
subjects were collected. Peripheral blood mononuclear cells (PBMCs)
were separated by Ficoll-Hypaque gradient centrifugation (d=1.077
g/ml; Haoyang Biological Products Technology Co., Ltd., Tianjin,
China). PBMCs were harvested and washed twice in phosphate-buffered
saline (PBS), and cell viability was assessed by Trypan Blue dye
assay (>95%). Serum samples were stored at −70°C for subsequent
use. Isolated PBMCs were seeded (final concentration,
1×106/ml) in RPMI 1640 media (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). Cells
were activated with 50 ng/ml phorbol 12-myristate 13-acetate (PMA)
and 500 ng/ml ionomycin (both Sigma-Aldrich; Merck Millipore,) for
2 h at 37°C in a humidified atmosphere containing 5%
CO2, and then further cultured for 4 h following the
addition of 3 µg/ml brefeldin A (eBioscience, San Diego, CA, USA).
Finally, cells were harvested and washed prior to flow cytometry
assay.
Flow cytometry
The percentage of Tc9 cells was determined by
surface molecule and intracellular cytokine staining using a flow
cytometer. Briefly, cells were incubated with a cocktail of
phycoerythrin (PE)-Cy5 anti-human CD3 (cat. no. 15-0038-42) and
fluorescein isothiocyanate anti-human CD8 (cat. no. 11-0086-42)
(eBioscience) for 30 min in the dark at 4°C. To analyze
intracellular IL-9 production, cells were further fixed,
permeabilized with fixation and permeabilization buffer, and then
stained with PE-conjugated anti-human IL-9 (cat. no. 12-7098-42;
eBioscience) for 30 min in the dark at room temperature. Following
treatment, all stained cells were analyzed by flow cytometry using
Expo32 software (version 1696954304; Beckman Coulter, Fullerton,
CA, USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from PBMCs using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
subsequently synthesized using the ReverTra Ace® qPCR RT
kit (Toyobo Co., Ltd., Osaka, Japan), according to the
manufacturer's instructions. mRNA expression levels of target genes
were examined by qPCR using SYBR Premix Ex Taq™ (Takara Bio,
Inc., Otsu, Japan). Relative expression levels of each gene were
normalized to the expression of glyceraldehyde phosphate
dehydrogenase (GAPDH) using the 2−∆∆Cq method (18). qPCR was performed on a Bio-Rad iQ5
real-time PCR detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) under the following conditions: Initial
denaturation at 95°C for 30 sec, followed by 35 cycles at 95°C for
5 sec, 58°C for 15 sec and 72°C for 15 sec. Primer sequences were
used as described previously (19–21) and
are shown in Table II.
 | Table II.Primer sequences for quantitative
polymerase chain reaction analysis. |
Table II.
Primer sequences for quantitative
polymerase chain reaction analysis.
| Gene | Primer sequences
(5′→3′) |
|---|
| IL-9 | F:
GTGCCACTGCAGTGCTAATGT |
|
| R:
CTCTCACTAAGCATGGTCTGG |
| STAT6 | F:
CCTCGTCACCAGTTGCTT |
|
| R:
TCCAGTGCTTTCTGCTCC |
| IRF4 | F:
TGGACATCTCAGACCCGTACAAAG |
|
| R:
ATGGACATCTGCGGGTCCTC |
| GAPDH | F:
GGTGTGAACCATGAGAAGTATGACA |
|
| R:
GTCCTTCCACGATACCAAAGTTGT |
Detection of serum IgE and IL-9
levels
Serum IgE and IL-9 levels were measured by
enzyme-linked immunosorbent assay (ELISA) according to the
manufacturer's protocols (IgE and IL-9 ELISA kits; cat. no. BMS2097
and BMS2081, respectively; eBioscience). All samples were tested in
duplicate.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). All
data were expressed as mean ± standard deviation. Differences
between groups were assessed using unpaired Student's t-test or
factorial analysis of variance. Correlation analysis was performed
by Pearson's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinical characteristics of studied
subjects
Table I shows the
detailed clinical characteristics of subjects. A total of 28
asthmatic patients and 20 age- and gender-matched healthy controls
were recruited. The FEV1% of the predicted value was
significantly decreased in asthmatics compared with controls
(P<0.001). Total serum IgE levels were significantly higher in
asthmatic patients than in healthy controls (P<0.001). Moreover,
blood eosinophil counts and fractioned exhaled nitric oxide (FeNO)
levels were significantly increased in asthmatic patients
(P<0.001).
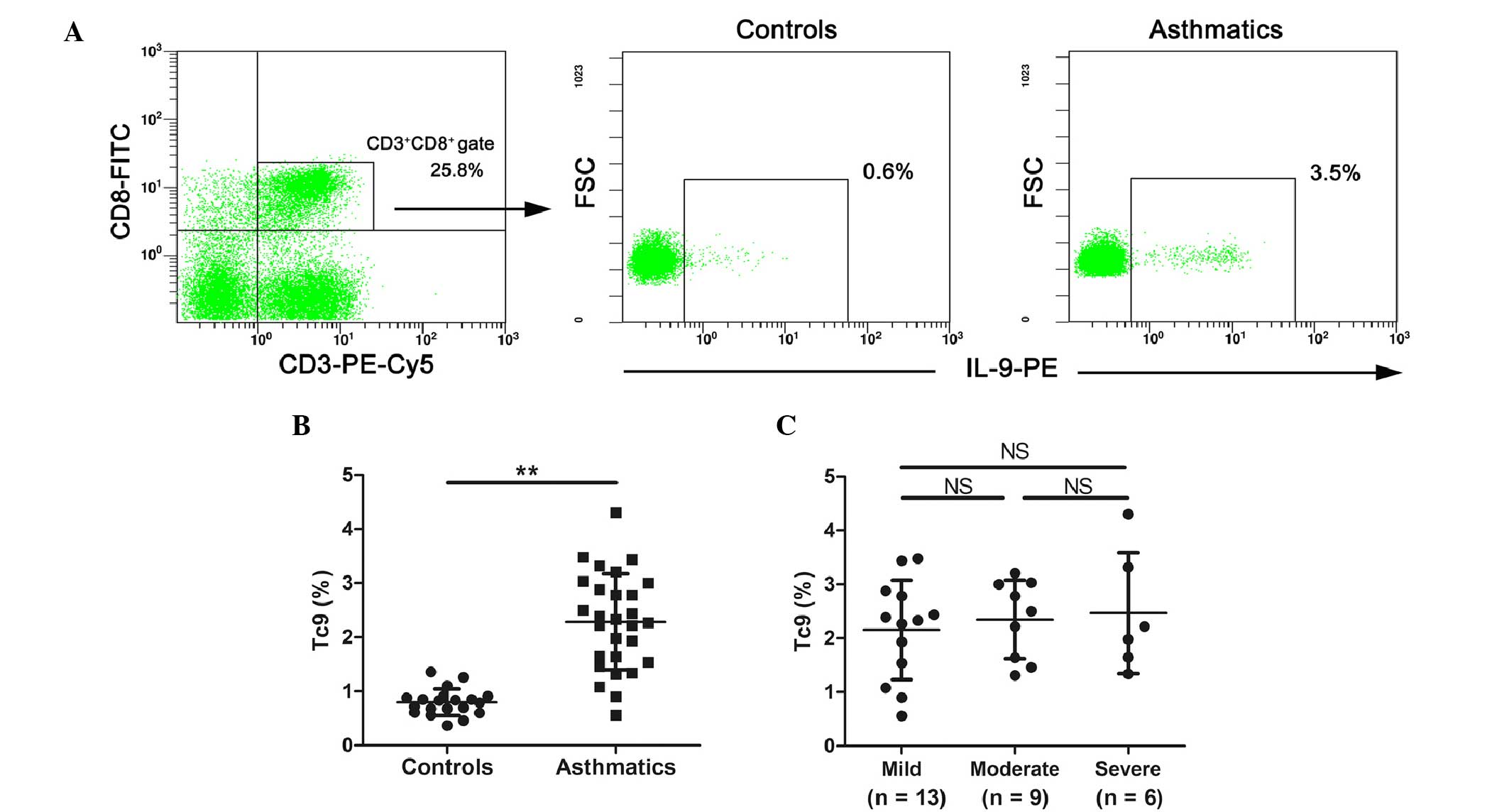
Tc9 cells are increased in patients
with allergic asthma
The frequency of Tc9 cells, which are IL-9-producing
CD8+ T cells, in PBMCs from healthy controls and
asthmatic patients was measured by flow cytometry. Representative
cytometric profiles of Tc9 from healthy controls and allergic
asthmatics are shown in Fig. 1A. As
depicted in Fig. 1B, significantly
increased numbers of Tc9 cells were detected in peripheral blood
from patients with allergic asthma (P<0.01). The frequency of
Tc9 cells were subsequently analyzed in asthmatic patients with
differing severities. The results revealed that there was no
correlation between the numbers of Tc9 cells and asthma severity
(Fig. 1C).
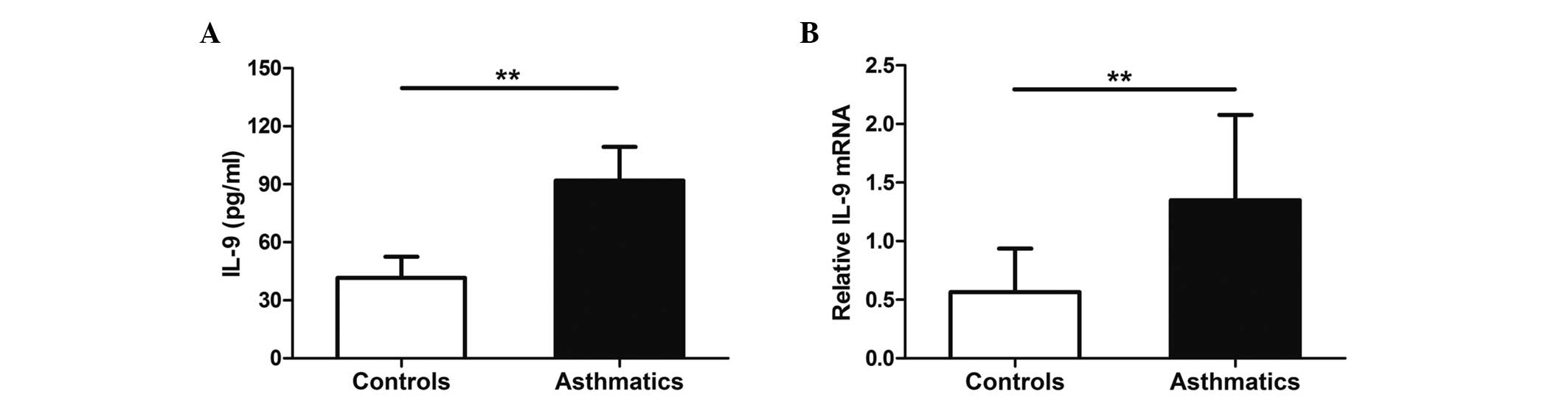
IL-9 expression levels are increased
in asthmatic patients
ELISA and RTqPCR were used to examine IL-9
expression levels in healthy controls and asthmatic patients. The
results showed that serum IL-9 protein levels in asthmatics were
increased compared with healthy controls (P<0.01; Fig. 2A). Similarly, the mRNA expression
levels of IL-9 in PBMCs were significantly higher in asthmatic
patients than in healthy controls (P<0.01; Fig. 2B).
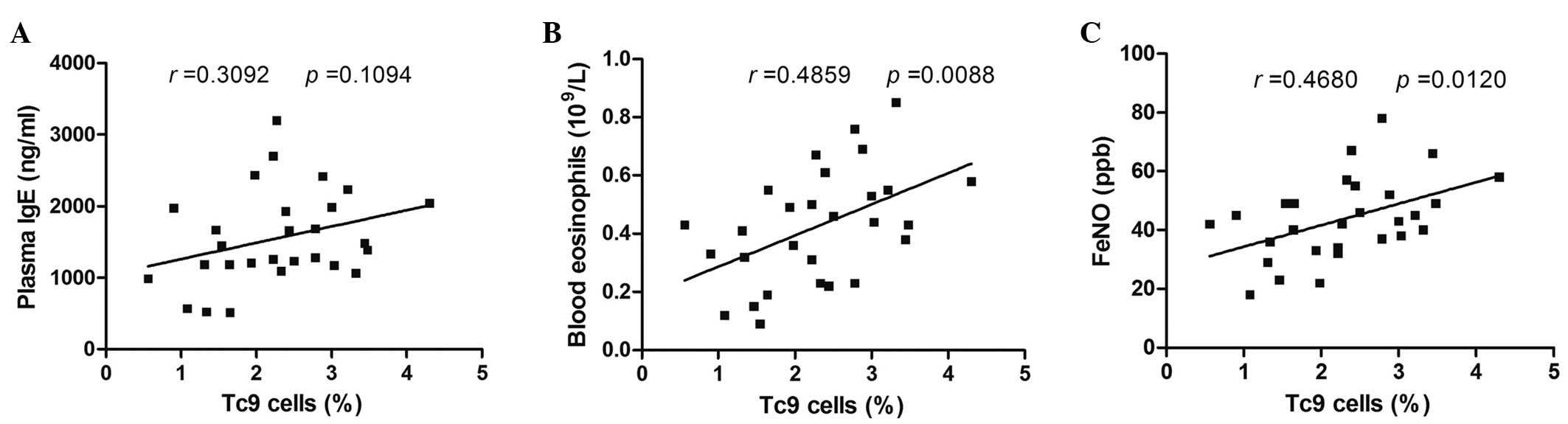
Correlations between the frequency of
circulating Tc9 cells and clinical features of asthmatics
To determine the potential role of Tc9 cells in
allergic asthmatic patients, whether the frequency of circulating
Tc9 cells was correlated with clinical features (serum IgE levels,
blood eosinophil counts and FeNO levels) was investigated in
allergic asthmatics. There was no significant correlation between
the frequency of Tc9 cells and serum IgE levels (Fig. 3A). However, circulating numbers of
Tc9 cells were positively correlated with blood eosinophil counts
or FeNO levels (P=0.0088, r=0.4859 and P=0.0120, r=0.4680,
respectively; Fig. 3B and C).
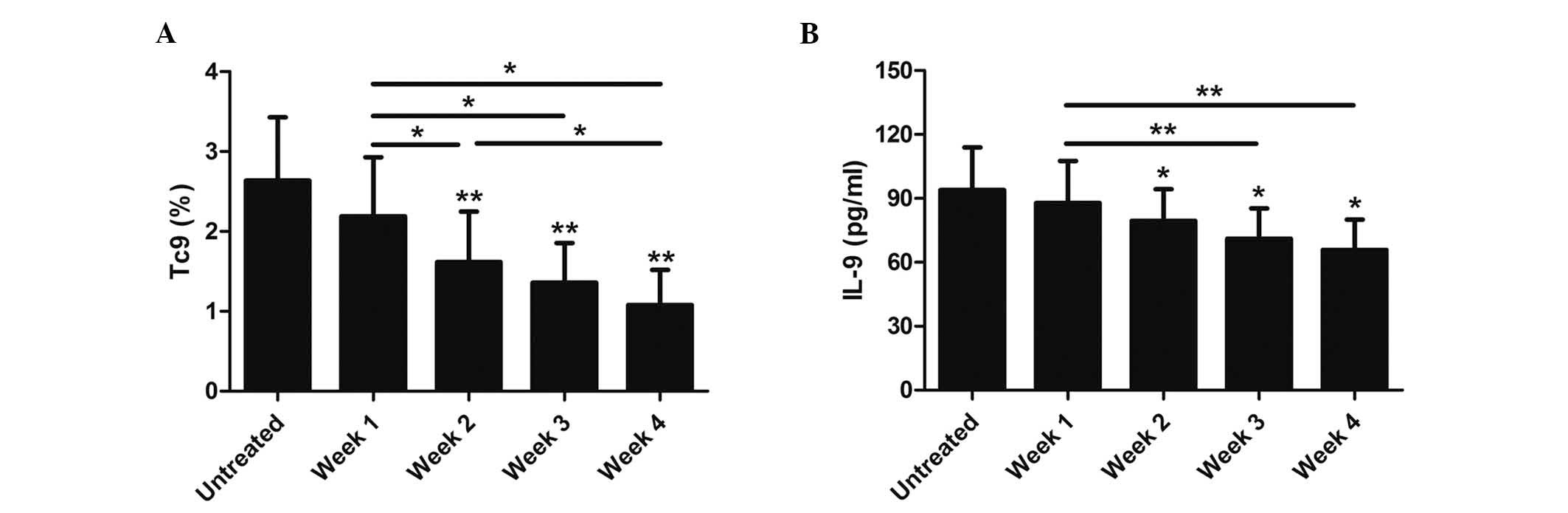
Glucocorticoids significantly decrease
the number of Tc9 cells and IL-9 protein levels in asthmatic
patients
Glucocorticoids can greatly reduce cytokine
synthesis and alleviate allergic airway inflammation. Therefore,
the effects of glucocorticoids on Tc9 cells and IL-9 protein levels
were analyzed in asthmatics. Accordingly, 15 patients in the
present study received treatment with inhaled corticosteroid (ICS;
160 µg budesonide, twice daily) according to asthma severity. The
results showed that the numbers of Tc9 cells and IL-9 protein
levels in these patients were significantly decreased after two
weeks of inhaled corticosteroid treatment (P<0.05; Fig. 4). Moreover, the percentage of Tc9
cells and IL-9 protein levels in these asthmatic patients gradually
declined at 2, 3, and 4 weeks following glucocorticoids treatment
(P<0.05; Fig. 4).
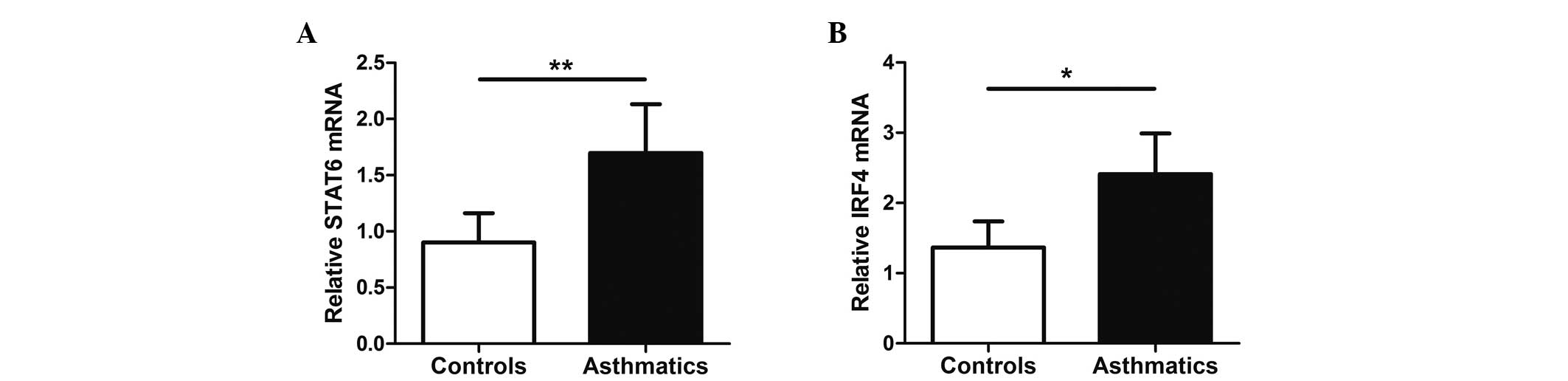
mRNA expression of STAT6 and IRF4 is
upregulated in asthmatics
STAT6 and IRF4 are key transcription factors for Tc9
cell differentiation. To assess whether STAT6 and IRF4 may
contribute to regulate Tc9 development, the mRNA expression levels
of STAT6 and IRF4 in PBMCs from peripheral blood were detected by
RT-qPCR. Notably, the results revealed elevated STAT6 and IRF4 mRNA
levels in PBMCs from allergic asthmatics (P<0.05; Fig. 5).
Discussion
CD8+ T cells are thought to contribute to
asthma pathology (8). Tc9 cells,
which are newly discovered IL-9-producing CD8+ T cells,
have been reported to promote the subsequent onset of allergic
airway inflammation mediated by pathogenic Th2 immunity in mice
(12). Nevertheless, it is not clear
whether Tc9 cells are involved in the immunopathogenesis of
asthmatic patients. In the present study, the frequency of Tc9
cells was markedly increased in asthmatic patients, and IL-9
protein and mRNA expression levels were significantly elevated in
asthmatics. It was also observed that the percentage of Tc9 cells
and IL-9 protein levels in asthmatics gradually declined following
glucocorticoids therapy. Notably, circulating numbers of Tc9 cells
were positively correlated with blood eosinophil counts and FeNO
levels. These findings suggest that Tc9 cells may contribute to the
pathogenesis of allergic asthmatics.
IL-9, a pleiotropic cytokine, has been reported to
have an important role in atopic asthma (22,23).
Studies have found that naïve CD8+ T cells are also able
to differentiate into Tc9 cells under similar Th9-polarizing
conditions (12,24). It has been demonstrated that
tumor-specific Tc9 cells may elicit antitumor responses dependent
on IL-9 production (13). Moreover,
Tc9 cells promote Th2-mediated airway inflammation in mice
(12). In the present study,
significantly increased numbers of Tc9 cells were detected in the
peripheral blood of allergic asthmatics, which was consistent with
the results of Visekruna et al (12). Furthermore, elevated serum IL-9 and
IL-9 mRNA expression levels were detected in asthmatic patients,
suggesting that Tc9 cells may have a crucial role in the
pathogenesis of allergic asthma. However, the exact mechanisms of
increased Tc9 cells in allergic asthma remain unclear. It has been
demonstrated that STAT6 and IRF4 are key transcription factors for
Tc9 cells differentiation (12,24).
They are essential for IL-9 production at the protein and mRNA
levels (12). Herein, it was
observed that allergic asthmatics had elevated STAT6 and IRF4 mRNA
levels, indicating that increased Tc9 cells exist in asthmatics at
least at the transcriptional level. Previous studies have shown
that Th9 cells increase mucus production, airway hyperreactivity
and peribronchial fibrosis mediated by IL-9 (25–27),
which is considered deleterious to lung function in asthmatics and
increases the severity of asthma. Accordingly, the percentage of
Tc9 cells in asthmatics with differing severities were analyzed. No
significant difference was found in the numbers of Tc9 cells
between them. This indicates that Th9 cells, but not Tc9 cells, may
be associated with airway remodelling.
IgE overproduction and increased eosinophils have
been recognized as cardinal features of Th2-mediated allergic
asthma. FeNO is an indirect biomarker of airway inflammation in
asthmatic patients (28). FeNO is
often increased in steroid-sensitive asthmatics, and is correlated
with eosinophilia (29,30). In particular, consistent with these
previous studies, the present study exhibited increased serum IgE
levels in asthmatics, as compared with healthy controls. Blood
eosinophil counts and FeNO levels were also significantly increased
in asthmatic patients. Notably, the present study found that
circulating numbers of Tc9 cells were positively correlated with
blood eosinophil counts and FeNO levels. Therefore, we hypothesize
that increased circulating Tc9 cells may be an important feature of
allergic asthma, and that Tc9 cells may activate Th2-mediated
allergic inflammation, including eosinophilia and high FeNO,
directly or/and through innate immune cells in asthmatics, which is
in combination with the previous findings that Tc9 cells may elicit
key features of allergic airway inflammation, such as eosinophilia
(12).
Glucocorticoids are powerful anti-inflammatory
factors used to treat asthma that can induce immune effects and
affect cytokine production by activating receptors. It has been
reported that glucocorticoids greatly reduced cytokine synthesis
and alleviated allergic airway inflammation in asthmatics (31). Therefore, the effects of
glucocorticoids on Tc9 cells and IL-9 protein levels were
investigated in asthmatics. Notably, the number of Tc9 cells and
serum IL-9 protein levels in asthmatics were significantly
decreased after two weeks of inhaled corticosteroid treatment.
Moreover, the percentage of Tc9 cells and IL-9 protein levels
gradually declined with therapy. Thus, similar to the effects of
glucocorticoids on eosinophils, glucocorticoids may downregulate
Tc9 cell populations as well as inhibit IL-9 expression in
asthmatics. Accordingly, we hypothesize that glucocorticoids may
dampen the subsequent onset of allergic asthma by inhibiting the
immunopathogenic role of Tc9 cells, which should be further
investigated in future studies.
In conclusion, the present results demonstrated
increased circulating Tc9 cells, IL-9-producing CD8+ T
cells, in allergic asthmatics, which are paralleled with
eosinophilia and high FeNO. Elevated mRNA expression levels of
transcription factors STAT6 and IRF4 may contribute to the abnormal
Tc9 immunity in asthmatics. These findings suggest that targeting
Tc9 cells in human asthma may be a promising therapy in the
future.
Acknowledgements
We would like to thank all the faculty in the Key
Laboratory of Allergy and Immune Diseases at Wuhan University
(Wuhan, China). The study was supported by the National Natural
Science Foundation of China (grant no. 81670021).
References
|
1
|
Kim HY, DeKruyff RH and Umetsu DT: The
many paths to asthma: Phenotype shaped by innate and adaptive
immunity. Nat Immunol. 11:577–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shalaby KH and Martin JG: Overview of
asthma; the place of the T cell. Curr Opin Pharmacol. 10:218–225.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Noelle RJ and Nowak EC: Cellular sources
and immune functions of interleukin-9. Nat Rev Immunol. 10:683–687.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaplan MH: Th9 cells: Differentiation and
disease. Immunol Rev. 252:104–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang HC, Sehra S, Goswami R, Yao W, Yu Q,
Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, et al: The
transcription factor PU.1 is required for the development of
IL-9-producing T cells and allergic inflammation. Nat Immunol.
11:527–534. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Faulkner H, Renauld JC, Van Snick J and
Grencis RK: Interleukin-9 enhances resistance to the intestinal
nematode Trichuris muris. Infect Immun. 66:3832–3840.
1998.PubMed/NCBI
|
|
7
|
Sehra S, Yao W, Nguyen ET, Glosson-Byers
NL, Akhtar N, Zhou B and Kaplan MH: TH9 cells are required for
tissue mast cell accumulation during allergic inflammation. J
Allergy Clin Immunol. 136:433–440.e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Sullivan S, Cormican L, Faul JL,
Ichinohe S, Johnston SL, Burke CM and Poulter LW: Activated,
cytotoxic CD8(+) T lymphocytes contribute to the pathology of
asthma death. Am J Respir Crit Care Med. 164:560–564. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miyahara N, Swanson BJ, Takeda K, Taube C,
Miyahara S, Kodama T, Dakhama A, Ott VL and Gelfand EW: Effector
CD8+ T cells mediate inflammation and airway
hyper-responsiveness. Nat Med. 10:865–869. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marsland BJ and Le Gros G: CD8+
T cells and immunoregulatory networks in asthma. Springer Semin
Immunopathol. 25:311–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Betts RJ and Kemeny DM: CD8+ T
cells in asthma: Friend or foe? Pharmacol Ther. 121:123–131. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Visekruna A, Ritter J, Scholz T, Campos L,
Guralnik A, Poncette L, Raifer H, Hagner S, Garn H, Staudt V, et
al: Tc9 cells, a new subset of CD8(+) T cells, support Th2-mediated
airway inflammation. Eur J Immunol. 43:606–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu Y, Hong B, Li H, Zheng Y, Zhang M, Wang
S, Qian J and Yi Q: Tumor-specific IL-9-producing CD8+
Tc9 cells are superior effector than type-I cytotoxic Tc1 cells for
adoptive immunotherapy of cancers. Proc Natl Acad Sci USA.
111:2265–2270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Global initiative for asthma, . GINA
report, global strategy for asthma management and prevention.
Available from. http://www.ginasthma.org2015.
|
|
15
|
Bousquet J, Heinzerling L, Bachert C,
Papadopoulos NG, Bousquet PJ, Burney PG, Canonica GW, Carlsen KH,
Cox L, Haahtela T, et al: Practical guide to skin prick tests in
allergy to aeroallergens. Allergy. 67:18–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chung KF, Wenzel SE, Brozek JL, Bush A,
Castro M, Sterk PJ, Adcock IM, Bateman ED, Bel EH, Bleecker ER, et
al: International ERS/ATS guidelines on definition, evaluation and
treatment of severe asthma. Eur Respir J. 43:343–373. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
ATS/ERS recommendations for standardized
procedures for the online and offline measurement of exhaled lower
respiratory nitric oxide and nasal nitric oxide, 2005, . Am J
Respir Crit Care Med. 2005.171(8): 912–930. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ouyang H, Shi Y, Liu Z, Feng S, Li L, Su
N, Lu Y and Kong S: Increased interleukin-9 and
CD4+IL-9+ T cells in patients with systemic
lupus erythematosus. Mol Med Rep. 7:1031–1037. 2013.PubMed/NCBI
|
|
20
|
Aoudjehane L, Pissaia A Jr, Scatton O,
Podevin P, Massault PP, Chouzenoux S, Soubrane O, Calmus Y and
Conti F: Interleukin-4 induces the activation and collagen
production of cultured human intrahepatic fibroblasts via the
STAT-6 pathway. Lab Invest. 88:973–985. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Gao YD and Yang J: Elevated
interferon regulatory factor 4 levels in patients with allergic
asthma. J Asthma. 49:441–449. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McLane MP, Haczku A, van de Rijn M, Weiss
C, Ferrante V, MacDonald D, Renauld JC, Nicolaides NC, Holroyd KJ
and Levitt RC: Interleukin-9 promotes allergen-induced eosinophilic
inflammation and airway hyperresponsiveness in transgenic mice. Am
J Respir Cell Mol Biol. 19:713–720. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Erpenbeck VJ, Hohlfeld JM, Volkmann B,
Hagenberg A, Geldmacher H, Braun A and Krug N: Segmental allergen
challenge in patients with atopic asthma leads to increased IL-9
expression in bronchoalveolar lavage fluid lymphocytes. J Allergy
Clin Immunol. 111:1319–1327. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huber M and Lohoff M: IRF4 at the
crossroads of effector T-cell fate decision. Eur J Immunol.
44:1886–1895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Louahed J, Toda M, Jen J, Hamid Q, Renauld
JC, Levitt RC and Nicolaides NC: Interleukin-9 upregulates mucus
expression in the airways. Am J Respir Cell Mol Biol. 22:649–656.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van den Brûle S, Heymans J, Havaux X,
Renauld JC, Lison D, Huaux F and Denis O: Profibrotic effect of
IL-9 overexpression in a model of airway remodeling. Am J Respir
Cell Mol Biol. 37:202–209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soroosh P and Doherty TA: Th9 and allergic
disease. Immunology. 127:450–458. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wadsworth S, Sin D and Dorscheid D:
Clinical update on the use of biomarkers of airway inflammation in
the management of asthma. J Asthma Allergy. 4:77–86. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Payne DN, Adcock IM, Wilson NM, Oates T,
Scallan M and Bush A: Relationship between exhaled nitric oxide and
mucosal eosinophilic inflammation in children with difficult
asthma, after treatment with oral prednisolone. Am J Respir Crit
Care Med. 164:1376–1381. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dweik RA, Boggs PB, Erzurum SC, Irvin CG,
Leigh MW, Lundberg JO, Olin AC, Plummer AL and Taylor DR: American
Thoracic Society Committee on Interpretation of Exhaled Nitric
Oxide Levels (FENO) for Clinical Applications: An official ATS
clinical practice guideline: Interpretation of exhaled nitric oxide
levels (FENO) for clinical applications. Am J Respir Crit Care Med.
184:602–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Richards DF, Fernandez M, Caulfield J and
Hawrylowicz CM: Glucocorticoids drive human CD8(+) T cell
differentiation towards a phenotype with high IL-10 and reduced
IL-4, IL-5 and IL-13 production. Eur J Immunol. 30:2344–2354. 2000.
View Article : Google Scholar : PubMed/NCBI
|