Introduction
Regeneration of periodontal tissue requires the
exclusion of the epithelium and, in some cases, the gingival
connective tissue from the root surface. In addition, previous
studies (1–3) strongly support the hypothesis that
enamel matrix proteins (EMPs), known for their impact on the
structural organization of tooth enamel, may serve an important
role in periodontal tissue formation (1). It is believed that enamel matrix
derivative (EMD), the active component of Emdogain (Straumann), has
odontogenic effects through inducing the proliferation and
differentiation of connective tissue progenitor cells, stimulating
bone growth and arresting gingival epithelial cell migration
(2,3).
EMD is comprised primarily of amelogenins (AMELs), a
family of proline-rich peptides synthesized from the AMEL gene by
alternative splicing and post-translational modifications. This
family includes full-length AMEL (25 kDa), which is processed into
a 20 kDa protein, and then into a tyrosine-rich AMEL peptide (TRAP)
and a leucine-rich AMEL peptide (LRAP) (4,5). Only a
small number of studies have directly described the effect of EMD
and full-length recombinant AMEL in vitro (6–8).
Furthermore, alternatively spliced products and degraded forms of
AMEL have biochemical properties that are distinct from full-length
AMEL that are critical for function (9,10), as
well as between amelogenins with different molecular mass (11). Previous studies that have analyzed
the influence of EMD on gingival epithelial cells are rare and the
results ambiguous. A number of studies have demonstrated that EMD
inhibits epithelial cell proliferation (12–15),
while another indicated no effect (16) and another observed acceleration of
epithelialization following EMD stimulation (17). Moreover, it is unclear which
component of EMD is a direct inhibitor of epithelial cell growth.
In previous studies, full-length recombinant AMEL was indicated to
be the active component (18,19).
The aim of present study was to investigate the
influence of commercial lyophilized EMD, porcine recombinant prAMEL
and TRAP on the adherence, proliferation and migration of human
epithelial cells. Real-time cell analysis (RTCA; xCELLigence) was
used to facilitate label-free and operator-independent
investigation of cell behavior, through monitoring the cells in
physiologically relevant conditions.
Materials and methods
Experimental proteins
Lyophilized EMD was provided by the Straumann AG
Institute (Basel, Switzerland). Porcine recombinant AMEL (49 KDa)
and TRAP (5.3 kDa) were synthesized, as described below. Cells were
stimulated with protein extracts of 12.5, 25 and 50 µg/ml.
Porcine recombinant AMEL
synthesis
Construction of pGex4T-1-AMEL-GST
AMEL protein was provided by BLIRT S.A. (Gdańsk,
Poland). The protein sequence of Sus scrofa AMEL was
obtained from the UniProt database (accession no. Q861X0;
uniprot.org/). This sequence, with an added
glutathione S-transferase (GST) tag to increase protein solubility,
is the following: ENFLYQGSMPLPPHPGHPGYINFYEDLYLEAIRIDRTAF
VLTPLKWYQNMIRHPYTSYGYEPMGGWLHHQIIPVVS
QQTPQSHALQPHHHIPMVPAQQPGIPQQPMMPLPGQH
SMTPTQHHQPNLPLPAQQPFQPQPVQPQPHQPLQPQSP
MHPIQPLLPQPPLPPMFSMQSLLPDLPLEAWPAT. The amelogenin construct
contains prAMEL (21.3 kDa) and GST, yielding a molecular mass of
~49 kDa.
The DNA sequence encoding the AMEL-GST protein was
synthesized using the GeneArt service (Thermo Fisher Scientific,
Inc.; Waltham, MA, USA). The sequence obtained was cloned into the
pGex4T-1 vector (Addgene, Inc., Cambridge, MA USA) with NdeI
and BamHI enzymes. The pGex4T-1-AMEL-GST construct was
transformed into ArcticExpress (DE3) E. coli (Agilent
Technologies, Inc., Santa Clara, CA, USA) using a chemical method.
Plasmid DNA was added to 100 µl competent cells on ice. The whole
mixture was incubated on ice for 30 min. The bacteria were shocked
at 42°C and cooled on ice. lysogeny broth (LB) medium was added and
the culture was grown at 37°C for 45 min. The transformation mix
was transferred on LB agar supplemented with ampicillin (100
µg/ml). The resulting clones were sequenced using an automated ABI
Prism 3130xl Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.) to confirm that cloning had been performed
correctly. The amelogenin construct included amelogenin (21.3 kDa)
and GST, yielding a final molecular mass of 43 kDa.
Overexpression of AMEL-GST in E. coli
ArcticExpress (DE3) E. coli containing the
pGex4T-1-AMEL-GST construct were cultured overnight in LB media,
supplemented with ampicillin (100 µg/ml) and gentamicin (40 µg/ml).
Cultures were then diluted to a 1:100 ratio in the same media and
cultured at 30°C until they reached an optical density reading of
0.6 at a wavelength of 600 nm. The cultures were then cooled to
10°C and protein expression induced with 0.1 mM isopropyl
β-D-1-thiogalactopyranoside (IPTG). Cultivation was performed for
~40 h, prior to centrifugation at 3,500 × g for 30 min at
4°C (Heraeus Multifuge 3 S-R; Thermo Fisher Scientific, Inc.). And
freezing of the resulting pellet.
Purification of the AMEL-GST fusion protein
The cell pellet (~15 g) was suspended in 200 ml of
buffer A (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 8), with
protease inhibitor (1:100; P8340; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) lysed by sonication (10 times for 20 sec, 60
sec rest, with total energy 200 J/ml) and the resulting lysate
centrifuged at 3,500 × g for 30 min at 4°C. As expected,
most protein remained in the soluble fraction. Following
centrifugation the protein was bound via its GST tag to a 5 ml
conditioned Glutathione Sepharose 4B (GE Healthcare Bio-Sciences
AB, Uppsala, Sweden) for 2 h at 4°C, rinsed twice with buffer A 4X
column volume (CV) and eluted with buffer B (50 mM Tris, 50 mM
NaCl, 20 mM GSH, pH 8) 3×1.5 CV. The fractions obtained were
analyzed by 4–12% SDS-PAGE to confirm the presence of AMEL and the
quantity from images of the gels analyzed using TotalLab Quant 1.2
software (Cleaver Scientific Ltd., Warwickshire, United Kingdom) to
determine the purity. Protein concentration was determined by the
Bradford assay (Thermo Fisher Scientific, Inc.) and a NanoDrop
instrument (Thermo Fisher Scientific, Inc.).
Lyophilization of the AMEL-GST fusion
protein
AMEL-GST and GST were lyophilized using the
freeze-dry technique. Proteins (1 mg) in buffer A were aliquoted,
frozen in liquid nitrogen and lyophilized overnight.
Reconstitution of the AMEL-GST fusion
protein
Lyophilized AMEL-GST and GST were reconstituted in 1
ml of buffer A to concentration of 1 mg/ml. Reconstituted samples
were centrifuged at 3,500 × g for 30 min at 4°C to remove
any aggregated protein and separated by 4–12% SDS-PAGE to confirm
reconstitution. Samples of AMEL-GST and GST were analyzed using the
Bradford assay to confirm a concentration of 1 mg/ml and estimate
the quantity of protein lost during reconstitution. Protein
concentration prior to lyophilization and following reconstitution
was 1 mg/ml, indicating that no protein was lost.
TRAP synthesis
Construction of pET-22b-TRAP
A TRAP gene construct was obtained by polymerase
chain reaction (PCR) amplification of the clone containing human
AMEL cDNA using the following modified primers: Forward, 5′-TTT CAT
ATG CAT CAC CAT CAC CAT CAC GAT GAC GAT GAC AAG ATG CCT CTA CCA CCT
CAT CC-3′ and reverse, 5′-TTT AAG CTT CAC CAT CCA CCC ATG GGT TCG
TAC CCA TAG GAA GTG TAC GGA TGT CTT ATC ATG TTC TG-3′. Human AMEL
cDNA clone was used as a template and modified primers converted
the human TRAP coding sequence to the pig coding sequence of TRAP.
These were capable of converting human TRAP coding sequence to pig
coding sequence of TRAP, and contained a histidine tag and
enterokinase recognition site. PCR was performed in a 25-µl total
reaction volume containing 5 ng plasmid DNA (human AMEL cDNA
clone), 1X KAPA2G Robust HotStart DNA Polymerase (KK5702; Robust
HotStart ReadyMix PCR kit; Kapa Biosystems, Inc., Wilmington, MA,
USA) and 0.125 µM of each primer. PCR thermal cycling conditions
were as follows: 35 cycles of 30 sec at 94°C; 30 sec at 58°C; and
45 sec at 72°C. Subsequently, the PCR product was purified using a
StrataPrep PCR Purification kit (400773; Agilent Technologies,
Inc.) and sequenced using an automated 3130xl Genetic Analyzer
(Applied Biosystems; Thermo Fisher Scientific, Inc.) to confirm
correct cloning. The TRAP fragment was digested with NdeI
and HindIII enzymes and ligated into the pET-22b(+)
expression vector (Novagen; Merck & Co., Inc., Whitehouse
Station, NJ, USA) digested with the same endonucleases.
General procedures of handling DNA were performed
according to Sambrook and Russel (20). Plasmid DNA was isolated using the
StrataPrep Plasmid Miniprep kit (400761; Agilent Technologies,
Inc.). PCR reagents, restriction enzymes and T4DNA ligase were
purchased from Kapa Biosystems, Inc., (Wilmington, MA, USA), Thermo
Fisher Scientific, Inc., and New England Biolabs, Inc., (Ipswich,
MA, USA), respectively.
TRAP overexpression in E. coli
This construct was transformed into Rosetta 2 (DE3)
pLysS E. coli (Novagen; Merck & Co., Inc.) using a
chemical method. Plasmid DNA was added to 100 µl competent cells on
ice. The whole mixture was incubated on ice for 30 min. The
bacteria were shocked at 42°C and cooled on ice. LB medium was
added and culture was grown at 37°C for 45 min. The transformation
mix was transferred onto LB agar supplemented with ampicillin (100
µg/ml) and chloramphenicol (34 µg/ml). E. coli was routinely
grown overnight at 37°C, with standard antibiotic plate selection
performed according to the manufacturer's instructions. Transformed
E. coli were grown overnight in LB media supplemented with
ampicillin (100 µg/ml) and chloramphenicol (34 µg/ml). Cultures
were then diluted in a 1:100 ratio in the same media and cultured
at 37°C until they reached an optical density reading of between
0.6 and 0.8 at a wavelength of 600 nm. Then, protein expression was
induced with 1 mmol/l IPTG and cultures were grown for 16 h at
37°C.
Immobilized metal affinity chromatography of
TRAP
Pellets were collected by centrifugation at 3,500 ×
g for 30 min at 4°C (Heraeus Multifuge 3 S-R; Thermo Fisher
Scientific, Inc.). Pellets were suspended in 2 ml of modified
phosphate buffer solution (50 mmol/l phosphate buffer disodium
hydrogen phosphate and potassium dihydrogen phosphate, pH 8.0, 300
mmol/l NaCl, 10% glycerol) and disrupted using a 10 ml tissue
grinder at 4°C. The resulting lysate was centrifuged at 14,000 ×
g for 20 min at 4°C. The supernatant from this was bound
overnight at 4°C onto 2 ml of cobalt resin (TALON Metal Affinity
Resin; Clontech; Takara Biotechnology Co., Ltd., Dalian, China)
equilibrated with phosphate buffer. Then, the suspension was placed
in a PD-10 column (Sigma-Aldrich, St. Louis, MO, USA) and washed
with 30 ml of phosphate buffer. Unbound proteins were washed away
using a stepwise pH gradient consisting of 10 ml of phosphate
buffer at pH 7.0, 10 ml of phosphate buffer at pH 6.0 and 10 ml of
phosphate buffer at pH 5.7. The recombinant protein was eluted with
pH 6.0 phosphate buffer, where 2 ml fractions were collected and
frozen at −20°C. Samples were subjected to 16% SDS-PAGE and stained
with Coomassie Brilliant Blue to visualize proteins. The
concentration of purified protein was measured using the Bradford
method and found a yield of ~4.7 mg/ml.
Cell culture
All experiments were performed on the human tongue
squamous cell carcinoma cell line (SCC-25; cat. no. CRL-1628;
American Type Culture Collection, Manassas, VA, USA). SCC-25 cells
were transferred under aseptic conditions from freezing medium
[Dulbecco's modified Eagle medium/Nutrient Mixture F-12 (DMEM/F12),
10% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (all Gibco;
Thermo Fisher Scientific, Inc.), and 400 ng/ml of hydrocortisone],
to a 90 mm sterile petri dish (Sarstedt AG & Co., Nümbrecht,
Germany) containing 10 ml of growth medium [DMEM/F12, 10% FBS, 100
µg/ml penicillin, 100 µg/ml streptomycin and 2 mmol/l L-glutamine
(all Gibco; Thermo Fisher Scientific, Inc.), and 400 ng/ml of
hydrocortisone]. Cells were grown at 37°C, under 5% CO2
and at 100% relative humidity. Cells were cultured until 90%
confluency, washed with phosphate buffered saline and trypsinized
(0.25% trypsin containing 0.01% EDTA). Following 5 min incubation
at room temperature, complete growth medium was added at a ratio of
1:10, and the cell suspension was transferred to new petri
dishes.
Cell adherence and monitoring
Cell adherence and proliferation was monitored in
real-time using the xCELLigence system and E-Plate 96 insert (ACEA
Biosciences, Inc., San Diego, CA, USA). Instrument measures the
electrical resistance of the sensor electrodes that is proportional
to the number of cells attached to the sensors, which allows real
time measurements by probing cell growth at different time
intervals. The electrical impedance value of each well was
automatically monitored by the xCELLigence system and expressed as
a cell index (CI) value. Each experiment was performed five times.
The external control plate contained cells that were not exposed to
the experimental proteins.
For cell adherence measurements, after reaching 90%
confluency, SCC-25 cells were passaged with 0.25% trypsin solution
and seeded into wells of the E-plate 96 at 10,000 cells/well.
Immediately, 96 wells were stimulated with the protein extracts
(EMD, AMEL and TRAP at final concentrations of 12.5, 25 and 50
µg/ml, respectively), released by the metallic alloy material, and
monitored every 15 min for 14 h.
For cell proliferation measurements, after reaching
confluence, SCC-25 cells were passaged with 0.25% trypsin and
seeded into wells of the E-plate 96 at 10,000 cells/well. Then,
cells were left to obtain cell a CI value equal to ~1. Afterwards,
cells were treated with EMD, prAMEL and TRAP (12.5, 25 and 50
µg/ml, respectively), released by the metallic alloy material, and
monitored every 15 min for 48 h. Evaluation was performed 12, 24,
48, 60 and 77 h after stimulation.
Monitoring of cell migration
The rate of cell migration was monitored in
real-time with the xCELLigence system and the CIM-plate 16 insert
(ACEA Biosciences, Inc.). Cells were passaged and placed in the
upper chamber of CIM-plate 16 in FBS-free media. The lower chamber
of the plate contained 160 µl of media with 10% of FBS as an
attractant. Cell migration was measured by electrodes located
between the lower and upper chambers. Immediately following seeding
at 20,000 cells/well, cells were treated with EMD, prAMEL and TRAP
(12.5, 25 and 50 µg/ml, respectively), and monitored every 15 min
for 49 h. The control plate contained cells not exposed to the
proteins.
Statistical analysis
Statistical analysis was performed using Statistica
software (version 10; StatSoft, Inc., Tulsa, OK, USA). The
Shapiro-Wilk test of normality was used on continuous variables.
The results are described as the mean ± standard deviation. One-way
analysis of variance with the multiple comparisons Tukey's test was
applied. P<0.05 was considered to indicate a statistically
significant difference.
Results
Cell morphology
None of the analyzed proteins affected SCC-25 cell
morphology, regardless of the dose (Fig.
1). SCC-25 cells, characterized by a spindle-like shape. Only
changes to a more circular shape with increasing cell density were
observed. In addition, characteristic proliferation in clusters was
observed (Fig. 1).
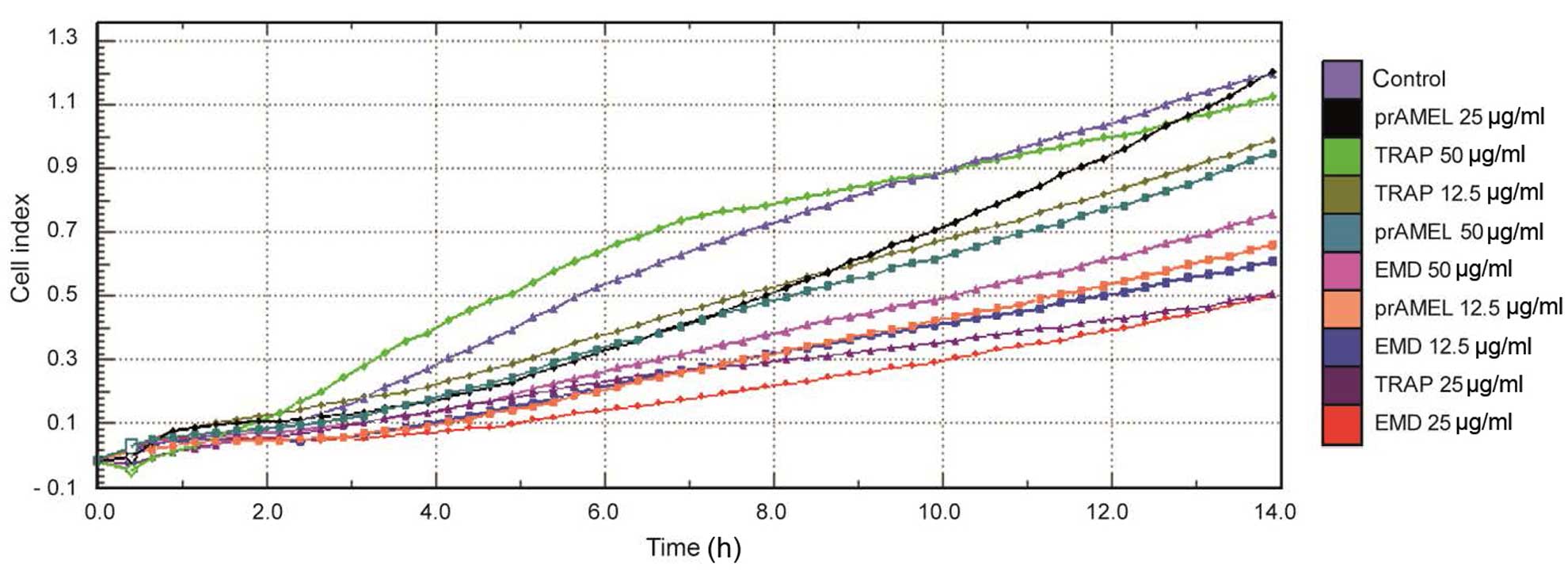
Cell adherence
The effect of experimental proteins on SCC-25 cell
adhesion was monitored over 14 h in real-time. A representative
graph comparing the rate of cell adherence, in terms of CI, when
incubated with EMD, prAMEL or TRAP protein is shown in Fig. 2. No significant difference in cell
adherence was observed among all the groups, regardless of dose
(Table I).
 | Table I.Effect of EMPs on the rate of
adherence of SCC-25 cells. |
Table I.
Effect of EMPs on the rate of
adherence of SCC-25 cells.
|
| Cell index value
mean ± standard deviation |
|---|
|
|
|
|---|
| EMP added,
µg/ml | 4-h incubation | 8-h incubation | 12-h
incubation |
|---|
| Control | 0.80±0.9 | 1.49±1.3 | 1.94±1.6 |
| EMD, 12.5 | 0.57±0.5 | 0.93±0.8 | 1.22±0.9 |
| EMD, 25 | 0.56±0.5 | 0.94±0.8 | 1.23±1.0 |
| EMD, 50 | 0.55±0.3 | 0.99±0.4 | 1.36±0.4 |
| prAMEL, 12.5 | 0.53±0.4 | 0.91±0.6 | 1.24±0.7 |
| prAMEL, 25 | 0.62±0.2 | 1.14±0.3 | 1.59±0.3 |
| prAMEL, 50 | 0.70±0.3 | 1.25±0.4 | 1.67±0.5 |
| TRAP, 12.5 | 0.77±0.5 | 1.28±0.8 | 1.66±0.9 |
| TRAP, 25 | 0.52±0.4 | 0.84±0.6 | 1.08±0.7 |
| TRAP, 50 | 1.07±0.6 | 1.65±0.7 | 1.95±0.8 |
| P-value | >0.05 | >0.05 | >0.05 |
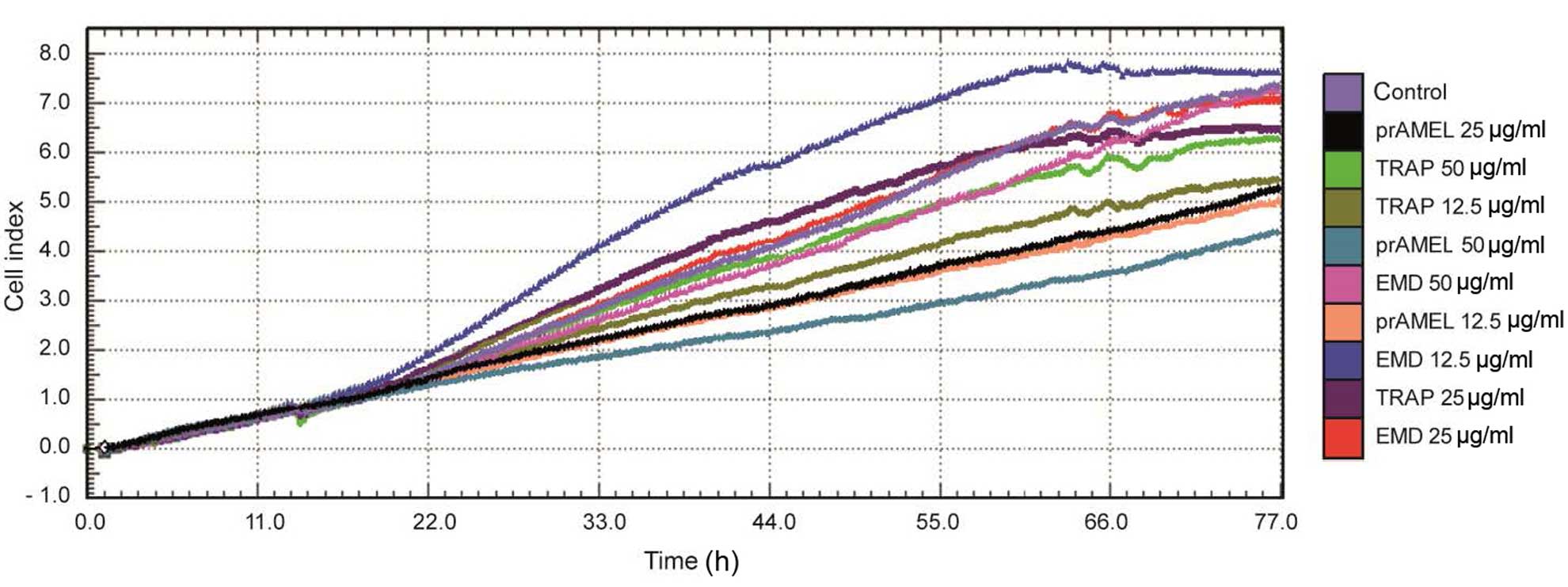
Cell proliferation
Cell proliferation was monitored using RTCA over a
period of 77 h after EMD, prAMEL or TRAP stimulation. A
representative graph comparing the rate of CI of SCC-25 cells is
shown in Fig. 3. No significant
difference in the rate of proliferation was observed after 12-h
incubation (Table II). RTCA
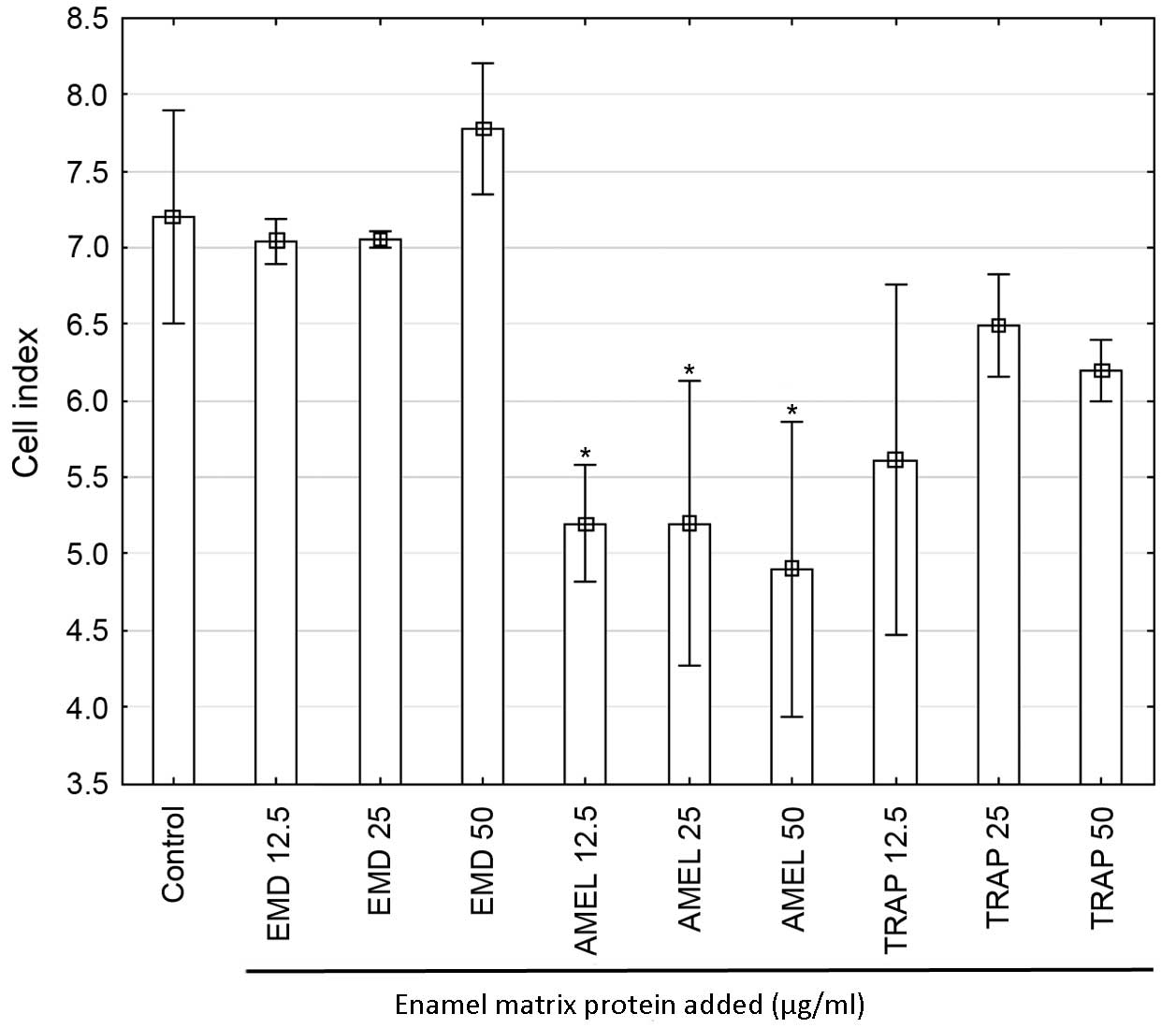
analysis performed after 24-h incubation showed a significant
decrease of CI in prAMEL (12.5 µg/ml) compared with cells
stimulated with EMD 12.5 µg/ml (P=0.02) and 25 µg/ml-stimulated
cells (P=0.02). Moreover, all doses of AMEL (12.5, 25 and 50 µg/ml)
administered for 48-h caused a significant decrease in the
proliferation rate in comparison with both control cells
(P<0.001 for all doses) and all EMD doses (P<0.001 for all
comparisons; Fig. 4) and EMD (50
µg/ml)-stimulated cells (P=0.005; Fig.
4).
 | Table II.Effect of EMPs on the rate of
proliferation of SCC-25 cells. |
Table II.
Effect of EMPs on the rate of
proliferation of SCC-25 cells.
|
| Cell index value
mean ± standard deviation |
|---|
|
|
|
|---|
| EMP added,
µg/ml | 12-h
incubation | 24-h
incubation | 48-h
incubation |
|---|
| Control | 3.9±0.9 | 5.0±1.3 | 7.2±0.6 |
| EMD, 12.5 | 5.6±1.1 | 6.8±1.1 | 7.0±0.1 |
| EMD, 25 | 5.4±0.8 | 6.7±0.5 | 7.1±0.1 |
| EMD, 50 | 4.4±0.1 | 5.7±0.1 | 7.8±0.4 |
| prAMEL, 12.5 | 3.0±0.5 | 3.7±0.4 | 5.1±0.3 |
| prAMEL, 25 | 4.5±1.2 | 5.0±0.8 | 5.2±0.9 |
| prAMEL, 50 | 3.5±1.3 | 4.4±0.7 | 4.9±0.9 |
| TRAP, 12.5 | 3.6±0.7 | 4.4±0.9 | 5.6±1.1 |
| TRAP, 25 | 5.1±0.3 | 6.2±0.4 | 6.5±0.3 |
| TRAP, 50 | 4.0±0.4 | 5.0±0.4 | 6.2±0.2 |
| P-value | 0.07 | 0.02a | 0.005b |
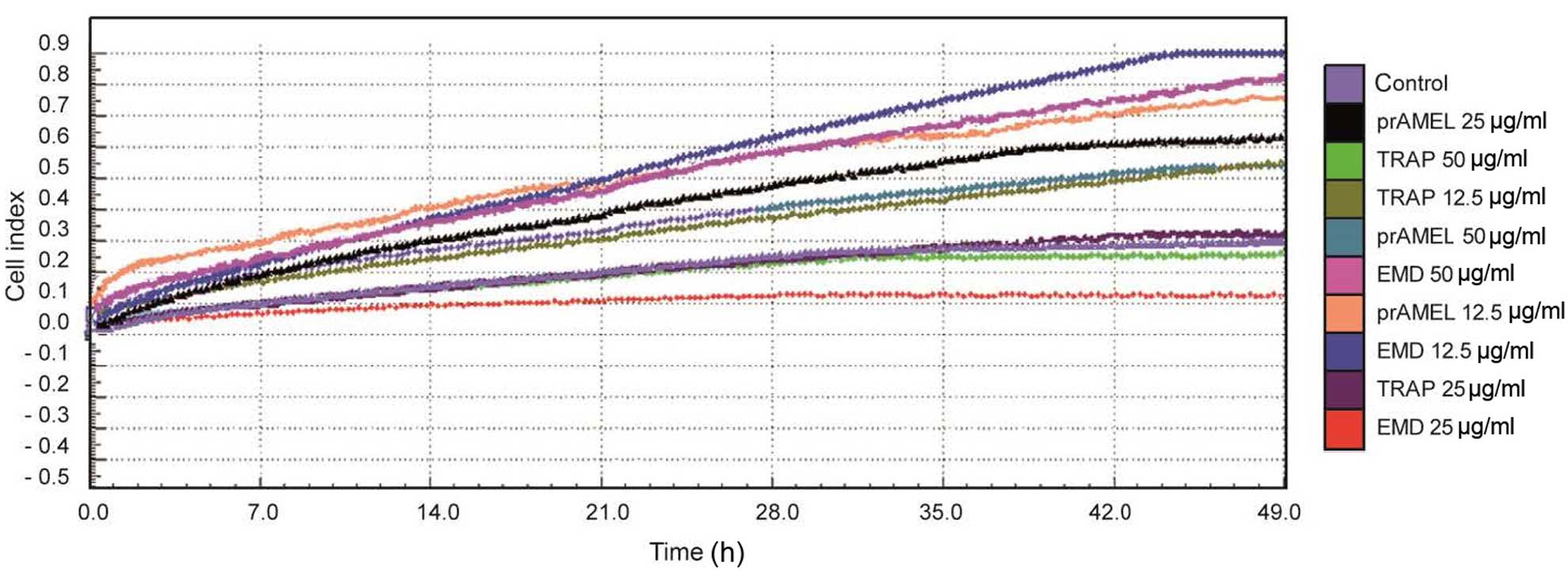
Cell migration
Regardless of the type of ligand, dose and time
following stimulation, no significant differences in SCC-25 cell
migration were observed (Table
III). A graph showing the rate of migration of SCC-25 cells
when incubated with EMD, prAMEL or TRAP protein is shown in
Fig. 5.
 | Table III.Effect of EMPs on the rate of
migration of SCC-25 cells. |
Table III.
Effect of EMPs on the rate of
migration of SCC-25 cells.
|
| Cell index value
mean ± standard deviation |
|---|
|
|
|
|---|
| EMP added,
µg/ml | 12-h
incubation | 24-h
incubation | 28-h
incubation |
|---|
| Control | 1.4±2.1 | 1.8±2.0 | 2.6±1.8 |
| EMD, 12.5 | 2.0±1.6 | 2.2±1.8 | 3.2±2.6 |
| EMD, 25 | 3.4±2.6 | 3.7±2.6 | 3.8±1.7 |
| EMD, 50 | 3.7±3.0 | 3.9±3.1 | 3.7±2.7 |
| prAMEL, 12.5 | 1.1±1.4 | 2.0±1.8 | 3.9±2.4 |
| prAMEL, 25 | 1.3±1.8 | 2.1±1.4 | 3.9±2.6 |
| prAMEL, 50 | 0.9±0.9 | 2.0±1.1 | 4.0±2.0 |
| TRAP, 12.5 | 1.7±2.1 | 2.5±2.3 | 3.7±2.0 |
| TRAP, 25 | 2.1±2.0 | 3.3±2.0 | 4.9±0.4 |
| TRAP, 50 | 2.1±2.5 | 2.9±2.1 | 4.0±2.2 |
| P-value | >0.05 | >0.05 | >0.05 |
Discussion
Previous studies on the effects of EMD, conducted on
a variety of research models, have been inconclusive (21,22). A
number of ambiguities have made it hard to compare the results
between studies and have impeded the characterization of the
functions of the different components of EMD. Firstly, previous
in vitro studies have used various cell types (epithelial,
tongue carcinoma, gingival fibroblast, periodontal ligament, bone
marrow-mesenchymal stem cells) obtained from different species
(such as, rat, pig and human) (18,19,21–24).
Secondly, the studies used a number of different EMPs, such as
commercial lyophilized EMD and different fractions isolated from it
(<6 kDa, mainly TRAP; >6 kDa, LRAP, sheathing peptides and
the full-length AMEL) (21), or
numerous recombinants, such as full-length AMEL (22) and chemically synthesized 5.3 kDa TRAP
(23). Furthermore, there are marked
differences in the concentration of EMPs used; from between 10
ng/ml and 100 µg/ml. Finally, different techniques were applied in
order to measure the biological effects of the EMPS. Conventional
cell-based assays may be more prone to artifacts, due to
considerable manipulation of the cell by labeling or
over-expression of target or reporter proteins (25).
Numerous studies concerning the effects of EMD focus
on periodontal tissue, including its stimulation (14,15,24).
These have shown that the effects of EMD are different in
mesenchymal and epithelial cells (14,18,19,24).
Results concerning the influence of EMD on oral epithelial cells
are particularly ambiguous; EMD was determined to have an
anti-proliferative effect on epithelial cells (12,13), but
numerous clinicians have observed accelerated epithelial
soft-tissue healing upon intrasurgical application of EMD (26–29). It
has been suggested that EMD may induce alterations in malignant
mucosal tissue, which implies that patients with pre-malignant or
malignant mucosal lesions should not be treated with EMD (27).
The aim of the present study was to determine the
influence of EMD, AMEL and TRAP on human tongue carcinoma cells
using a cell-based, label-free and real-time platform technology
(xCELLigence). Label-free technologies have the advantage of being
non-invasive. The real-time monitoring of cells provides important
information regarding their biological status, such as cell growth,
arrest and morphological changes. The qualities of this system made
it possible to obtain physiologically relevant results.
The results of the present study indicate that EMD
does not influence the morphology of SCC-25 significantly. Kawase
et al (13) observed that
SCC-25 cell cultures treated with 100 µg/ml EMD for 3 days became
more flattened and had a slightly lower cell density. In the
present study, cells were stimulated with EMD at concentrations of
12.5, 25 and 50 µg/l, which may explain the differences in the
results obtained.
Real-time tests performed using the xCELLigence
system indicate no significant effects of EMD on adhesion,
proliferation and migration of SCC-25 cells. These results
contradict the observations of Kawase et al (13), that EMD (in a dose-dependent manner)
inhibited oral epithelial cell division and concomitantly arrested
cell cycling at the G1 phase, although no apoptosis was observed.
Kawase et al (13) concluded
that EMD acts as a cytostatic, rather than cytotoxic, agent on
epithelial cells. In other studies the same group of researchers
showed that EMD reduced DNA synthesis in a dose-dependent manner
(12,14). Evidence from the literature suggests
that the suppression of epithelial cell growth observed may be
mediated by transforming growth factor β1 (TGF-β1) (14). Porcine TGF-β1 up-regulates p21
(WAF/CIP1) expression and inhibits epithelial proliferation
(14). In addition, TGF-β1
phosphorylates the mitogen-activated protein kinase (MAPK) family,
similar to EMD. Anti-TGF-β antibody completely blocks the
up-regulation of p21 protein and anti-proliferative action by EMD
or TGF-β in epithelial cells (14).
In addition, anti-TGF-β antibody blocks other actions of EMD in
epithelial cells, p38-MAPK and inhibition of DNA synthesis
(14).
Anti-TGF-β antibody blocks TGF-β1- and EMD-induced
SMAD family member 2 (SMAD2) translocation (14). Kawase et al (14) concluded that TGF-β1, as a principal
bioactive factor in EMD, likely inhibits epithelial cell
proliferation by a SMAD2-mediated, p21-dependent mechanism.
Moreover, Kawase et al (12)
showed that 50 µg/ml EMD promoted SCC-25 cell adherence and
stimulated cytoskeletal actin polymerization. However, Laaksonen
et al (30) did not confirm
the inhibitory effects of EMD on tongue squamous cell carcinoma
proliferation, no differences were found between the control and
the EMD-treated (100 and 200 µg/ml) cells after 12, 24, 48, 72 and
96 h of incubation. Furthermore, Gestrelius et al (16) did not observe any statistically
significant changes in rat tongue epithelial cell proliferation
after exposure to 100 µg/ml EMD. In addition, Mirastschijski et
al (17) revealed significant
epithelization after EMD treatment in vivo in rabbits.
Moreover, a previous study indicated that EMD promotes
re-epithelialization and neovascularization in full-thickness
surgical wounds in rat oral mucosa (28). Maymon-Gil et al (29) observed that EMD had no effect on
epithelial gap closure of an oral mucosa surgical wound in
vivo in rats. The differences in the results of these studies
are likely associated with the method of EMD application; directly
on the wound, or underneath the soft tissues.
Previous studies conducted on cervical cancer cells
indicated that EMD has an inhibitory effect on epithelial cells
(13,27). Lyngstadaas et al (15) showed that HeLa cells growing in the
presence of EMD exhibited a highly increased intracellular level of
cyclic adenosine monophosphate compared with controls. EMD
primarily contains glycoproteins, and AMEL to non-AMEL proteins
(such as, ameloblastin and enamelin) at a ratio of ~9:1 (AMEL:rest
of proteins) (31,32). Full-length AMEL induces proliferation
in periodontal cells, such as mesenchymal stem cells (33), cementoblasts (34), periodontal fibroblasts (18,34) and
gingival fibroblasts (18).
The present study observed a dose-dependent
inhibitory effect of porcine recombinant AMEL (21.3 kDa) on SCC-25
cell proliferation. These results are consistent with observations
made by a previous study that indicated that recombinant AMEL
inhibits the growth rate, adhesion and migration of gingival
epithelial cells (18). Li et
al (19) identified that
recombinant 25 kDa porcine AMEL (5, 10 and 20 µg/ml) inhibited
human gingival epithelial cell attachment, migration and growth
rate in a time- and dose-dependent manner. In addition, a previous
study demonstrated that recombinant AMEL inhibits epithelial cell
proliferation in vitro. The results of Kuramitsu-Fujimoto
et al (35) suggest that
ameloblastin is the primary bioactive factor of EMD in regards to
inhibition of epithelial cell proliferation. It has been suggested
that EMPs, such as AMELs and ameloblastin, are required for enamel
biomineralization and have synergistic cellular functions (36).
The present study examined the effects of
recombinant 5.3 kDa TRAP on SCC-25 cells. No significant
differences were found between the control and TRAP-treated (12.5,
25 and 50 µg/ml) cells in terms of adhesion, migration and
proliferation. Villa et al (28) observed increased migration of
epithelial cells following EMD treatment compared with recombinant
TRAP stimulation in palatal wounds in rats. Numerous previous
studies have analyzed the effect of the EMD protein fraction with a
molecular weight of ~5 kDa, which is presumably composed by TRAP
(4,5,37,38).
These studies performed the following TRAP preparation methods:
TRAP isolated from EMD; recombinant peptide TRAP; and synthetic
TRAP, which resulted in different observations concerning their
biological effects (4,5,37,38).
This suggests that the method of TRAP preparation may be an
important factor in influencing its biological activity. Jonke
et al (37) demonstrated that
TRAP isolated from EMD and synthetic TRAP (100 µg/ml) significantly
decreased human umbilical vein endothelial cell proliferation and
viability. No statistically significant decrease in proliferation
of TRAP-treated cells was observed in the present study, although
this was recombinant TRAP, 50 µg/ml was the highest concentration
used and was on different cell line. EMPs are conserved as well as
the TRAP cleavage site in humans and other mammals (1), however, because EMD contains porcine
AMELs, porcine AMEL and porcine TRAP were used in the present study
to minimize any differences. To the best of our knowledge, no
previous studies used a similar research model of TRAP synthesis,
which impeded the verification of results obtained.
In conclusion, the aim of the present study was to
investigate effects of EMD, porcine recombinant AMEL and TRAP on
SCC-25 cells using a real-time cell analysis platform
(xCELLigence). The results demonstrated that EMD and its active
components did not increase the tongue cancer cell viability, and
that porcine recombinant AMEL inhibited epithelial cell
proliferation and migration. To the best of our knowledge, no
previous EMD studies concerning SCC-25 cells were conducted with
the use of real-time monitoring. Thus, differences between the
results of the present study and those obtained by previous studies
are likely due to differences in the measurement technique used and
the structure of applied ligands (AMEL and/or EMD). The amelogenin
construct was coding a protein with a mass of 21.3 kDa; however, a
GST tag was added in order to increase the protein solubility. The
final product used in the present research, comprising of
amelogenin and GST, had a molecular mass of 49 kDa. The xCELLigence
system enables better reproducibility than other instruments, which
is an argument in favor of its use in real-time analysis of SCC-25
cells.
References
|
1
|
Fincham AG, Belcourt AB, Termine JD,
Butler WT and Cothran WC: Amelogenins. Sequence homologies in
enamel-matrix proteins from three mammalian species. Biochem J.
211:149–154. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miron RJ, Caluseru OM, Guillemette V,
Zhang Y, Gemperli AC, Chandad F and Sculean A: Influence of enamel
matrix derivative on cells at different maturation stages of
differentiation. PLoS One. 8:e710082013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miron RJ, Wei L, Yang S, Caluseru OM,
Sculean A and Zhang Y: Effect of enamel matrix derivative on
periodontal wound healing and regeneration in an osteoporotic
model. J Periodontol. 85:1603–1611. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fincham AG, Belcourt AB, Termine JD,
Butler WT and Cothran WC: Dental enamel matrix: Sequences of two
amelogenin polypeptides. Biosci Rep. 1:771–778. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fincham AG, Hu YY, Pavlova Z, Slavkin HC
and Snead ML: Human amelogenins: Sequences of ‘TRAP’ molecules.
Calcif Tissue Int. 45:243–250. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grayson RE, Yamakoshi Y, Wood EJ and Agren
MS: The effect of the amelogenin fraction of enamel matrix proteins
on fibroblast-mediated collagen matrix reorganization.
Biomaterials. 27:2926–2933. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoang AM, Klebe RJ, Steffensen B, Ryu OH,
Simmer JP and Cochran DL: Amelogenin is a cell adhesion protein. J
Dent Res. 81:497–500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsuzawa M, Sheu TJ, Lee YJ, Chen M, Li
TF, Huang CT, Holz JD and Puzas JE: Putative signaling action of
amelogenin utilizes the Wnt/beta-catenin pathway. J Periodontal
Res. 44:289–296. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Z, Fan D, Fan Y, Du C and
Moradian-Oldak J: Enamel proteases reduce amelogenin-apatite
binding. J Dent Res. 87:1133–1137. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan J, Leung W, Moradian-Oldak J,
Zeichner-David M and Fincham AG: Quantitative analysis of
amelogenin solubility. J Dent Res. 77:1388–1396. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamakoshi Y: Porcine Amelogenin:
Alternative splicing, proteolytic processing, protein-protein
interactions, and possible functions. J Oral Biosci. 53:275–283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawase T, Okuda K, Momose M, Kato Y,
Yoshie H and Burns DM: Enamel matrix derivative (EMDOGAIN) rapidly
stimulates phosphorylation of the MAP kinase family and nuclear
accumulation of smad2 in both oral epithelial and fibroblastic
human cells. J Periodontal Res. 36:367–376. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawase T, Okuda K, Yoshie H and Burns DM:
Cytostatic action of enamel matrix derivative (EMDOGAIN) on human
oral squamous cell carcinoma-derived SCC25 epithelial cells. J
Periodontal Res. 35:291–300. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawase T, Okuda K, Yoshie H and Burns DM:
Anti-TGF-beta antibody blocks enamel matrix derivative-induced
upregulation of p21WAF1/cip1 and prevents its inhibition of human
oral epithelial cell proliferation. J Periodontal Res. 37:255–262.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lyngstadaas SP, Lundberg E, Ekdahl H,
Andersson C and Gestrelius S: Autocrine growth factors in human
periodontal ligament cells cultured on enamel matrix derivative. J
Clin Periodontol. 28:181–188. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gestrelius S, Andersson C, Lidström D,
Hammarström L and Somerman M: In vitro studies on periodontal
ligament cells and enamel matrix derivative. J Clin Periodontol.
24:685–692. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mirastschijski U, Konrad D, Lundberg E,
Lyngstadaas SP, Jorgensen LN and Agren MS: Effects of a topical
enamel matrix derivative on skin wound healing. Wound Repair Regen.
12:100–108. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Shu R, Liu D and Jiang S: Different
effects of 25-kDa amelogenin on the proliferation, attachment and
migration of various periodontal cells. Biochem Biophys Res Commun.
394:581–586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li XT, Shu R, Song ZC and Zhou YB: The
effects of recombinant porcine amelogenin on human gingival
epithelial cells. Shanghai Kou Qiang Yi Xue. 21:257–261. 2012.(In
Chinese). PubMed/NCBI
|
|
20
|
Sambrook J and Russel DW: Molecular
cloning a laboratory manual. Cold Spring Harbor Laboratory Press;
New Yourk: Cold Spring Harbor Laboratory Press. 2001
|
|
21
|
Amin HD, Olsen I, Knowles JC and Donos N:
Differential effect of amelogenin peptides on osteogenic
differentiation in vitro: Identification of possible new drugs for
bone repair and regeneration. Tissue Eng Part A. 18:1193–1202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Frasheri I, Ern C, Diegritz C, Hickel R,
Hristov M and Folwaczny M: Full-length amelogenin influences the
differentiation of human dental pulp stem cells. Stem Cell Res
Ther. 7:102016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Amin HD, Olsen I, Knowles J, Dard M and
Donos N: A tyrosine-rich amelogenin peptide promotes
neovasculogenesis in vitro and ex vivo. Acta Biomater.
10:1930–1939. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grandin HM, Gemperli AC and Dard M: Enamel
matrix derivative: A review of cellular effects in vitro and a
model of molecular arrangement and functioning. Tissue Eng Part B
Rev. 18:181–202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xi B, Yu N, Wang X, Xu X and Abassi YA:
The application of cell-based label-free technology in drug
discovery. Biotechnol J. 3:484–495. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanz M, Tonetti MS, Zabalegui I, Sicilia
A, Blanco J, Rebelo H, Rasperini G, Merli M, Cortellini P and Suvan
JE: Treatment of intrabony defects with enamel matrix proteins or
barrier membranes: Results from a multicenter practice-based
clinical trial. J Periodontol. 75:726–733. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laaksonen M, Sorsa T and Salo T: Emdogain
in carcinogenesis: A systematic review of in vitro studies. J Oral
Sci. 52:1–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Villa O, Wohlfahrt JC, Mdla I, Petzold C,
Reseland JE, Snead ML and Lyngstadaas SP: Proline-rich peptide
mimics effects of enamel matrix derivative on rat oral mucosa
incisional wound healing. J Periodontol. 86:1386–1395. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maymon-Gil T, Weinberg E, Nemcovsky C and
Weinreb M: Enamel matrix derivative promotes healing of a surgical
wound in the rat oral mucosa. J Periodontol. 87:601–609. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Laaksonen M, Suojanen J, Nurmenniemi S,
Läärä E, Sorsa T and Salo T: The enamel matrix derivative
(Emdogain) enhances human tongue carcinoma cells gelatinase
production, migration and metastasis formation. Oral Oncol.
44:733–742. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schwartz Z, Carnes DL Jr, Pulliam R,
Lohmann CH, Sylvia VL, Liu Y, Dean DD, Cochran DL and Boyan BD:
Porcine fetal enamel matrix derivative stimulates proliferation but
not differentiation of pre-osteoblastic 2T9 cells, inhibits
proliferation and stimulates differentiation of osteoblast-like
MG63 cells, and increases proliferation and differentiation of
normal human osteoblast NHOst cells. J Periodontol. 71:1287–1296.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shimizu-Ishiura M, Tanaka S, Lee WS,
Debari K and Sasaki T: Effects of enamel matrix derivative to
titanium implantation in rat femurs. J Biomed Mater Res.
60:269–276. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang YC, Tanimoto K, Tanne Y, Kamiya T,
Kunimatsu R, Michida M, Yoshioka M, Yoshimi Y, Kato Y and Tanne K:
Effects of human full-length amelogenin on the proliferation of
human mesenchymal stem cells derived from bone marrow. Cell Tissue
Res. 342:205–212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hatakeyama J, Philp D, Hatakeyama Y,
Haruyama N, Shum L, Aragon MA, Yuan Z, Gibson CW, Sreenath T,
Kleinman HK and Kulkarni AB: Amelogenin-mediated regulation of
osteoclastogenesis, and periodontal cell proliferation and
migration. J Dent Res. 85:144–149. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuramitsu-Fujimoto S, Ariyoshi W, Saito N,
Okinaga T, Kamo M, Ishisaki A, Takata T, Yamaguchi K and Nishihara
T: Novel biological activity of ameloblastin in enamel matrix
derivative. J Appl Oral Sci. 23:49–55. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hatakeyama J, Fukumoto S, Nakamura T,
Haruyama N, Suzuki S, Hatakeyama Y, Shum L, Gibson CW, Yamada Y and
Kulkarni AB: Synergistic roles of amelogenin and ameloblastin. J
Dent Res. 88:318–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jonke E, Gemperli AC, Zhang T, Özdemir B,
Dard M, Rausch-Fan X and Andrukhov O: Effect of tyrosine-rich
amelogenin peptide on behavior and differentiation of endothelial
cells. Clin Oral Investig. Feb 12–2016.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ravindranath RM, Tam WY, Nguyen P and
Fincham AG: The enamel protein amelogenin binds to the
N-acetyl-D-glucosamine-mimicking peptide motif of cytokeratins. J
Biol Chem. 275:39654–39661. 2000. View Article : Google Scholar : PubMed/NCBI
|