Introduction
As a highly specialized cell in mature animals, the
vascular smooth muscle cell (VSMC) has a principal function of
contraction; however, production of matrix components of the blood
vessel wall and proliferation becomes the primary function of VSMCs
during vasculogenesis (1). Abnormal
contraction of SMC is a major incentive of vasospasm of the
cerebral and coronary arteries, as well as hypertension (2). VSMC accumulation and hypertrophy are
common in vascular disorders, such as atherosclerosis,
hypertension, restenosis (3,4) and inflammation, which can be induced by
hypoxia and has crucial roles in the development of these diseases
(5,6). Thus, there is an urgency to elucidate
the effect of inflammation on the neurotransmission of VSMCs.
Several pharmacological agents are capable of
inducing inflammation. In human aortic smooth muscle cells,
lipopolysaccharide (LPS) promotes the production of nitric oxide
(NO) and Toll-like receptor 4 (TLR4) expression, inducing
inflammatory responses (4). A
previous study demonstrated that propranolol has a negative
chronotropic effect on the expression levels of pro-inflammatory
cytokines after myocardial infarction (MI) in rats (7). In rat aorta, β-adrenoceptors were
overstimulated by the agonist, isoproterenol, which resulted in an
increase of vascular inflammatory mediators, such as interleukin
(IL)-1β, IL-6 and nuclear factor κB (NF-κB) (8). At a concentration of 20 µg/ml, the
non-selective β-adrenergic receptor agonist, propranolol, was
revealed to suppress cell growth of infantile hemangioma
endothelial cells (IHECs) in vitro once the proliferation
stage of IHECs has been affected for between 72 and 96 h, whereas
isoproterenol yielded the opposite results (9).
Previous reports have been conducted to survey the
effect of inflammation on VSMC. As an adipocytokine, extracellular
pre-B cell colony-enhancing factor/nicotinamide
phosphoribosyltransferase/visfatin (ePBEF/NAMPT/visfatin)
functions as a direct contributor to vascular inflammation via its
NAMPT activity (10). Through
regulating vascular cell activation and inflammatory cell
recruitment, the adhesion protein, cluster differentiation (CD) 44,
has an important role in the development of atherosclerotic
diseases (11). By inhibiting the
activation of hypoxia-inducible factor-1α, transcription factors,
NF-κB and activator protein-1 (AP-1), and
3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase
inhibitors have anti-proliferative and anti-inflammatory effects on
human endothelial and vascular smooth muscle cells; thus, statins
can be used to treat atherosclerosis (12).
In the present study, next generation sequencing was
conducted to obtain sequence data. Differentially expressed genes
(DEGs) between the LPS treatment group and the control group were
screened and their functions were predicted by enrichment analyses.
Moreover, a protein-protein interaction (PPI) network was
constructed to investigate the interaction relationships between
these DEGs.
Materials and methods
Cell cultivation
Rat VSMC cell line A7r5, which was purchased from
Shanghai enzyme research Biotechnology Co., Ltd. (Shanghai, China),
was cultivated in Dulbecco's modified Eagle medium (DMEM) (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and maintained at
37°C using an incubator (Thermo Fisher Scientific, Inc). DMEM was
discarded, and A7r5 cells were digested with pancreatin (Gibco;
Thermo Fisher Scientific, Inc) for 5 min. Subsequently, 6-fold DMEM
was added to terminate digestion and cells were centrifuged at room
temperature with 157 × g for 5 min, and the supernatant and
resuspension was discarded. A7r5 cells were cultured in new culture
flasks in a 5% CO2 incubator at 37°C (Thermo Fisher
Scientific, Inc.). Using the frozen stock solution made of 10%
dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) and 90% fetal bovine serum (FBS) (Gibco; Thermo
Fisher Scientific, Inc.), A7r5 cells were resuspended at a density
of 5×106 cells/ml. Following stewing at 4°C for 10 min,
cells were cryopreserved at −20°C for 2 h and stored at −80°C
overnight.
Calcium detection
A7r5 cells were inoculated into confocal plates
(Wohong Biotechnology Co., Ltd., Shanghai, China) (2×104
cells/plate) and cultivated overnight. Cells were washed with
phosphate-buffered saline (PBS) twice with 100 µl PBS containing 5
µmol/l Fluo-4/AM and lucifugally incubated at 37°C in a 5%
CO2 incubator for 45 min. Subsequently, cells were
washed with PBS twice again and induced by LPS (100 µg/ml) for 30
min. In the control groups, PBS was used instead of LPS. Under
confocal laser scanning microscope, A7r5 cells were observed and
images were captured before and after treatment with isoprenaline
(10 µmol/l, Melone pharmaceutical, Co., Ltd., Dalian, China) or
propranolol (10 µmol/l, Melone pharmaceutical, Co., Ltd.).
Cell hypoxia treatment
Cells were inoculated with 0.5% FBS in confocal
plates (4×105 cells/plate). After being cultivated for
24 h, the 0.5% FBS medium was replaced with 10% FBS and cells were
treated with cobalt dichloride (200 µmol/l) for 24 h. Subsequently,
cells were induced by LPS (100 µg/ml) for 30 min; however, cells in
the control group received PBS. After cell digestion by pancreatin
(Gibco; Thermo Fisher Scientific, Inc.) for 5 min, the mixture was
centrifuged at room temperature at 157 × g for 5 min. Then,
cells were washed three times with DMEM at room temperature and
further centrifuged at room temperature at 157 × g for 5
min. The supernatant was discarded and A7r5 cells were preserved in
TRIzol (Invitrogen; Thermo fisher Scientific, Inc.) at −80°C. Each
assay was performed in triplicate.
RNA isolation and RNA-sequence library
construction
Total RNA of the LPS treatment group and the control
group were extracted using a TRIzol total RNA extraction kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The integrity and purity of total RNA
was detected using 2% agarose gel electrophoresis and a
spectrophotometer (Merinton Instrument, Ltd., Beijing, China),
respectively. The RNA-sequence library was constructed using
methods described in a previous study (13). Subsequently, DNA cluster
amplification was performed and high-throughput sequencing was
conducted for the library, using Illumina Hiseq 2000 100PE
(Illumina Inc., San Diego, CA, USA). After the raw data was
obtained, sequences containing an adaptor, >50% low quality
bases and >3% unknown bases were filtered out.
DEGs screening
Following filtering, the sequences were mapped to
the rat genome (rn5), using bowtie1 in TopHat software (version
2.1.0, accessible at http://ccb.jhu.edu/software/tophat/index.shtml)
(14). The maximum read mismatch
number was set at 2, and the parameter ‘max-multihits’ was set at 1
(15). And the other parameters were
set to defaults. Combining with annotation information of rn5 in
ensemble, expression of each sample was annotated using Cufflinks
software (version 2.2.1, accessible at http://cole-trapnell-lab.github.io/cufflinks/)
(16) under the default parameter
values. The NOISeq package (version 2.18.0, accessible at
http://www.bioconductor.org/packages/release/bioc/html/NOISeq.html)
(17) in R was used to screen the
DEGs between the LPS treatment group and the control group. The
probability of a gene being DEGs (q) was set to 0.99.
Functional and pathway enrichment
analysis
Gene Ontology (GO), which consists of three
categories, including biological process (BP), molecular function
(MF) and cellular component (CC), is used to generate the
vocabulary that can be applied to all eukaryotes (18). As a database, Kyoto Encyclopedia of
Genes and Genomes (KEGG) includes information of known genes and
their biochemical functionalities (19). Using the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) online tool
(20), GO and KEGG pathway
enrichment analyses were conducted for the upregulated and
downregulated genes between the LPS treatment and control groups,
respectively. P<0.01 was used as the cut-off criterion.
PPI network construction
Using STRING online software (version 10.0,
accessible at http://www.string-db.org/) (21), interaction relationships of the
proteins encoded by the DEGs were searched, and the required
confidence (combined score) >0.1 was used as the cut-off
criterion. Subsequently, PPI network was visualized using Cytoscape
(version 3.2.0, accessible at http://www.cytoscape.org/) (22). Proteins in the network were named as
nodes and the degree of a node was equal to the number of nodes
interacted with it. Moreover, the nodes with degrees higher than 20
were defined as hub nodes.
Results
Calcium detection
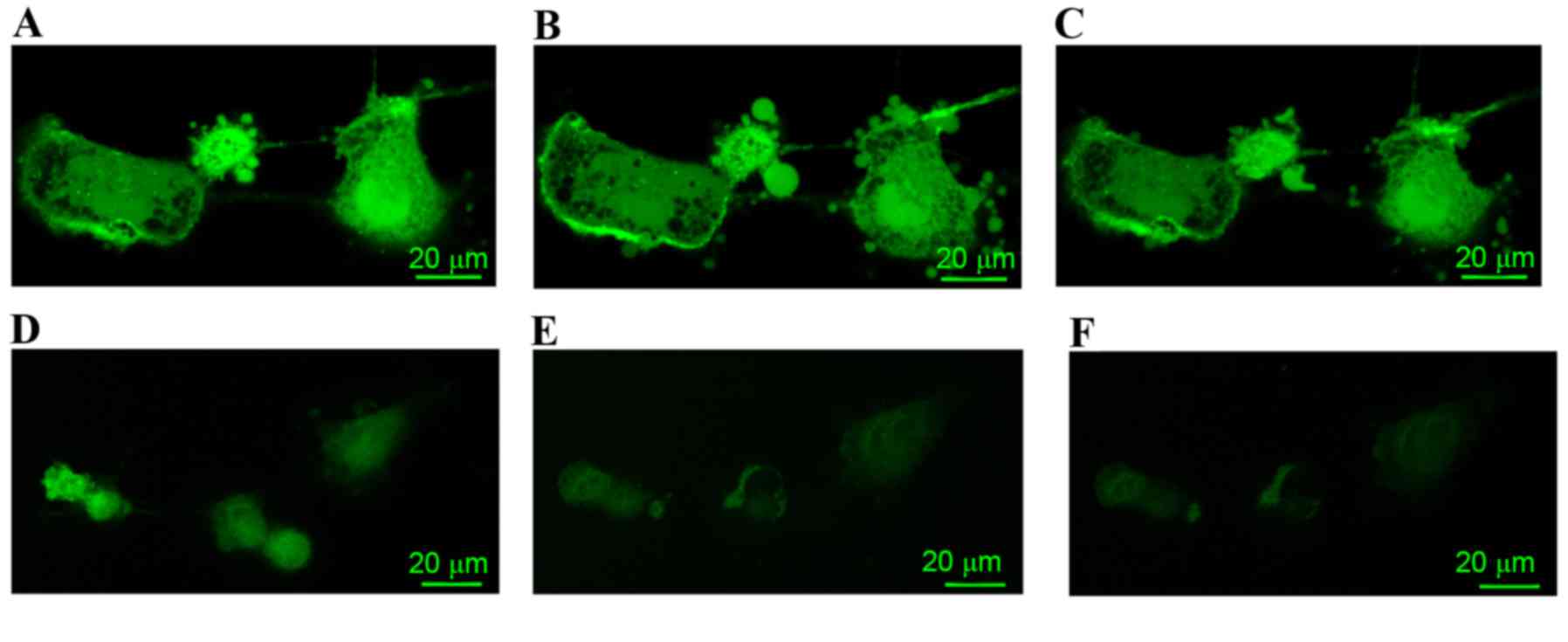
In the PBS control group, A7r5 cells prior to
treatment with isoprenaline or propranolol are indicated in
Fig. 1A. In A7r5 cells treated with
isoprenaline, the concentration of calcium decreased (Fig. 1B); however, the level of calcium in
propranolol-treated A7r5 cells increased (Fig. 1C).
The LPS-treated group (Fig. 1D-F) exhibited a reduced calcium
signal prior to treatment with isoprenaline or propranolol when
compared with the PBS control group (Fig. 1D). In the A7r5 cells that were
treated with isoprenaline, the concentration of calcium decreased
(Fig. 1E). However, the level of
calcium in the A7r5 cells treated with propranolol had no change
when compared with the LPS group cells treated with isoprenaline
(Fig. 1F).
DEGs analysis
Compared with the control PBS group, a total of
2,038 DEGs, including 1,094 upregulated and 944 downregulated genes
were identified in the LPS-treated group. These findings indicated
that the number of upregulated genes was markedly higher in
comparison to the number of downregulated genes.
Functional and pathway enrichment
analysis
The enriched GO functions for upregulated genes are
listed in Table I. The enriched
functions in the BP category included chemical homeostasis
(P=4.81E-04), mitogen-activated protein kinase kinase kinase
cascade (P=7.85E-04) and nucleoside monophosphate metabolic process
(P=8.39E-04). The enriched functions in the CC category included
condensed chromosome kinetochore (P=1.33E-04), chromosome,
centromeric region (P=1.63E-04) and plasma membrane (P=2.96E-04).
Enriched functions in the MF category included hydrolase activity,
acting on carbon-nitrogen (but not peptide) bonds in linear
amidines (P=1.40E-04), quaternary ammonium group binding
(P=8.28E-04) and phosphatidylcholine binding (P=9.81E-04).
 | Table I.Enriched GO functions and Kyoto
Encyclopedia of Genes and Genomes pathways for upregulated genes in
the lipopolysaccharide treatment group compared with the phosphate
buffered saline control group. |
Table I.
Enriched GO functions and Kyoto
Encyclopedia of Genes and Genomes pathways for upregulated genes in
the lipopolysaccharide treatment group compared with the phosphate
buffered saline control group.
| Category | Term | Description | Gene no. | Gene symbol | P-value |
|---|
| BP | GO:0048878 | Chemical
homeostasis | 43 | C7, Uts2, Fgf7,
Hnf1a, Slc9a4, Grik2, Pde3b, Oxtr, Cacnb4, Aqp2, Best2, Apoa2,
Apoe, Slc1a6, Apoa5, Galr2, Il1b, Eif2b2, Guca2b, Avp, Gip, Fech,
Epas1, Cckbr, Mal, Pfkm, Sod2, Slc34a2, Alox15, P2rx7, Abcg5, Cntf,
Gck, Avpr1b, Pgm1, Nab1, F2, Fabp4, Stc1, Uts2r, Chrnb1, Cp,
Chrng | 4.81E-04 |
| BP | GO:0000165 | MAPKKK cascade | 18 | Ret, Cckbr,
Tgfbr1, Muc20, Oxtr, Itpkb, Smad1, Mapk10, Avpi1, P2rx7, Wnt7b,
Myd88, Mdfic, Map3k2, Lax1, Map3k1, Il1b, Spred1 | 7.85E-04 |
| BP | GO:0009123 | Nucleoside
monophosphate metabolic process | 12 | Gucy2g, Adss,
Tyms, Adcy9, Entpd8, Nppc, Pde3b, Cacnb4, Guca2b, Gucy2c, Ppat,
Ampd1 | 8.39E-04 |
| BP | GO:0009124 | Nucleoside
monophosphate biosynthetic process | 10 | Gucy2g, Adss,
Tyms, Adcy9, Entpd8, Nppc, Guca2b, Gucy2c, Ppat, Ampd1 | 8.54E-04 |
| BP | GO:0042592 | Homeostatic
process | 56 | Uts2, Rab9a,
Hnf1a, Fgf7, Slc9a4, Grik2, Pde3b, Aqp2, Lilrb3l, Apoa2, Apoe,
Slc1a6, Galr2, Apoa5, Il1b, Glp2r, Eif2b2, Guca2b, Avp, Fech, Gip,
Cckbr, Lyn, Pfkm, Mecom, Slc34a2, Bbs1, Alox15, Rps17, Pgm1, F2,
Nab1, Stc1, C7, Blm, Vpreb2, Oxtr, Cacnb4, Best2, Sh2b2, Tinf2,
Epas1, Loc690948, Mal, Smad1, Sod2, P2rx7, Abcg5, Cntf, Gck,
Avpr1b, Fabp4, Uts2r, Cp, Chrnb1, Sash3, Chrng | 8.82E-04 |
| CC | GO:0000777 | Condensed
chromosome kinetochore |
9 | Spc25, Cenpa,
Bub1, Nuf2, Cenpf, Ska2, Pmf1, Mis12, Zw10 | 1.33E-04 |
| CC | GO:0000775 |
Chromosome/centromeric region | 14 | Nuf2, Cenpf,
Pmf1, Cenpi, Mis12, Spc25, Cdca8, Mad2l1, Cenpa, Ppp2cb, Bub1,
Ska2, Tigd5, Zw10 | 1.63E-04 |
| CC | GO:0005886 | Plasma
membrane | 159 | Rab9a, Gabrb2,
Grik2, Slc9a4, Syt9, Cd52, Cspg5, Aqp3, Aqp2, Sctr, Cdh20, Apoe,
Galr2, Glp2r, Ddah1, Chrna2, F10, Scn2b, Ncf4, Ptprr, Actn2, Myh7,
F7, Pdyn, Pkd2l1, Slc34a2, Gabrr3, Egflam, Rasgrf2, Htr6, F2,
Cd300lf, Slc38a1, Car4, Sh3gl2, Slc38a4, Cav2, Lppr4, Gpr149,
Aldob, Olr1469, Mme, Nostrin, Cacnb4, Ubac1, Itgam, Oscp1, Hcrtr1,
Gorasp1, Folr1, Entpd8, Zap70, Gucy2g, Pard6a, Vav3, Pth2r, Acy3,
Tgfbr1, Slc6a13, Slc6a19, Kctd7, Kctd6, P2rx7, Abcg5, Pkp3, Slc7a1,
Avpr1b, Cacna1h, Rheb, S100g, Uts2r, Chrnb1, Abl1, Pdzd2, Slc5a11,
Mtnr1a, Chrng, Gnaz, Rt1-m10-1, Cldn6, Susd2, Gja1, Slc23a1, Kcnq4,
Cxcr6, Mc5r, Taar1, Tpo, Tie1, Slc43a1, Scn10a, Cd200r1, Ptger2,
Rxfp1, Lyn, Cckbr, Noxo1, Zp3, Cmklr1, Slc7a10, Cftr, Pfkm, Ncr1,
Proc, Ncr3, Stom, Ambp, Slc26a3, Alox15, Taf12, Lax1, Nab1, Grm6,
Trem1, Ngfr, Otoa, Faim2, Ptafr, Slc27a5, Rasd2, Rab3a, C7, Vpreb2,
Oxtr, Rt1-m1-4, Gpr1, Cdh5, Gpr4, Igsf11, Pex19, Tmed1, Syn2, Krt1,
Col6a2, Cd22, Tgm3, Sh2b2, Npffr1, Htr3a, Ehd3, Kl, Muc20, Gjb3,
Mapk10, Gjb6, Cish, Agtrap, Iyd, Wnt7b, Ppp1r9a, P2ry10, Cd19,
Slc6a7, Golph3, Gng10, Zmynd19, Cp, Perp, Chrna10 | 2.96E-04 |
| CC | GO:0000779 | Condensed
chromosome/ centromeric region |
9 | Spc25, Cenpa,
Bub1, Nuf2, Cenpf, Ska2, Pmf1, Mis12, Zw10 | 5.98E-04 |
| CC | GO:0000776 | Kinetochore | 10 | Spc25, Mad2l1,
Cenpa, Bub1, Nuf2, Cenpf, Ska2, Pmf1, Mis12, Zw10 | 6.78E-04 |
| MF | GO:0016813 | Hydrolase activity,
acting on carbon-nitrogen (but not peptide) bonds, in linear
amidines |
6 | Arg1, Padi4,
Agmat, Ddah1, Padi1, Allc | 1.40E-04 |
| MF | GO:0050997 | Quaternary ammonium
group binding |
5 | Apoa2, Apoa5,
Aldob, Pctp, Acpp | 8.28E-04 |
| MF | GO:0031210 | Phosphatidylcholine
binding |
4 | Apoa2, Apoa5,
Aldob, Pctp | 9.81E-04 |
| MF | GO:0015171 | Amino acid
transmembrane transporter activity | 10 | Slc38a4,
Slc7a15, Slc6a7, Slc7a1, Slc1a6, Slc6a13, Slc7a10, Slc38a1,
Slc43a1, Slc6a19 | 1.69E-03 |
| MF | GO:0005275 | Amine transmembrane
transporter activity | 11 | Slc38a4,
Slc7a15, Slc6a7, Slc7a1, Slc1a6, Slc6a13, Slc22a4, Slc7a10,
Slc38a1, Slc43a1, Slc6a19 | 2.19E-03 |
The enriched KEGG pathways for upregulated genes are
also listed in Table I, including
purine metabolism (P=3.13E-04), neuroactive ligand-receptor
interaction (P=0.003996) and pyrimidine metabolism
(P=0.007245).
The enriched GO functions for downregulated genes
are presented Table II. The
enriched functions in the BP category included oxidation reduction
(P=1.19E-05), cofactor metabolic process (P=0.001298) and
erythrocyte homeostasis (P=0.0013). Enriched functions in the CC
category included mitochondrion (P=6.87E-07), organelle membrane
(P=6.03E-06) and mitochondrial part (P=1.67E-05). The enriched
functions in the MF category included nicotinamide adenine
dinucleotide or nicotinamide adenine dinucleotide hydride binding
(P=3.63E-04), protein homodimerization activity (P=4.08E-04) and
iron ion binding (P=4.97E-04).
 | Table II.Enriched GO functions and Kyoto
Encyclopedia of Genes and Genomes pathways for the down-regulated
genes in the lipopolysaccharide treatment group compared with the
phosphate-buffered saline control group. |
Table II.
Enriched GO functions and Kyoto
Encyclopedia of Genes and Genomes pathways for the down-regulated
genes in the lipopolysaccharide treatment group compared with the
phosphate-buffered saline control group.
| Category | Term | Description | Gene number | Gene symbol | P-value |
|---|
| BP | GO:0055114 | Oxidation
reduction | 51 | Uqcrc2, Acox2,
Ldha, Kcnab1, Prdx1, Bbox1, Hibadh, Fdft1, Ero1lb, Ndufs4, Hmox1,
Loc688320, Alox12b, Cat, Srd5a2, Gfod2, Hpd, Pcyox1l, Cyp2e1,
Grhpr, Morc1, Dhrs4, Aldh9a1, Mdh1, Me1, Bcmo1, Tyrp1, Adhfe1,
Adh5, Loc685351, Gclm, Aldh3a1, Hsd17b6, Hsd17b4, Rgd1562758,
Hsd17b7, Fgfbp3, Ndufa9, Fads1, Maoa, Scd, Idh3b, Cyp4v3, Ido1,
Idh3a, Adi1, Cyp17a1, Lepre1, Cyp4f18, Sdhd, Ndufv2 | 1.19E-05 |
| BP | GO:0051186 | Cofactor metabolic
process | 20 | Ldha, Aco1,
Ireb2, Idh3b, Gstt1, Ido1, Gclm, Hibadh, Idh3a, Mthfd1l, Gstm1,
Hagh, Mthfs, Acss1, Tpi1, Hmox1, Sdhd, Qprt, Urod, Mdh1 | 1.30E-03 |
| BP | GO:0034101 | Erythrocyte
homeostasis |
9 | Hmox1, Ireb2,
Dyrk3, Bcl6, Tcea1, Rb1, Klf1, Prdx1, Mb | 1.30E-03 |
| BP | GO:0006732 | Coenzyme metabolic
process | 17 | Ldha, Aco1,
Idh3b, Gstt1, Ido1, Gclm, Hibadh, Idh3a, Mthfd1l, Gstm1, Hagh,
Mthfs, Acss1, Tpi1, Sdhd, Qprt, Mdh1 | 1.63E-03 |
| BP | GO:0046496 | Nicotinamide
nucleotide metabolic process |
8 | Tpi1, Ldha,
Idh3b, Qprt, Ido1, Hibadh, Idh3a, Mdh1 | 1.76E-03 |
| CC | GO:0005739 | Mitochondrion | 94 | Mrps36, Uqcrc2,
Atp5e, Ldha, Tspo, Cmc1, Pdp2, Timm17a, Rgd1566320, Fam110b, Lemd3,
Bnip3, Mipep, Cox5a, Ndufaf1, Cox5b, Prdx1, Hibadh, Bbox1, Mthfd1l,
Acss1, Ndufs4, Cisd1, Ctu1, Slc25a24, Slc25a23, Slc25a29, Atp5o,
Cat, Gng5, Wwox, Mrpl34, Rgd1309676, Acaa2, Rpusd4, Slc25a4, Aco1,
Cox4i2, Lypla1, Mrps7, Cyp2e1, Mcart1, Ndufa12, Clpx, Hagh,
Tmem186, Dhrs4, Pebp1, Aldh9a1, Gatc, Mdh1, Tufm, Me1, Tshz3,
Mrps16, Ndufb5, Adhfe1, Mtx2, Ndufb9, Adh5, Myg1, Fam136a, Afap1l1,
Ccdc58, Mthfs, Rgd1303003, Nudt9, Hk3, Rnaset2, Oxct1, Hspe1,
Hsd17b4, Pptc7, Ndufa9, Ndufa6, Maoa, Phb, Ireb2, Gars, Idh3b,
Ndfip2, Vdac2, Idh3a, Atad1, Armc1, Mrpl22, Cyp17a1, Chchd10, Ucp3,
Dusp26, Ucp2, Ndufv2, Sdhd, Comtd1 | 6.87E-07 |
| CC | GO:0031090 | Organelle
membrane | 73 | Uqcrc2, Atp5e,
Clstn3, Timm17a, Lemd3, Tlr3, Bnip3, Anpep, Cd1d1, Cox5a,
Cox5b, Spink5, Fdft1, Ero1lb, Ndufs4, Cisd1, Slc2a4,
Map1lc3a, Slc25a24, Slc25a23, Slc25a29, Atp5o, Cat, Hpd,
Scamp1, Acaa2, Slc25a4, Cox4i2, Cacng4, Cyp2e1, Mcart1, Clpx,
Sacm1l, Rnf180, Zfyve28, Pebp1, Trappc3, Tufm, Ndufb5, Stx8,
Tyrp1, Mtx2, Ndufb9, Drd4, Slc35a5, Rer1, Atp6v1g2,
Loc685351, Atp6v1g1, Afap1l1, Slc11a1, Serinc1, Gp1ba,
Hspa5, Hsd17b7, Soat2, Vps18, Ndufa9, Ndufa6, Phb, Maoa, Scd,
Fig4, Vdac2, Gjb2, Coro1a,
Cyp17a1, Ucp3, Cyp4f18, Ucp2, Faah, Ndufv2, Sdhd | 6.03E-06 |
| CC | GO:0044429 | Mitochondrial
part | 46 | Tufm, Mrps36,
Uqcrc2, Atp5e, Mrps16, Ndufb5, Pdp2, Mtx2, Ndufb9, Timm17a, Bnip3,
Afap1l1, Mipep, Cox5a, Prdx1, Cox5b, Acss1, Cisd1, Ndufs4,
Slc25a24, Oxct1, Slc25a23, Slc25a29, Atp5o, Hspe1, Cat, Mrpl34,
Acaa2, Slc25a4, Ndufa9, Ndufa6, Phb, Maoa, Cox4i2, Gars, Idh3b,
Mcart1, Vdac2, Idh3a, Clpx, Hagh, Ucp3, Ucp2, Ndufv2, Sdhd,
Pebp1 | 1.67E-05 |
| CC | GO:0005625 | Soluble
fraction | 31 | Me1, Ldha, Mvd,
Adh5, Asns, Anpep, Gstm5, Gclm, Ctsl1, Camkk1, Tpi1, Slc2a4, Hdc,
Eno2, Aspg, Cpa1, Rgd1562758, Eif2b3, Actc1, Aco1, Maoa, Pde3a,
Ido1, Fuca1, Pitpnm2, Blmh, Prkar1b, Pebp1, Sytl1, Cryba4,
Mdh1 | 3.48E-05 |
| CC | GO:0005743 | Mitochondrial inner
membrane | 27 | Uqcrc2, Tufm,
Atp5e, Ndufb5, Timm17a, Ndufb9, Afap1l1, Cox5a, Cox5b, Ndufs4,
Slc25a24, Slc25a23, Slc25a29, Atp5o, Acaa2, Slc25a4, Ndufa9, Phb,
Ndufa6, Cox4i2, Vdac2, Mcart1, Clpx, Ucp3, Ucp2, Sdhd,
Ndufv2 | 4.90E-04 |
| MF | GO:0051287 | NAD or NADH
binding | 13 | Me1, Ldha, Adh5,
Idh3b, Grhpr, Hibadh, Idh3a, Cryl1, Ndufv2, Parp1, Rgd1562758,
Aldh9a1, Mdh1 | 3.63E-04 |
| MF | GO:0042803 | Protein
Homodimerization | 28 | Tyrp1, Mvd,
Adh5, Bnip3, Asns, Pdx1, Sdcbp2, Prdx1, Mthfd1l, Tgfb1, Gstm1,
activity Slc11a1, Cryl1, Cep57, Hdc, Gtf2a2, Eno2, Cda, Cat,
Hsp90aa1, Cr2, Trpm8, Tbx1, Coro1a, Faah, Qprt, Add2,
Aldh9a1 | 4.08E-04 |
| MF | GO:0005506 | Iron ion
binding | 26 | Bcmo1, Acp5,
Mipep, Cox5a, Prdx1, Bbox1, Slc11a1, Cisd1, Hmox1, Alox12b,
Cat,Nenf, Hpd, Mb, Aco1, Scd, Fads1, Ireb2, Cyp4v3, Ido1, Cyp2e1,
Cyp17a1, Lepre1, Cyp4f18, Sdhd, Ndufv2 | 4.97E-04 |
| MF | GO:0046914 | Transition metal
ion binding | 106 | Gda, Pdp2,
Apobec4, Lemd3, Mipep, Dnase1l2, Cox5a, Cox5b, Bbox1, Zfp786,
Ighmbp2, Cisd1, Rgd1309534, Cpa1, Mb, Nudt18, Zhx1, Bsn, Spire2,
Cyp2e1, Clpx, Tesk1, Pias1, Adamts4, Me1, Acp5, Loc680200, Slc11a1,
Rnf166, Zdhhc9, Fbxo43, Cda, Rnf168, Osgep, Scd, Ireb2, Idh3b,
Csrp2, Idh3a, Rnf8, Cyp17a1, Lepre1, Nr1i2, Rnf208, Ndufv2, Cpb1,
Parp1, Abo, Klf1, Uqcrc2, Zcchc24, Zfp438, Rnf185, Rnf187, Anpep,
Cbfa2t3, Prdx1, Hmox1, Setmar, Dpep3, Alox12b, Cat, Hpd, Zfp365,
Zdhhc4, Aco1, Prkci, Prkch, Topors, Morc1, Tut1, Hagh, Rnf180,
Acvr2b, Myrip, Ppm1e, Zfyve28, Adam17, Vps29, Bcmo1, Tyrp1,
Trim47l, Adh5, Rpl37, Cpn1, Tcea3, Nudt9, Tcea1, Bcl6, Nenf,
Galnt13, Dtx4, Zbtb7c, Rbm20, Fads1, Cyp4v3, Ido1, Isl1, Adi1,
Rnf112, Cyp4f18, Rbak, Sdhd, Mep1a, Ace2, Chn2 | 8.40E-04 |
| MF | GO:0050662 | Coenzyme
binding | 21 | Me1, Acox2,
Soat2, Ldha, Ndufa9, Maoa, Adh5, Idh3b, Loc685351, Grhpr, Cyp2e1,
Hibadh, Idh3a, Cryl1, Ero1lb, Ndufv2, Cat, Parp1, Rgd1562758,
Aldh9a1, Mdh1 | 1.27E-03 |
The enriched KEGG pathways for downregulated genes
are also listed in Table II,
including oxidative phosphorylation (P=2.49E-04), Alzheimer's
disease (P=0.00105), Parkinson's disease (P=0.00299), Huntington's
disease (P=0.00397) and proteasome (P=0.008983). Furthermore, NADH
dehydrogenase ubiquinone flavoprotein 2 (NDUFV2) was
revealed to be involved in several neurological diseases, including
oxidative phosphorylation, Alzheimer's disease, Parkinson's disease
and Huntington's disease.
PPI network analysis
Pyrimidine metabolism, oxidative phosphorylation,
Alzheimer's disease and Parkinson's disease were all metabolic
pathways associated with neurological diseases. DEGs enriched in
these pathways and adrenergic receptor genes α1d (ADRA1D)
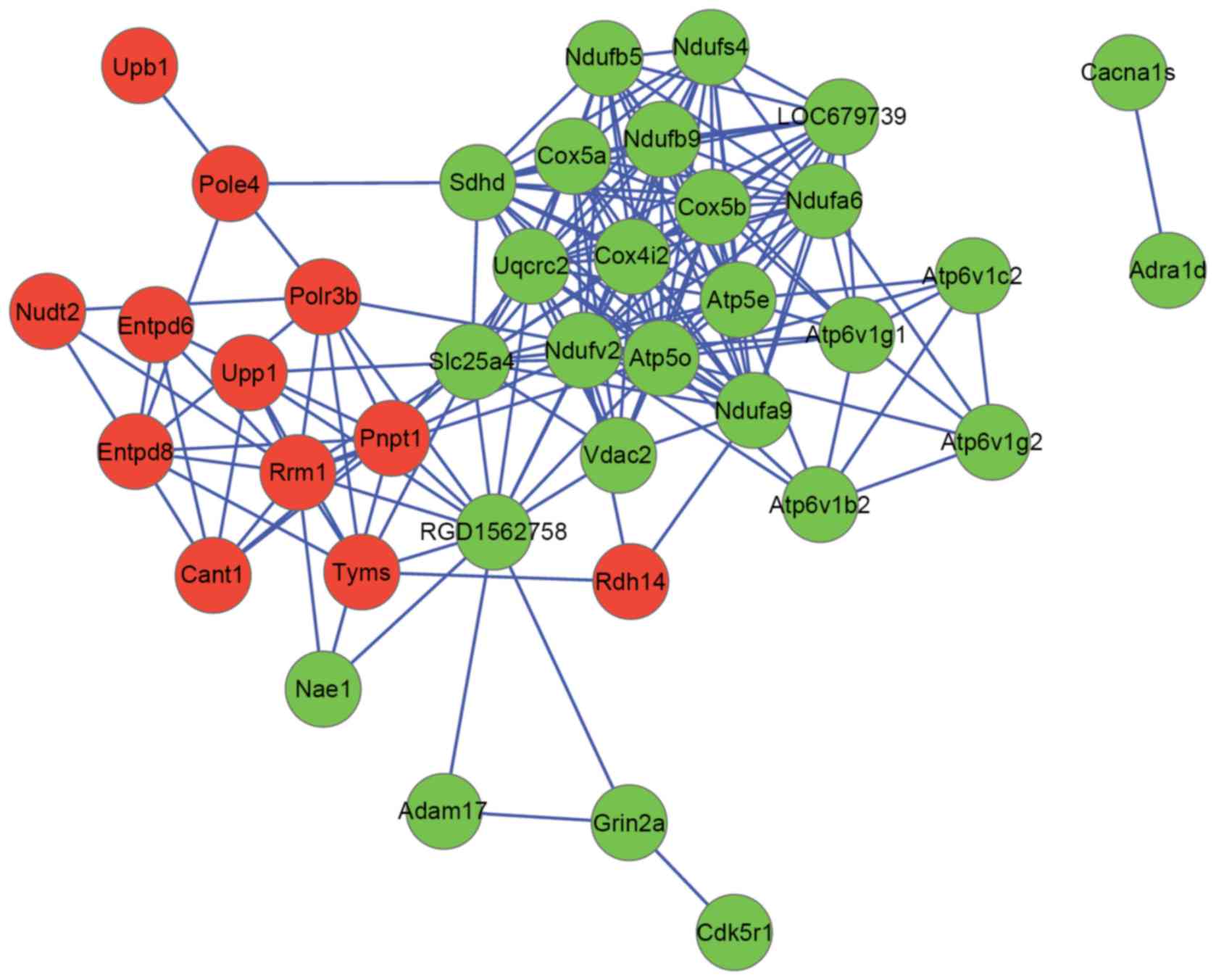
were used to construct a PPI network. The PPI network consisted of
39 nodes and 183 interactions (Fig.
2). ATP synthase, mitochondrial F1 complex, O subunit (ATP5O;
degree, 22) and NDUFV2 (degree, 20) were hub nodes in the
PPI network.
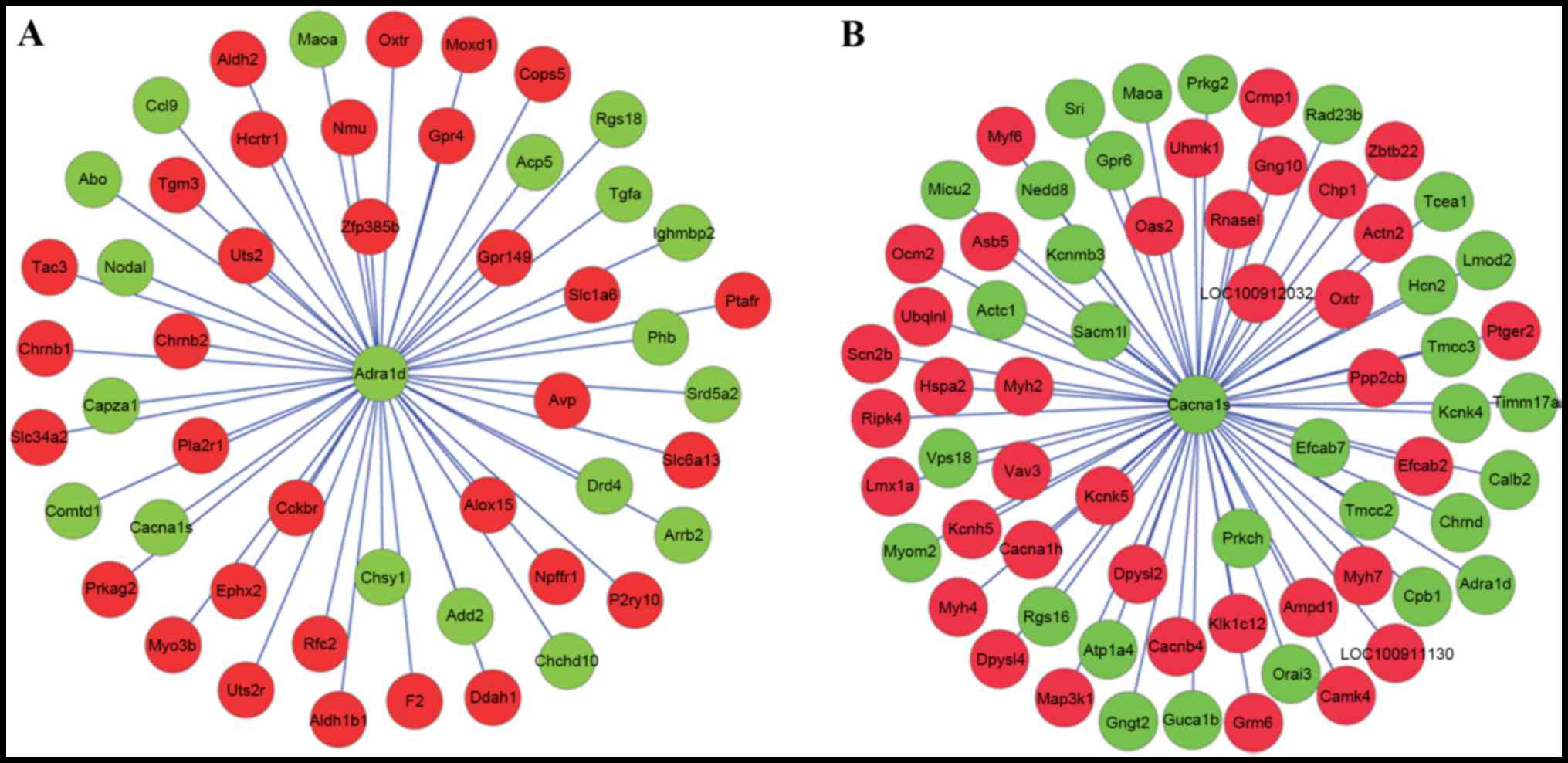
With the required confidence threshold of >0, the
DEGs which had interaction relationships with ADRA1D and
voltage-dependent L-type calcium channel subunit α-1S
(ACNA1S) were indicated in Fig.
3A and B, respectively. DEGs which had interaction
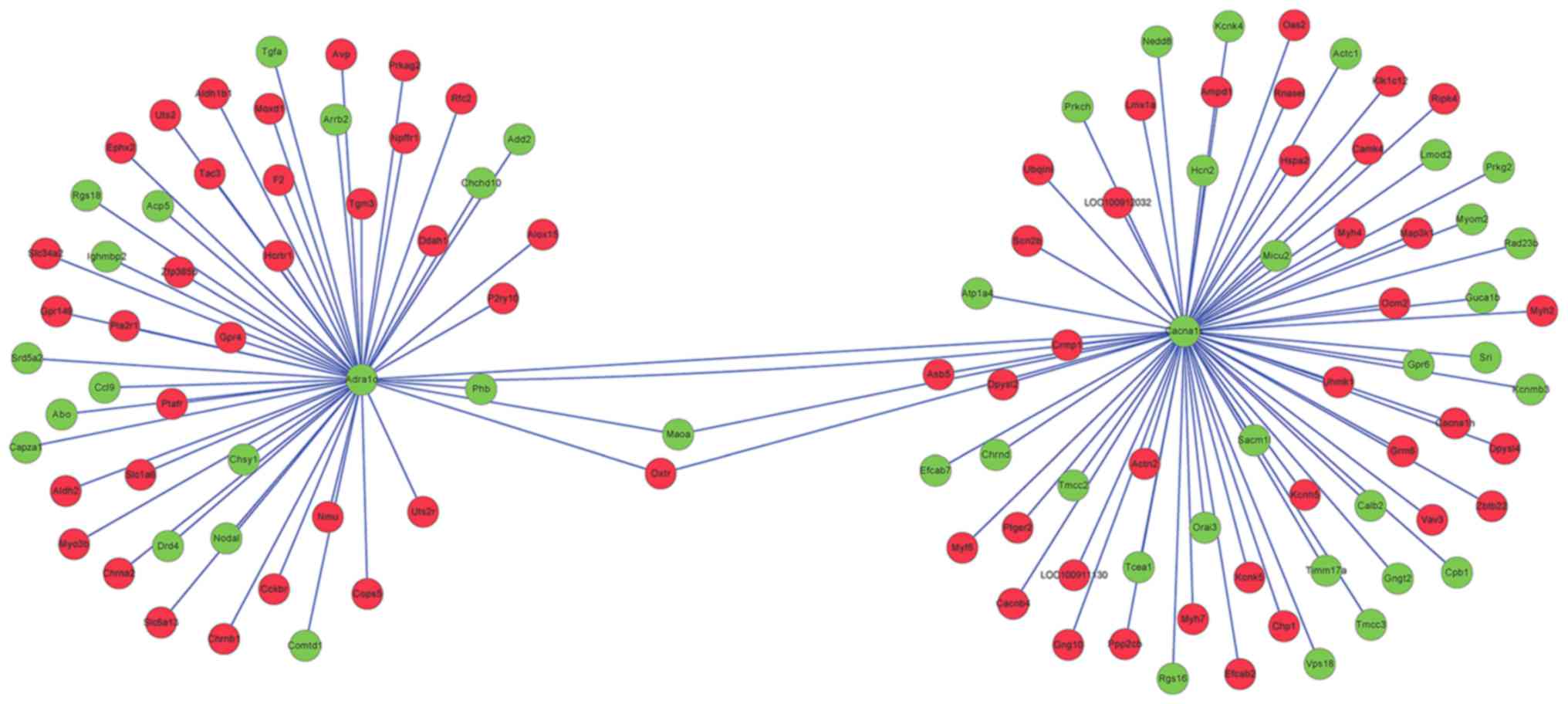
relationships with ADRA1D and CACNA1S were merged in
Fig. 4. In the PPI network,
prohibitin (PHB), oxytocin receptor (OXTR), collapsin
response mediator protein 1 (CRMP1) and
dihydropyrimidinase-like 2b (DPYSL2) exhibited an
interaction relationship with both ADRA1D and
CACNA1S.
Discussion
The present study indicated that calcium signals in
A7r5 cells treated with LPS were weaker when compared with
PBS-treated cells (the control group). It has been reported that
calcium oxalate crystals are able to trigger inflammation through
regulating IL-1β (23). Calcium
pyrophosphate has been revealed to induce a novel acute
inflammation, pleurisy (23). Thus,
LPS induced inflammation in A7r5 cells. In the present study, a
total of 2,038 DEGs, including 1,094 upregulated and 944
downregulated genes, were identified in the LPS-treated and control
groups. Enrichment analyses indicated that NDUFV2 was
involved in several neurological diseases, including oxidative
phosphorylation, Alzheimer's disease, Parkinson's disease and
Huntington's disease. Furthermore, NDUFV2 (degree, 20)
exhibited a higher degree in the PPI network for DEGs enriched in
pathways associated with neurological diseases. NDUFV2,
which is located on chromosome 18p11.31-p11.2, has been named as a
causative gene in neurological diseases, such as schizophrenia,
Parkinson's disease, and bipolar disorder (24,25). A
human disease cell model has revealed that the injury of
mitochondrial localization of NDUFV2 is related to the
pathogenesis of early-onset hypertrophic cardiomyopathy and
encephalopathy (26). Therefore, the
expression of NDUFV2 may be involved in the effect of
inflammation on neurotransmission of VSMC.
In the PPI network for ADRA1D, CACNA1S
and the DEGs interacting with these components, PHB,
OXTR, CRMP1 and DPYSL2 were revealed to have
interaction relationships with both ADRA1D and
CACNA1S. PHB, a member of the Band-7 family of
proteins, has a neuro-damaging role following oxidative and
excitotoxic stress, and may serve as target for designing agents to
control neuronal death in brain injury, such as cerebral ischemia
(27). In cardiomyocytes,
overexpressed PHB contributes to the maintenance of the
mitochondrial membrane potential and improves cell survival during
hypoxia (28). It has been
speculated that the function of PHB in protecting against
oxidative and hypoxic stress may be correlated with its role in
mediating the electron transport chain enzyme, cytochrome C oxidase
(29). A previous report has
implicated oxytocin (OXT) in inflammatory processes
(30). As G-protein coupled
receptors, oxytocin receptors (OXTRs) are regulated by G-proteins,
which stimulate the phosphatidylinositol-calcium secondary
messenger system (31). In addition,
OXTRs are widely distributed in the central nervous system and
mediate various behaviors (32),
such as social memory and recognition, responses to stress and
anxiety, sexual and maternal behaviors, and bonding (33). These may indicate that the expression
levels of PHB and OXTR are correlated with the effect
of inflammation on neurotransmission of VSMC.
CRMP1 is affiliated with a cytoplasmic family
of proteins and is involved in the development of the central
nervous system (34). CRMP1,
which may be disturbed by Speedy A1 (Spy1) from interacting
with actin, has a role in the collapse and regeneration of growth
cones after sciatic nerve crush (35). Previous studies have identified that
within hypertrophic cells of a brain that has suffered a stroke,
overexpression of SPNA2 and DPYSL2 (also known as
CRMP2) was revealed to be correlated with neurite outgrowth
and plasticity, which suggests an early activation of neuronal
regeneration, repair and development (36,37).
Meanwhile, in the neonatal rat brain, the Akt/glycogen synthase
kinase-3β/CRMP2 pathway modulates axonal injury following
hypoxia-ischemia (38,39). Thus, the expression levels of
CRMP1 and DPYSL2 may be associated with the effect of
inflammation on neurotransmission of VSMC.
In conclusion, we screened 2,038 DEGs, including
1,094 upregulated and 944 downregulated genes in the LPS group
compared with the control group. The present study identified that
NDUFV2, PHB, OXTR, CRMP1 and
DPYSL2 may have key roles in the effect of inflammation on
the neurotransmission of VSMC. However, the results were
speculations following bioinformatics analysis and require further
experimental validation.
Acknowledgments
The present study was supported by Shanghai Science
and Technology Commission Fund (grant no. 15495810202) and the
Medical Engineering Cross Research Fund, Shanghai Jiaotong
University (grant nos. YG2013MS08 and YG2014QN09).
References
|
1
|
Owens GK: Regulation of differentiation of
vascular smooth muscle cells. Physiol Rev. 75:487–517.
1995.PubMed/NCBI
|
|
2
|
Fukata Y, Amano M and Kaibuchi K:
Rho-Rho-kinase pathway in smooth muscle contraction and
cytoskeletal reorganization of non-muscle cells. Trends Pharmacol
Sci. 22:32–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arita Y, Kihara S, Ouchi N, Maeda K,
Kuriyama H, Okamoto Y, Kumada M, Hotta K, Nishida M, Takahashi M,
et al: Adipocyte-derived plasma protein adiponectin acts as a
platelet-derived growth factor-BB-binding protein and regulates
growth factor-induced common postreceptor signal in vascular smooth
muscle cell. Circulation. 105:2893–2898. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heo SK, Yun HJ, Noh EK, Park WH and Park
SD: LPS induces inflammatory responses in human aortic vascular
smooth muscle cells via Toll-like receptor 4 expression and nitric
oxide production. Immunol Lett. 120:57–64. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Irani K: Oxidant signaling in vascular
cell growth, death, and survival a review of the roles of reactive
oxygen species in smooth muscle and endothelial cell mitogenic and
apoptotic signaling. Circ Res. 87:179–183. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eltzschig HK and Carmeliet P: Hypoxia and
inflammation. N Engl J Med. 364:656–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deten A, Volz HC, Holzl A, Briest W and
Zimmer HG: Effect of propranolol on cardiac cytokine expression
after myocardial infarction in rats. Mol Cell Biochem. 251:127–137.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davel AP, Fukuda LE, De Sá LL, Munhoz CD,
Scavone C, Sanz-Rosa D, Cachofeiro V, Lahera V and Rossoni LV:
Effects of isoproterenol treatment for 7 days on inflammatory
mediators in the rat aorta. Am J Physiol Heart Circ Physiol.
295:H211–H219. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu Y, Tuerxun A, Hui Y and Abliz P:
Effects of propranolol and isoproterenol on infantile hemangioma
endothelial cells in vitro. Exp Ther Med. 8:647–651.
2014.PubMed/NCBI
|
|
10
|
Romacho T, Azcutia V, Vázquez-Bella M,
Matesanz N, Cercas E, Nevado J, Carraro R, Rodríguez-Mañas L,
Sánchez-Ferrer CF and Peiró C: Extracellular PBEF/NAMPT/visfatin
activates pro-inflammatory signalling in human vascular smooth
muscle cells through nicotinamide phosphoribosyltransferase
activity. Diabetologia. 52:2455–2463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cuff CA, Kothapalli D, Azonobi I, Chun S,
Zhang Y, Belkin R, Yeh C, Secreto A, Assoian RK, Rader DJ and Puré
E: The adhesion receptor CD44 promotes atherosclerosis by mediating
inflammatory cell recruitment and vascular cell activation. J Clin
Invest. 108:10312001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dichtl W, Dulak J, Frick M, Alber HF,
Schwarzacher SP, Ares MP, Nilsson J, Pachinger O and Weidinger F:
HMG-CoA reductase inhibitors regulate inflammatory transcription
factors in human endothelial and vascular smooth muscle cells.
Arterioscler Thromb Vasc Biol. 23:58–63. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Christodoulou DC, Gorham JM, Herman DS and
Seidman J: Construction of normalized RNA-seq libraries for
next-generation sequencing using the crab duplex-specific nuclease.
Curr Protoc Mol Biol Chapter. 4:Unit4.12. 2011. View Article : Google Scholar
|
|
14
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
D'Antonio M, Picardi E, Castrignanò T,
D'Erchia AM and Pesole G: Exploring the RNA editing potential of
RNA-Seq data by ExpEdit. Methods Mol Biol. 1269:327–328. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tarazona S, Furio-Tari P, Ferrer A and
Conesa A: NOISeq: Exploratory analysis and differential expression
for RNA-seq data. R package version 200. 2012.
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nature genetics. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
21
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Willoughby D, Dunn C, Yamamoto S, Capasso
F, Deporter D and Giroud J: Calcium pyrophosphate-induced pleurisy
in rats: A new model of acute inflammation. 1975. Agents Actions.
43:221–224. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishioka K, Vilariño-Güell C, Cobb SA,
Kachergus JM, Ross OA, Hentati E, Hentati F and Farrer MJ: Genetic
variation of the mitochondrial complex I subunit NDUFV2 and
Parkinson's disease. Parkinsonism Relat Disord. 16:686–687. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Washizuka S, Kametani M, Sasaki T, Tochigi
M, Umekage T, Kohda K and Kato T: Association of mitochondrial
complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the
Japanese population. Am J Med Genet B Neuropsychiatr Genet.
141B:301–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu HY, Liao PC, Chuang KT and Kao MC:
Mitochondrial targeting of human NADH dehydrogenase (ubiquinone)
flavoprotein 2 (NDUFV2) and its association with early-onset
hypertrophic cardiomyopathy and encephalopathy. J Biomed Sci.
18:292011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teoh J, Boulos S, Chieng J, Knuckey NW and
Meloni BP: Over-expressing prohibitin (PHB) in neuronal cultures
exacerbates cell death following hydrogen peroxide and L-glutamic
acid induced injury. Neurosci Med. 5:149–160. 2014. View Article : Google Scholar
|
|
28
|
Muraguchi T, Kawawa A and Kubota S:
Prohibitin protects against hypoxia-induced H9c2 cardiomyocyte cell
death. Biomed Res. 31:113–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsutsumi T, Matsuda M, Aizaki H, Moriya K,
Miyoshi H, Fujie H, Shintani Y, Yotsuyanagi H, Miyamura T, Suzuki T
and Koike K: Proteomics analysis of mitochondrial proteins reveals
overexpression of a mitochondrial protein chaperon, prohibitin, in
cells expressing hepatitis C virus core protein. Hepatology.
50:378–386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amrani Y, Syed F, Huang C, Li K, Liu V,
Jain D, Keslacy S, Sims MW, Baidouri H, Cooper PR, et al:
Expression and activation of the oxytocin receptor in airway smooth
muscle cells: Regulation by TNFalpha and IL-13. Respir Res.
11:1042010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kimura T, Tanizawa O, Mori K, Brownstein
MJ and Okayama H: Structure and expression of a human oxytocin
receptor. Nature. 356:3561992. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heinrichs M, Baumgartner T, Kirschbaum C
and Ehlert U: Social support and oxytocin interact to suppress
cortisol and subjective responses to psychosocial stress.
Biological Psychiatry. 54:1389–1398. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thompson RJ, Parker KJ, Hallmayer JF,
Waugh CE and Gotlib IH: Oxytocin receptor gene polymorphism
(rs2254298) interacts with familial risk for psychopathology to
predict symptoms of depression and anxiety in adolescent girls.
Psychoneuroendocrinology. 36:144–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li K, Qi Y, Xia T, Yao Y, Zhou L, Lau KM
and Ng HK: CRMP1 inhibits proliferation of medulloblastoma and is
regulated by HMGA1. PLoS One. 10:e01279102015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao L, Liu YH, Li X, Ji YH, Yang XJ, Hang
XT, Ding ZM, Liu F, Wang YH and Shen AG: CRMP1 interacted with Spy1
during the collapse of growth cones induced by Sema3A and acted on
regeneration after sciatic nerve crush. Mol Neurobiol. 53:879–893.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Indraswari F, Wong PT, Yap E, Ng YK and
Dheen ST: Upregulation of Dpysl2 and Spna2 gene expression in the
rat brain after ischemic stroke. Neurochem Int. 55:235–242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanaka H, Morimura R and Ohshima T: Dpysl2
(CRMP2) and Dpysl3 (CRMP4) phosphorylation by Cdk5 and DYRK2 is
required for proper positioning of Rohon-Beard neurons and neural
crest cells during neurulation in zebrafish. Dev Biol. 370:223–236.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiong T, Tang J, Zhao J, Chen H, Zhao F,
Li J, Qu Y, Ferriero D and Mu D: Involvement of the
Akt/GSK-3β/CRMP-2 pathway in axonal injury after hypoxic-ischemic
brain damage in neonatal rat. Neuroscience. 216:123–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang CY, Feng SJ, Xu L, Bu XN, Zhang N,
Zheng YX, Yuan XW, Li XG and Li JF: CRMP-2 is involved in hypoxic
preconditioning-induced neuroprotection against cerebral ischemic
injuries of mice. Chin J Basic Clin Med. 11:0072009.(In
Chinese).
|