Introduction
Asthma is a chronic disease of airway inflammation
that presents with varying and recurrent symptoms, including cough,
chest tightness and shortness of breath (1). Progress has been made in understanding
the pathogenesis, diagnosis and treatment of asthma (2), and novel drugs, formulations and dosing
instruments have been applied to treat asthma (3,4).
However, asthma still presents a considerable threat to health, due
to its high morbidity and mortality rates (5). Therefore, the development of novel
drugs and complementary therapies, possibly in the form of
traditional medicine, such as traditional Uighur medicine, is
urgently required.
Uighur medicine is a well-established branch of
medicine comprised of unique types of medicines, including munziq
and mushily of abnormal savda. It is currently practiced by
physicians and clinicians from the Xinjiang Uighur autonomous
region in China (6,7). Uighur medicine shares an origin with
Greco-Arab medicine and describes the incidence of illnesses
associated with abnormal Hilits (representing syndromes), which are
caused by an imbalance amongst four normal Hilits (representing
humors), known as Safra, Kan, Phlegm and Savda (6,8).
Quantitative or qualitative changes in any Hilit and the resulting
disturbance of dynamic homeostasis of these Hilits may result in
the development of corresponding symptoms, including increased
quantity of urine, facial edema and weak pulse. Among these,
abnormal Savda is the dominant syndrome in disease progression and
often develops in conjunction with other abnormal Hilits (8). In traditional Uighur medicine, Savda is
the major syndrome responsible for almost all complex diseases,
including asthma, type II diabetes mellitus, cancer, and various
cardiovascular and neurodegenerative illnesses (8). Previous studies have demonstrated that
abnormal Savda is associated with relatively consistent biological
changes in complex diseases that are manifested holistically within
a population (9–11). Abnormal Savda may be treated with a
unique Uighur prescription, comprised of ten herbal ingredients
mixed in specific proportions. These are C. dichotoma
(10.6%), A. italic (10.6%), G. Uralensis (7.1%),
A. capillus-veneris (4.9%), E. humifusa (4.9%), Z.
jujuba (4.9%), L. angustifolia (4.9%), F. vulgare
(4.9%), M. officinalis (4.9%) and A. pseudoalhagi
(42.3%) (7). A number of studies
have demonstrated that Uighur prescription may mitigate oxidative
stress associated with abnormal Savda, possibly by protecting cells
from mitochondrial oxidative damage (12). Uighur medicine may also modulate
abnormal changes in the neuroendocrine-immunity network and prevent
carcinogenesis in murine models (13). Furthermore, flavonoids isolated from
abnormal Savda Munziq are capable of inducing cell-cycle arrest and
apoptosis of tumor cells (14). The
results of these studies indicate that the biological basis of
Savda may change as abnormal Savda develops, but is restored
following Uighur prescription treatment.
In the disease state, protein expression is
dynamically altered in a spatiotemporal manner and modulated by
post-translational processing and chemical modifications. Along
with the application of emerging proteomics techniques,
serum/plasma has become a biomedium for the study of disease
etiology, diagnostic biomarkers and drug targets of asthma in
contemporary medicine (15). In
particular, proteomics analysis has improved knowledge on the
allergens of asthma and the effects of clinical therapy, and has
even guided personalized therapy (16). In addition, abnormal changes in the
whole regulatory network of protein expression are associated with
the overall pathological state of patients with asthma (13). This holistic concept of understanding
disease etiology through understanding of biological systems is
shared by the theories of traditional Uighur medicine.
The present study assessed the effect of Uighur
prescription of abnormal Savda on the regulatory network of
relevant plasma proteins in asthma patients using proteomics. It
also identified differentially expressed proteins that are
potentially targeted by Uighur prescription. The aim of the present
study was to provide evidence for the role of Uighur prescription
in treating abnormal Savda at the proteomics level and to
contribute to the scientific interpretation and application of
Uighur medicine.
Materials and methods
Uighur prescription
Abnormal Savda Munziq is a traditional Uyghur
medicinal herbal preparation that consists of the following
(10,12,13): 15
g Cordia dichotoma fruit and Ziziphus jujube fruit, 7
g Anchusa italic plant, Dracocephalum moldavica L,
Lavandula angustifolia, Adiantum capillus-veneris,
Foeniculum vulgare Mill, Euphorbia humifusa Willd, 10
g Glycyrrhiza uralensis Fisch and 15 g Alhagi
pseudalhagi. Abnormal Savda Mushil preparations contain: 45 g
Alhagi pseudalhagi and Fructus Cassiae fistulae, 15 g
Pogonatherum crinitum, Terminalia chebula Retz. and
Tenninlia chebula, 12 g Rosa rugosa, 6 g
Polypodium vulgare, Glycyrriza Uralensis Fisch. and
Foeniculum vuLgare Mill., 10 g Lavandula
angustifolia, Adiantum capillus-veneris, Euphorbia
humifusa Willd, Anchusa italic, Iberis pectilata
L., Viola tianschanica Maxim, Nymphaea L. and
Amygdalus communis L., 18 g Raisin, 30 g Cordia
dichotoma fruit and Qizil Guliqent and 21 g Cassia
angustifolia. All ten herbs were originally grown in Xinjiang,
China and collected by the Chi Kang Barbour Pharmaceutical Co.
(Xinjiang, China) by professional herbal growers.
Patient data
In total, 40 patients diagnosed with bronchial
asthma between August 2013 and January 2015 were selected for the
present study according to the diagnostic criteria of the Guide for
Prevention and Treatment of Bronchial Asthma established by the
Chinese Medical Association in 2013 (17) and the Global Initiative for Asthma in
2010 (revised in 2012) (18). The
present study consisted of 23 males and 17 females, with a mean age
of (36.93±12.14) years old (age range, 20 and 60 years old).
Abnormal Hilits, including abnormal Kan, Phlegm,
Safra or Savda, were identified by an independent diagnosis of two
experienced physicians specialized in Uighur medicine. The
classification outcome as abnormal Savda was further defined
according to the established criteria of Uighur medicine (6,7) and
using the assessment of symptom scores for abnormal Savda,
including slow pulse (<60 beats/min), bleary eyes, dark purple
lips, blue tongue, cool skin temperature (<36.6°C), dreams (or
nightmares), night sweating, turbid urine and dry stools lasting a
duration >1 month.
All enrolled individuals were orally administered
with an Uighur prescription of abnormal Savda (patent no. ZL
02130082.8, China), including abnormal Savda Munziq extract powder
at a dose of 3–6 g twice daily for 10–15 day, followed by Savda
Mushil powder at a dose of 3–6 g twice daily for 3–5 days based
upon the specific status of the asthma patients, as described
previously (7). The entire clinical
therapy involved completion of ≥3 courses of treatment, with each
course consisting of treatment with abnormal Savda Munziq at a dose
of 3–6 g for 10–15 days, followed by a 3–5 day therapy with
abnormal Savda Mushil at a dose of 3–6 g.
The study design was approved and monitored by the
Ethics Committee of the Xinjiang Medical University (Urumqi,
China). All procedures of the study were in accordance with the
Helsinki Declaration. Informed consent was obtained from all
patients, and patient data were analyzed anonymously throughout the
experiment.
Sample collection
Blood samples (2 ml) were obtained from 40 asthma
patients by venipuncture into evacuated blood collection tubes
containing EDTA-K2 anticoagulant (Promega Corporation, Madison, WI,
USA). Plasma samples were separated by centrifugation at 3,000 × g
for 10 min at 4°C and preserved at −80°C. Blood samples were
collected twice: Prior to treatment (at baseline level) and once
following final treatment with abnormal Savda Mushil.
Protein extraction and proteomic
analysis
Proteomics analysis was performed using 8-plex
isobaric tags for relative and absolute quantitation (iTRAQ,
Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Plasma samples were collected twice; once at baseline and
once following treatment, and were randomly and equally divided
into four subgroups (n=4 in each group). Up to five samples from
different individuals in each subgroup were mixed in equal volumes
to form a pooled sample for further testing. The resulting samples
were enriched for low-abundance proteins by depletion of medium-
and high-abundance proteins using a pre-packed 1-ml affinity liquid
chromatography (LC) column provided with a ProteoMiner™ low
abundance protein enrichment kit (catalogue no. 56-2588-44; Bio-Rad
Laboratories Inc., Hercules, CA, USA) according to the
manufacturer's instructions. Following enrichment, precipitated
proteins were dissolved in 300 µl lysis buffer consisting of 6 M
urea, 4% 3-cyclohexylamino propanesulfonic acid
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate,
2 mM EDTA (all from Promega Corporation) and 1 mM phenylmethane
sulfonyl fluoride (Sigma-Aldrich; Merk KGaA). Following reduction,
10 mM dithiothreitol and 100 mM NH4HCO3 (Sigma-Aldrich; Merck KGaA)
were added at 56°C for 45 min. Subsequently, alkylation was
performed with 55 mM iodoacetic acid (Promega Corporation) and 100
mM NH4HCO3 for 45 min. Samples were then precipitated in 80%
acetone at −20°C overnight and dissolved in 0.8 M urea and 500 mM
tetraethylammonium bromide (TEAB; pH 8.5). Each protein sample (30
µg) was digested by supplementing sequencing-grade modified trypsin
(Promega Corporation) at an enzyme/substrate ratio of 1:30 (w/w) in
dissociation buffer (0.1% in TEAB; Promega Corporation) at 37°C for
24 h. The tryptic peptides were then labeled with 8-plex iTRAQ
reagents (AB Sciex, Foster City, CA, USA) according to the
manufacturer's instructions (iTRAQ113, 114, 115 and 116 for samples
of the baseline, and 117, 118, 119 and 121 for samples following
treatment). The reaction solvents were removed by speed vacuum at
3,000 × g and the labeled peptides were dissolved in 20 mM NH4FA
(pH 10.0; Sigma-Aldrich; Merck KGaA) for subsequent analysis.
Peptide fractionation by strong cation
exchange chromatography and C18 column reversed-phase (RP)
chromatography
High-resolution strong cationic exchange (SCX)
chromatography was performed to remove redundant iTRAQ reagents and
any interfering substances that could affect mass spectrometry (MS)
analysis. Labeled peptides were loaded onto an SCX column (Luna
SCX, 4.6×250 mm; Phenomenex Inc., Torrance, CA, USA), and eluted by
a stepwise linear elution program as follows: 0–10 min
equilibration in Buffer A at pH 3.0 including 25% acetonitrile
(ACN), 20 mM KCl and 10 mM KH2PO4. A 10–15 min fast elution was
also completed using 0–5% Buffer B prepared with 25% ACN, 1 M KCl
and 10 mM KH2PO4, and a pH 3.0, 15–50 min linear elution with 5–30%
Buffer B, and 50–55 min washing elution with 30–80% Buffer B. For
desalting and further fractionation, peptide fractions were loaded
onto an RP column (Luna C18, 4.6 mm inner diameter ×250 mm length,
Phenomenex, Inc.), and eluted by a step linear elution program as
follows: 0–10 min equilibration in 100% solution A (2% ACN and 20
mM NH4FA, pH 10.0), 10–15 min fast elution with 0–12% solution B
(80% ACN and 20 mM NH4FA, pH 10.0), 15–50 min linear elution with
12–56% solution B, and 50–55 min washing elution with 56–80%
solution B. All procedures were performed using a prominence high
performance liquid chromatography (HPLC) system (Shimadzu
Corporation, Kyoto, Japan) with a flow rate of 1.0 ml/min and the
peptides were monitored at 214 nm. In addition, the fractions
containing the peptides were collected at a rate of 1 tube/min
during a linear elution period.
Peptide analysis by nano-liquid
chromatography coupled with Q-exactive MS
Peptide fractions were loaded onto a nano-RP column
(5 µm Hypersil C18 phenomenex Luna SCX 100A; 75 µm × 100 mm; Thermo
Fisher Scientific, Inc.) mounted in a Prominence Nano HPLC system
(Shimadzu Corporation). The peptides were then eluted with an ACN
gradient from 5–40% containing 0.1% formic acid (Sigma-Aldrich;
Merck KGaA) for 65 min at 400 nl/min. Elutes were then transferred
to a Thermo Scientific™ Q-exactive™ mass spectrometer (Thermo
Fisher Scientific, Inc.), which was run in positive ion mode and in
a data-dependent manner with a full MS scan of 350 to 6,000 m/z,
70,000 resolution, 320°C, 400 nl/min flow rate under a nubuliser
pressure of 1,800 V, MS/MS scan with minimum signal threshold
17,500 and isolation at 2 kDa. In order to evaluate the performance
of MS on iTRAQ-labeled samples, two MS/MS acquisition modes-higher
collision energy dissociation and collision induce
dissociation-were employed. In order to optimize the MS/MS
acquisition efficiency of higher collision energy dissociation,
normalized collision energy was systemically examined from 25 to
70%.
Database search and quantitative data
analysis
Raw MS/MS data were converted into Mascot generic
format (MGF) using Proteome Discoverer 1.3 (Thermo Fisher
Scientific Inc.). Exported MGF files were searched using Mascot 2.3
(Matrix Science, Inc., Boston, MA, USA) against the Uniprot Human
2009-12 database (www.uniprot.org) with a precursor mass tolerance set
at 15 ppm and product ion tolerance of 0.02 kDa. An automatic decoy
database search was performed. Carbamidomethylation of cysteines
was set as a fixed modification (C), and oxidation of methionines
(M), Gln to pyro-Glu (N-term Q) and 8-plex iTRAQ modifications of
N-term, K and Y were considered variable modifications. A maximum
of one miscleavage was acceptable.
A protein with at ≥1 unique peptide and a false
discovery rate <0.01 qualified for further quantification
analysis. Moreover, the fold change in protein abundance was
defined as the median ratio of all significantly matched spectra
with tag signals. Based on Proteome Discoverer 1.3 software
analysis (Thermo Fisher Scientific, Inc.), the coefficient of
variation distribution of all quantified proteins and quantitative
results derived from duplicated injections were compared in
parallel. The differential expression of all proteins was presented
as a fold change in iTRAQ ratios. In addition, the upregulation of
a protein was indicated by an increase of ≥1.2-fold, and
downregulation by a decrease of ≤0.83-fold (1.0/1.2).
Bioinformatics analysis by MetaCore™
software
Differentially expressed proteins were further
characterized using a the MetaCore™ 6.18 software package
(http://thomsonreuters.com/metacore,
Thomson Reuters Co., New York, NY, USA) and an online database
(https://portal.genego.com; Thomson
Reuters Co.) in order to understand the underlying signaling
pathways and protein-protein interaction networks, and to evaluate
candidate proteins as potential biomarkers.
Enzyme-linked immunosorbent assay
(ELISA)
The plasma level of candidate proteins was verified
for plasma samples collected at baseline and following ELISA using
commercially available reagents (USCN Life Science Inc., Wuhan,
China) according to the manufacturer's instructions. ELISA reagents
were used for the following candidate proteins: Perioxiredoxin 2
(PRDX2; catalogue no. SEF757Hu), protein S100A7 (catalogue no.
SEC035Hu), transforming growth factor-β1 (TGF-β1; catalogue no.
SEA124Hu), myeloperoxidase (MPO; catalogue no. SEA601Hu),
carboxypeptidase B2 (CPB2; catalogue no. SEA615Hu) and keratin type
II cytoskeletal 6A (KRT6A; catalogue no. SED234Hu). The final data
were confirmed by three independent measurements of each
protein.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
statistical software for windows (SPSS, Inc., Chicago, IL USA). All
P-values were two-sided, and P<0.05 was used to indicate a
statistically significant difference. The data of protein
expression derived from the ELISA experiment at baseline and
following treatment were statistically compared using a paired
samples t-test.
Results
Identification of proteins
differentially expressed by Nano-LC coupled with Q-Exactive MS
Therapeutic evaluation models for the treatment of
asthma patients with abnormal Savda receiving unique Uighur
prescription were established using proteomics analysis. Following
preparation of pooled samples at baseline and following treatment
by depletion of high-abundance proteins, enzymatic digestion and
iTRAQ labeling, resulting peptides were simultaneously analyzed by
nano-LC coupled with Q-Exactive MS, leading to the output of
peptide spectra with 95% confidence intervals representing the
relative and absolute quantitation of each sample. To analyze the
effect of Uighur treatment on proteomic profiles, peptide spectra
data of pooled samples corresponding to biological replicates at
baseline and following treatment were collectively analyzed using a
functional module of Proteome Discoverer software. Analysis of
peptide spectra set a fold change ≥1.2 or ≤0.83 (1.0/1.2) as the
cutoffs for quantitative differences, and 22 proteins were
identified as differentially expressed in response to the
corresponding treatment (Table I).
Among these, 16 proteins were upregulated whereas 6 were
downregulated.
 | Table I.Identification of differentially
expressed proteins in the plasma of patients with asthma and with
an abnormal Savda in response to the treatment with the unique
Uighur prescription. |
Table I.
Identification of differentially
expressed proteins in the plasma of patients with asthma and with
an abnormal Savda in response to the treatment with the unique
Uighur prescription.
|
|
|
|
|
|
| Peptide
detection |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Nos. | Protein
information | Rec. Symbol | Uniprot ID | MW (kDa) | Calc. pI | Peptide score | Peptide
coverage | Unique
peptides |
Fold-changea |
|---|
| 1 | Keratin, type II
cytoskeletal 6A | KRT6A | P02538 | 60 | 8 | 1,803.12 | 55.32 | 3 | 6.346 |
| 2 | Calmodulin-like
protein 3 | CALL3 | P27482 | 16.9 | 4.42 | 37.41 | 8.05 | 1 | 4.897 |
| 3 | Keratin, type II
cytoskeletal 1 | K2C1 | P04264 | 66 | 8.12 | 1,883.75 | 53.73 | 34 | 4.878 |
| 4 | Keratin, type II
cytoskeletal 72 | K2C72 | Q14CN4 | 55.8 | 6.89 | 177.81 | 7.24 | 1 | 4.511 |
| 5 | Keratin, type I
cytoskeletal 27 | K1C27 | Q7Z3Y8 | 49.8 | 5.05 | 44.26 | 6.54 | 1 | 3.927 |
| 6 | Annexin A1 | ANXA1 | P04083 | 38.7 | 7.02 | 65.14 | 8.96 | 2 | 3.499 |
| 7 | InaD-like
protein | INADL | Q8NI35 | 196.2 | 4.94 | 18.24 | 0.33 | 1 | 2.748 |
| 8 | Keratin, type I
cytoskeletal 10 | KRT10 | P13645 | 58.8 | 5.21 | 1,061.74 | 44.18 | 21 | 2.664 |
| 9 | Thioredoxin | THIO | P10599 | 11.7 | 4.92 | 56.05 | 8.57 | 1 | 2.065 |
| 10 | Carboxypeptidase
B2 | CPB2 | Q96IY4 | 48.4 | 7.71 | 54.12 | 3.55 | 2 | 1.841 |
| 11 | Serum amyloid A-2
protein | SAA2 | P0DJI9 | 13.5 | 9.14 | 161.27 | 31.97 | 1 | 1.697 |
| 12 |
Peroxiredoxin-2 | PRDX2 | P32119 | 21.9 | 5.97 | 48.54 | 9.09 | 2 | 1.502 |
| 13 |
Myeloperoxidase | MPO | P05164 | 83.8 | 8.97 | 30.51 | 1.61 | 1 | 1.501 |
| 14 | Keratin, type II
cytoskeletal 2 epidermal | KRT2 | P35908 | 65.4 | 8 | 718.83 | 28.01 | 10 | 1.390 |
| 15 | Zinc finger and BTB
domain-containing protein 18 | ZBT18 | Q99592 | 58.3 | 5.69 | 28.1 | 1.34 | 1 | 1.345 |
| 16 | Transforming growth
factor beta-1 | TGFβ1 | P01137 | 44.3 | 8.53 | 35.31 | 7.69 | 2 | 1.263 |
| 17 |
Thrombospondin-4 | TSP4 | P35443 | 105.8 | 4.68 | 142.57 | 6.35 | 4 | 0.764 |
| 18 | Protein
S100-A7 | S100A7 | P31151 | 11.5 | 6.77 | 50.31 | 10.89 | 1 | 0.701 |
| 19 | Retinol-binding
protein 4 | RBP4 | P02753 | 23 | 6.07 | 123.03 | 9.95 | 2 | 0.692 |
| 20 | Platelet factor 4
variant | PF4VL | P10720 | 11.5 | 9.1 | 193.28 | 48.08 | 2 | 0.679 |
| 21 | Lysozyme C | LYSC | P61626 | 16.5 | 9.16 | 115.72 | 18.24 | 3 | 0.642 |
| 22 | Protein CASC1 | CASC1 | Q6TDU7 | 83.1 | 5.29 | 24.76 | 1.40 | 1 | 0.581 |
Bioinformatics analysis of
differentially expressed proteins by MetaCore™ software and
ontology database
The role of the 22 identified proteins during
disease development was further evaluated by bioinformatics
analysis using MetaCore™ 6.16 software and an online database
(http://www.genego.com). Gene Ontology analysis
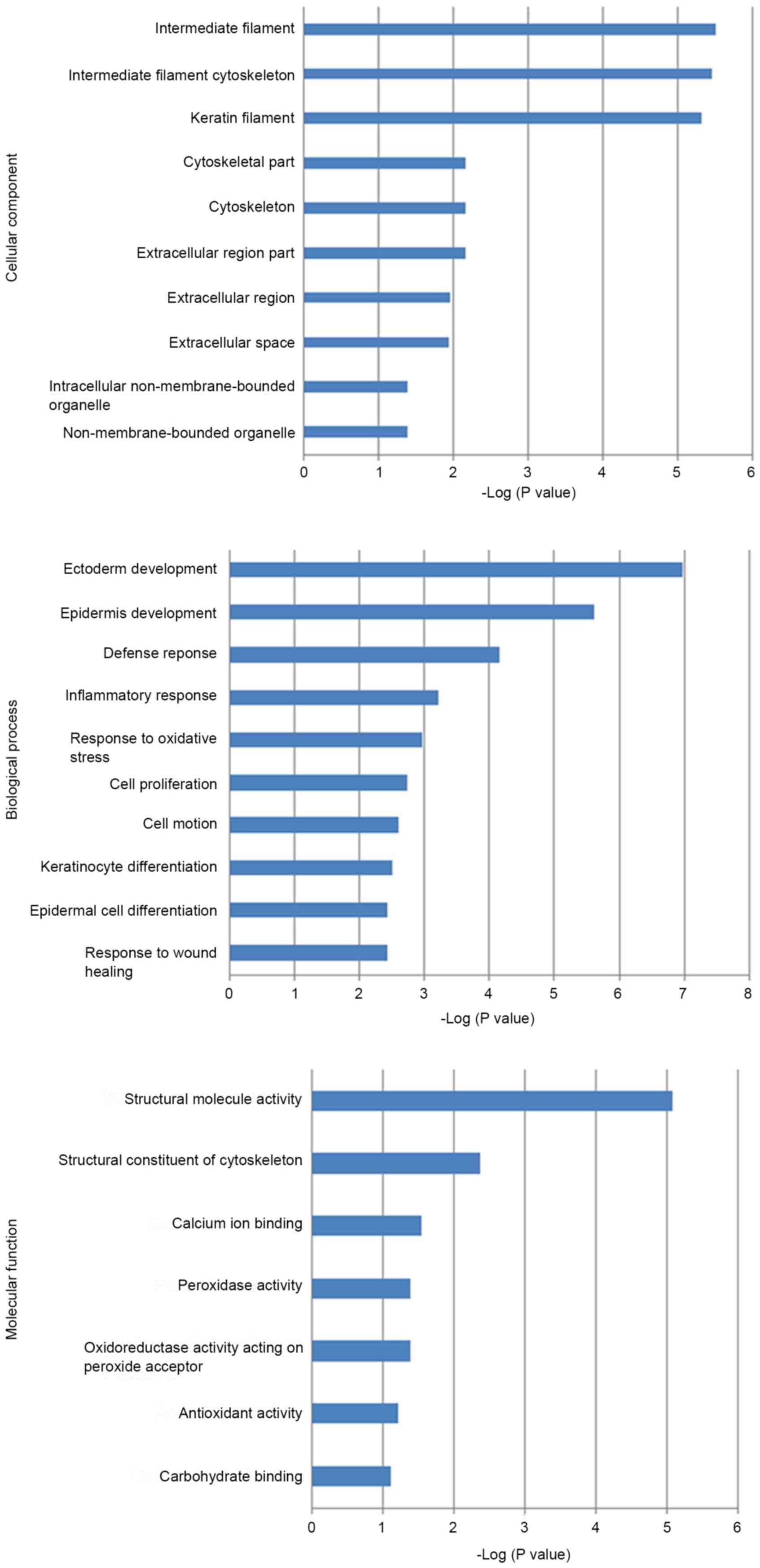
revealed that the majority of these proteins were localized in
intermediate and keratin filaments, as well as the cytoskeleton.
These proteins acted as peroxidases, oxidoreductases, acceptors of
peroxides or carbohydrate binding proteins, and participated in the
process of cytoskeleton remodeling, development, impaired lipoxin
A4 signaling and stimulating transforming growth factor beta (TGFβ)
signaling. Regarding disease pathology, these proteins were largely
involved in the defense and inflammatory response and the response
to oxidative stress and wound healing (Fig. 1). Biomarker Assessment analysis based
on the Disease Ontology database identified myeloperoxidase (MPO)
and TGFβ1 as potential biomarkers for asthma and chronic
obstructive pulmonary disease (COPD), keratin type II cytoskeletal
6A (KRT6A) as a biomarker for asthma and retinol-binding protein 4
(RBP4) for COPD (Fig. 1). The
results of the present study suggested that these proteins, which
were potentially targeted by the prescription for abnormal Savda,
were most likely associated with dysregulation of overall protein
interaction and signaling network during the development and
progression of asthma.
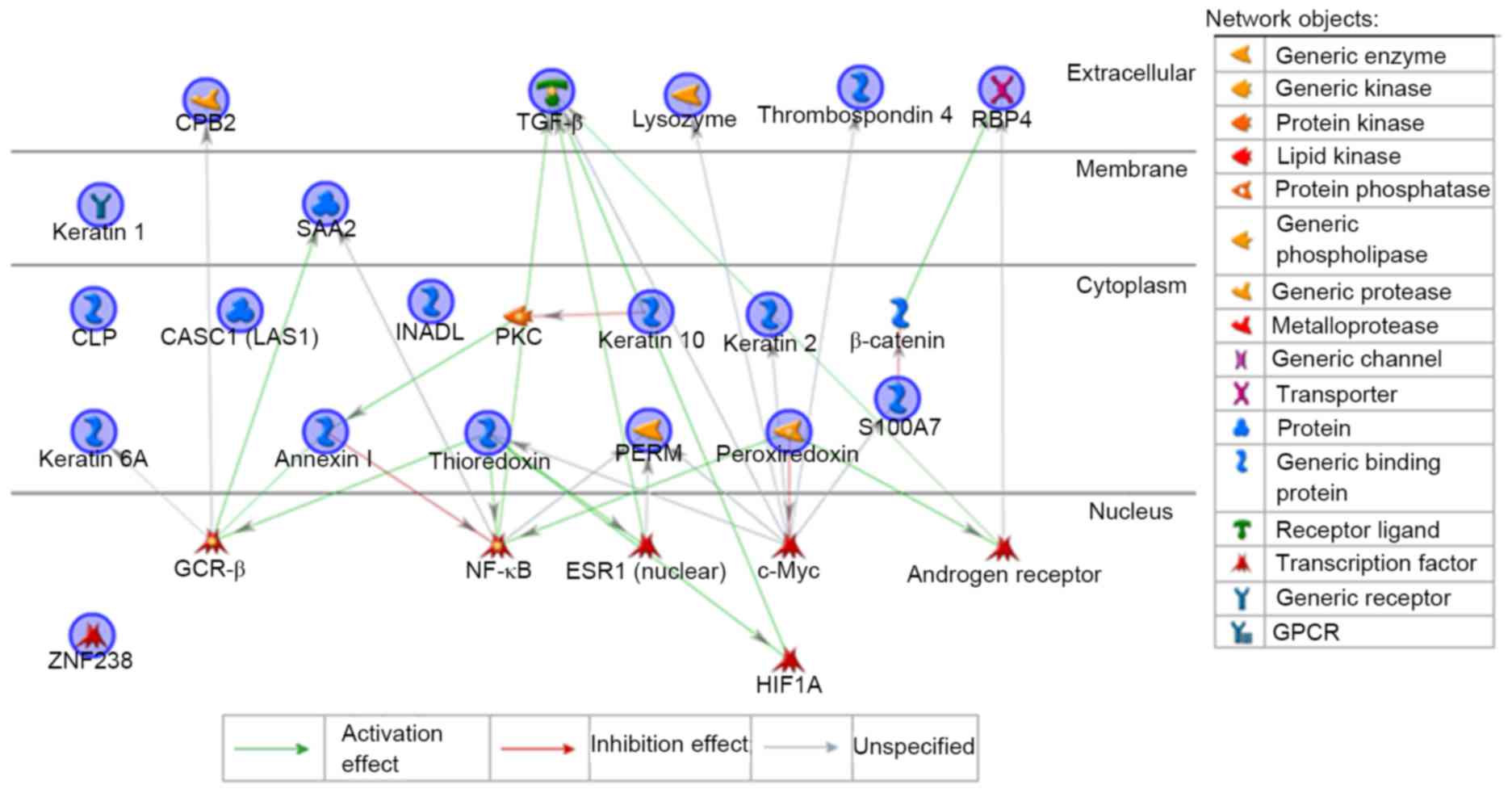
As presented in Table
I, 6 out of 22 proteins, namely PRDX2, CBP2, MPO, TGFβ1, S100
A7 and KRT6A, may be molecular targets of Uighur prescription for
abnormal Savda. Thus, these proteins served as pivotal candidate
biomarkers for abnormal Savda-type asthma. A potential interaction
and regulation network is associated with these proteins regarding
cellular signaling and gene expression (Fig. 2), which was distributed in the
extracellular space, membrane and cytoplasm. These proteins
interacted with diverse effectors, including endopeptidases, matrix
metalloproteinases and phosphatases, and were primarily regulated
by transcription factors of distinct downstream signaling
pathways.
Verification of changes in candidate
proteins by ELISA
To verify the data from the proteomics and
bioinformatics analysis, the plasma levels of the 6 candidate
proteins were determined using whole blood (plasma) samples
collected at baseline (before treatment) and after treatment by
ELISA (Table II). The analysis
demonstrated a significant upregulation of PRDX2 and CPB2, and
downregulation of S100A7 and KRT6A in the plasma of patients in
response to treatment (all P<0.05). However, no difference was
found for levels of MPO and TGFβ1 (P>0.05). Due to the
consistency between the results of iTRAQ proteomics for PRDX2, CPB2
and S100A7 (Table I) and the ELISA
data (Table II), these proteins may
be the potential targets of Uighur prescription for abnormal Savda.
However, the ELISA results for the expression of KRT6A were
inconsistent with those of iTRAQ proteomics, which indicated
upregulation in response to the corresponding treatment.
 | Table II.ELISA verification of candidate
protein expression in response to the treatment of abnormal Savda
in asthma patients with Uighur prescription. |
Table II.
ELISA verification of candidate
protein expression in response to the treatment of abnormal Savda
in asthma patients with Uighur prescription.
|
| Plasma protein
level (ng/ml)a |
|
|---|
|
|
|
|
|---|
| Protein | Prior to treatment
(mean ± SD) | Following treatment
(mean ± SD) | Paired t-test
P-value |
|---|
| PRDX2 | 379.170±40.978 | 419.180±48.579 | <0.001b |
| CBP2 |
1,693.570±114.878 |
1,783.030±194.455 | 0.030b |
| MPO | 253.900±46.917 | 273.606±46.263 | 0.065 |
| TGFβ1 | 0.410±0.314 | 0.445±0.306 | 0.531 |
| S100A7 | 1.747±0.115 | 1.686±0.111 | 0.026b |
| KRT6A | 0.592±0.128 | 0.557±0.131 | 0.027b |
Discussion
Recent proteomics studies of asthma have made
significant progress in the identification of biomarkers and drug
targets for asthma (10,12,13).
Proteomics analysis of bronchial epithelium of asthma patients who
were prescribed with budesonide (glucocorticoids) identified a
number of differentially expressed proteins, including fibronectin
1, secretoglobin 1, KRT6A, interleukin enhancer-binding factor 3,
dihydropyrimidinase like 5, cofilin 1, enolase 1 and vimentin as
potential drug targets (16).
Plasma-based proteomics revealed that heat-shock protein 70 and
eotaxin were upregulated whereas vitamin D binding protein 3 was
downregulated in patients diagnosed with asthma. These alterations
were reversed following treatment with glucocorticoids (19). In a mouse model of acute-phase
asthma, serum-based proteomics identified that immunomodulatory
proteins were differentially expressed following glucocorticoid
therapy (20). In addition,
proteomics analysis determined that expression of 78 kDa
glucose-regulated protein (GRP78) was upregulated in the lungs of
mice with acute-phase asthma, and bronchial perfusion of anti-GRP78
small interfering RNA may modulate the inflammatory response
induced by eosinocytes and bronchial hyper-responsiveness (21). Furthermore, increased expression of
glycoprotein 39 and intercellular adhesion molecule 1 was detected
in bronchoalveolar lavage fluid and lung tissues from asthma models
of mice and monkeys, which was reversed following glucocorticoid
treatment (22). Although different
drugs may have distinct molecular targets, the progression or
clinical therapy of asthma may be causally associated with overall
changes in the expression of proteins throughout the whole body,
which may be integrated into the blood plasma. Therefore, it is
necessary to investigate the entire regulatory network of protein
expression associated with abnormal Savda, which is potentially
targeted by Uighur prescription of abnormal Savda through a
holistic concept that is shared by both systems biology and Uighur
medicine.
In the present study, a therapeutic evaluation model
was established and was used to evaluate abnormal Savda in patients
with asthma receiving treatment with unique Uighur prescription. In
total, proteomics identified 22 differentially expressed proteins
in response to the corresponding treatment. Bioinformatics analysis
demonstrated that a majority of these proteins were localized in
intermediate filaments and the cytoskeleton, indicating that these
proteins may act as antioxidant enzymes and binding proteins, and
be crucial in the defense and inflammatory response, and the
response to oxidative stress or wound healing. A database search
identified that MPO, TGFβ1, KRT6A and RBP4 acted as biomarkers for
asthma or COPD, as reported previously (19). There was a discrepancy between the
iTRAQ and ELISA data collected in the present study, therefore the
association of these four proteins with the effect of the
prescription was not fully confirmed in the current study. The
majority of participants in the current study presented with no
severe side effects or adverse events following treatment with
Uighur prescription. A total of 4 patients suffered from a slight
degree of diarrhea, which may be controlled. Our group plans on
analyzing adverse events that occur following administration of
Munziq and Mushil of abnormal Savda in patients with asthma. It has
been demonstrated that abnormal Savda asthma can induce metabolic
disorder, and disrupt gluconeogenesis and host immunity (23). However, abnormal Savda Munziq is
capable of counteracting abnormal metabolism and is important in
upregulating gluconeogenesis and immune disorders (24). The present study aimed to identify a
novel medication target of applying the Munziq and Mushil in
treating abnormal Savda asthma from the perspective of protein
genomics.
The consistency between the iTRAQ and ELISA data for
PRDX2, CPB2 and S100A7 indicated that the expression of these
proteins may be associated with abnormal Savda in asthma patients
and that these proteins may be targeted by Uighur prescription. Of
these biomarkers, PRDX2 is one of the non-selenium-dependent
peroxidases with anti-oxidative activities and serves a potential
role in the modulation of oxidative stress-related signaling and
disease processes (25). PRDX2 may
participate in a variety of cellular signaling pathways, and may
reduce hyper-responsiveness during allergic airway inflammation by
eliminating cellular reactive oxygen species (26). CPB2 is a zinc-containing
carboxypeptidase that is involved in regulating the
coagulation-fibrinolysis balance in a variety of diseases,
including cancer and cardiovascular disease (27). It has been demonstrated that CPB2
regulates thrombin-mediated tissue inflammation and other
inflammatory responses by inactivating a variety of active
inflammatory mediators, including bradykinin, C3a, C5a and
thrombin-cleaved osteopontin (28).
S100A7/psoriasin is expressed in the airway epithelium, lung
epithelial cells and macrophages (29). S100A7 is induced by oxidative stress
and is involved in a variety of inflammatory diseases (30). The expression of S100A7/psoriasin is
increased in the nasal lavage fluid of allergic patients and the
S100A7 gene polymorphism is associated with allergic rhinitis
(31). In addition, S100A7 may be
important in the development of breast cancer by increasing
inflammatory cell infiltration, stimulating the pro-inflammatory
response, and promoting oxidative stress and angiogenesis (32,33).
These observations are in accordance with the results of the
present study, which suggest that treatment of abnormal Savda in
asthma patients with Uighur prescription upregulates the expression
of PRDX2 and CPB2 and thus the anti-oxidative and anti-inflammatory
response capacity of the body, whereas it downregulates the
expression of S100A7, which may reduce oxidative stress and
inflammatory responses.
In conclusion, the results of the current study
indicate that the therapeutic effect of Uighur prescription for
abnormal Savda in patients with asthma is achieved by predominantly
targeting the entire regulatory network of protein expression,
particularly of those proteins involved in responses to
inflammation and oxidative stress. Further in vitro and
in vivo studies are required to identify the underlying
mechanism of the therapeutic effect of the prescription for
abnormal Savda during asthma progression, which may be revealed by
verifying alternative identified proteins.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (grant no. 81273873).
References
|
1
|
Yoder M, Zhuge Y, Yuan Y, Holian O, Kuo S,
van Breemen R, Thomas LL and Lum H: Bioactive
lysophosphatidylcholine 16:0 and 18:0 are elevated in lungs of
asthmatic subjects. Allergy Asthma Immunol Res. 6:61–65. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohta K, Ichinose M, Nagase H, Yamaguchi M,
Sugiura H, Tohda Y, Yamauchi K, Adachi M and Akiyama K: Japanese
Society of Allergology: Japanese Guideline for Adult Asthma 2014.
Allergol Int. 63:293–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wells KE, Peterson EL, Ahmedani BK,
Severson RK, Gleason-Comstock J and Williams LK: The relationship
between combination inhaled corticosteroid and long-acting
β-agonist use and severe asthma exacerbations in a diverse
population. J Allergy Clin Immunol. 129:1274–1279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salpeter SR, Wall AJ and Buckley NS:
Long-acting beta-agonists with and without inhaled corticosteroids
and catastrophic asthma events. Am J Med. 123:322–328. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Global Initiative for Asthma: Global
strategy for asthma management and prevention. 2012, http://ginasthma.org/
|
|
6
|
Upur H, Yusufu A and Aimaiti N: New
interpretations of Abnormal Savda theory of traditional Uighur
Medicine. Xinjiang People's Press; Urumqi: 2009
|
|
7
|
Upur H, Maitisidike A, Aimaiti A
Feidasiyefu, Yiming Y and Dubrovin D: Diagnosis of abnormal humor
syndrome in traditional Uighur medicine and its prescriptions and
herbs. Xinjiang People's Press; Urumqi: 2013
|
|
8
|
Upur H, Dubrovin D, Amat N, Liu W, Moore
N, Bauer R, Suidre G, Gogol I, Dong J and Lapham JC:
Graeco-Arab-Uighur Medicine. Acupress; New York, NY: 2013
|
|
9
|
Halmurat U, Askar Y, Ilhamjan S, Obul K
and Roxangul S: Gene polymorphisms in Uighur patients with Abnormal
Savda. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 20:77–78. 2003.(In
Chinese). PubMed/NCBI
|
|
10
|
Tursun K, Asmtula D, Smayil M, Upur D and
Upur H: The relationship between asthma patients with abnormal
Savda in Uighur medicine and the gene polymorphism of IL-4.
Zhongguo Zhong Xi Yi Jie He Za Zhi. 33:1076–1080. 2013.(In
Chinese). PubMed/NCBI
|
|
11
|
Yusup A and Upur H, Abla A and Upur H:
Association study of gene polymorphisms and depression with
abnormal humor in traditional Uighur medicine. BMC Complement
Altern Med. 13:3322013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Upur H, Yusup A, Umar A and Moore N:
Uighur traditional medicine syndrome of Abnormal Savda in men is
associated with oxidative stress, which can be improved by Munziq
and Mushil of Abnormal Savda. Therapie. 59:483–484. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amat N, Upur H, Ablimit A, Matsidik A,
Yusup A and Kijjoa A: Immunomodulatory effects of Abnormal Savda
Munsiq, a traditional Uighur medicine, on the combined stress mice.
J Ethnopharmacol. 122:42–47. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang YX, Abliz G, Ye WJ, Mutalipu Z, Li
XW, Wang HQ, Buranjiang G and Upur H: Mechanisms of hela cell
apoptosis induced by abnormal Savda Munziq total phenolics combined
with chemotherapeutic agents. Asian Pac J Cancer Prev. 15:743–747.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park CS and Rhim T: Application of
proteomics in asthma research. Expert Rev Proteomics. 8:221–230.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
O'Neil SE, Sitkauskiene B, Babusyte A,
Krisiukeniene A, Stravinskaite-Bieksiene K, Sakalauskas R, Sihlbom
C, Ekerljung L, Carlsohn E and Lötvall J: Network analysis of
quantitative proteomics on asthmatic bronchi: Effects of inhaled
glucocorticoid treatment. Respir Res. 12:1242011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Diagnostic criteria of the guide for
prevention and treatment of Bronchial Asthma, Chinese medical
association. Chin J Tuberculosis Respir Disease. 36:52013.
|
|
18
|
Boulet LP, FitzGerald JM, Levy ML, Cruz
AA, Pedersen S, Haahtela T and Bateman ED: A guide to the
translation of the global initiative for asthma (GINA) strategy
into improved care. Eur Respir J. 39:1220–1229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang H, Zhang X, Chi X, Wang J, Wang J
and Dou J: The effect of inhaled glucocorticoid therapy on serum
proteomics of asthmatic patients. Zhonghua Jie He He Hu Xi Za Zhi.
37:274–278. 2014.(In Chinese). PubMed/NCBI
|
|
20
|
Roh GS, Shin Y, Seo SW, Yoon BR, Yeo S,
Park SJ, Cho JW and Kwack K: Proteome analysis of differential
protein expression in allergen-induced asthmatic mice lung after
dexamethasone treatment. Proteomics. 4:3318–3327. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calvo F Quesada, Fillet M, Renaut J,
Crahay C, Gueders M, Hacha J, Paulissen G, Foidart JM, Noel A,
Rocks N, et al: Potential therapeutic target discovery by 2D-DIGE
proteomic analysis in mouse models of asthma. J Proteome Res.
10:4291–4301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Louten J, Mattson JD, Malinao MC, Li Y,
Emson C, Vega F, Wardle RL, van Scott MR, Fick RB, McClanahan TK,
et al: Biomarkers of disease and treatment in murine and cynomolgus
models of chronic asthma. Biomarker Insights. 7:87–104. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Y, Batur M, Li W, Gao Z and Halmurat U:
Different syndromes in the same disease of bronchial asthma of
Uygur medicine abnormal savda syndrome based on metabonomics. J
Xinjiang Med Univ. 37:1441–1446. 2014.
|
|
24
|
Nazuk K, Batur M, Mavlanjan H, Denise D
and Halmurat U: Effect of Abnormal Savda Munziq on urine
metabolites from Abnormal Savda Syndrome carrying asthma rat model.
J Xinjiang Med University. 36:419–426. 2013.
|
|
25
|
Brinkmann C and Brixius K: Peroxiredoxins
and sports: New insights on the antioxidative defense. J Physiol
Sci. 63:1–5. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kwon HS, Bae YJ, Moon KA, Lee YS, Lee T,
Lee KY, Kim TB, Park CS, Moon HB and Cho YS: Hyperoxidized
peroxiredoxins in peripheral blood mononuclear cells of asthma
patients is associated with asthma severity. Life Sci. 90:502–508.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bajzar L, N esheim ME and Tracy PB: The
profibrinolytic effect of activated protein C in clots formed from
plasma is TAFI-dependent. Blood. 88:2093–2100. 1996.PubMed/NCBI
|
|
28
|
Leung LL, Myles T, Nishimura T, Song JJ
and Robinson WH: Regulation of tissue inflammation by
thrombin-activatable carboxypeptidase B (or TAFI). Mol Immunol.
45:4080–4083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Andresen E, Lange C, Strodthoff D,
Goldmann T, Fischer N, Sahly H, Branscheid D and Heine H:
S100A7/psoriasin expression in the human lung: Unchanged in
patients with COPD, but upregulated upon positive S. aureus
detection. BMC Pulm Med. 11:102011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carlsson H, Yhr M, Petersson S, Collins N,
Polyak K and Enerbäck C: Psoriasin (S100A7) and calgranulin-B
(S100A9) induction is dependent on reactive oxygen species and is
downregulated by Bcl-2 and antioxidants. Cancer Biol Ther.
4:998–1005. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bryborn M, Adner M and Cardell LO:
Psoriasin, one of several new proteins identified in nasal lavage
fluid from allergic and non-allergic individuals using
2-dimensional gel electrophoresis and mass spectrometry. Respir
Res. 6:1182005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nasser MW, Qamri Z, Deol YS, Ravi J,
Powell CA, Trikha P, Schwendener RA, Bai XF, Shilo K, Zou X, et al:
S100A7 enhances mammary tumorigenesis through upregulation of
inflammatory pathways. Cancer Res. 72:604–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shubbar E, Vegfors J, Carlström M,
Petersson S and Enerbäck C: Psoriasin (S100A7) increases the
expression of ROS and VEGF and acts through RAGE to promote
endothelial cell proliferation. Breast Cancer Res Treat. 134:71–80.
2012. View Article : Google Scholar : PubMed/NCBI
|