Introduction
Esophageal achalasia is motility disorder resulting
from incomplete relaxation of the lower esophageal sphincter (LES)
and the loss of normal peristalsis (1–3). The
predominant symptoms of esophageal achalasia are dysphagia and
regurgitation. Patients with achalasia are diagnosed by
self-reported symptoms, endoscopy and esophagography. Achalasia is
a rare and chronic disease that can occur at any age (2), and is considered as a risk factor of
esophageal cancer (1,3). The risk of developing squamous cell
carcinoma in achalasia patients was demonstrated to be increased by
33-fold compared with that in the general population (4). However, the etiology of esophageal
achalasia, particularly the process leading to carcinogenesis,
remains largely unknown.
Persistent esophageal distension with retention of
food and fluids, bacterial overgrowth, and impaired clearance of
regurgitated acid and gastric contents are known to lead to chronic
inflammation and to passively cause dysplasia and carcinoma
(5–7). Therapy for esophageal achalasia aims to
reduce food stagnation (8). Various
treatments have been devised for achalasia patients, including
medication, balloon dilatation and surgery. Endoscopic balloon
dilatation remains a widely performed treatment due to the relative
noninvasiveness and simplicity of the procedure; however, it has a
relatively low success rate and often requires multiple treatment
sessions (9). Peroral endoscopic
myotomy (POEM) has been recently established as a minimally
invasive procedure with high success rate (10,11).
Inoue et al (10) reported
that, in short-term results, there was no recurrence subsequent to
the POEM procedure in 17 cases of achalasia. POEM can be
successfully and safely performed by skilled endoscopists, and
effectively ameliorates dysphagia symptoms. Manometric pressure
studies have also demonstrated significant improvement in the lower
esophageal sphincter pressure following POEM (11). In addition, POEM substantially
decreased the Ki-67-positive and P53-positive ratios in esophageal
epithelia. Thus, POEM appears to reduce the risk of esophageal
carcinogenesis (11).
MicroRNAs (miRs) are small non-coding RNAs that
negatively regulate gene expression via translational repression or
messenger RNA degradation (12).
Over 2,800 miRs have been identified in humans, with each
individual miR predicted to target multiple genes based on the seed
sequence matching their 3′-untranslated regions (UTRs) (13). miRs are involved in biological and
pathological processes, including cell differentiation,
proliferation, apoptosis and metabolism (14), and they are emerging as highly
tissue-specific biomarkers with potential clinical applicability
for defining cancer type and origin (15,16).
Accumulating evidence has indicated that deregulation of miRs is
associated with human malignancies, and suggested that miRs may
have a causal role in tumor initiation and progression, since they
can function as oncogenes or tumor suppressors (17). Indeed, previous studies have
indicated distinct differences in miR expression patterns between
squamous cell carcinoma and adenocarcinoma in esophageal and other
cancer types (18–20). For instance, Kimura et al
(21) reported that the highest
expression of miR-205 was identified in both benign and malignant
squamous epithelia, including in esophageal squamous cell
carcinoma, whereas a lower expression was observed in cell lines
and tissues other than squamous epithelia. Additionally, miR-21,
which is an oncogenic miR in various malignancies, was also
upregulated in esophageal squamous cell carcinoma compared with its
expression in paired normal squamous epithelia (21). There is also growing evidence
regarding the pathogenic roles of miRs in immune and inflammatory
disorders, including esophagitis. For example, elevated miR-143,
miR-145 and miR-205 expression levels were observed in the
esophageal squamous mucosa of individuals with ulcerative
esophagitis, where they may be involved in regulating epithelial
restoration in response to injury caused by gastro-esophageal
reflux (22).
As mentioned earlier, it is generally accepted that
achalasia is a pre-malignant disorder that is possibly caused by
longstanding mucosal inflammation due to persistent stasis of food
(23). Nevertheless, there is little
information regarding the miR expression profile in achalasia.
Therefore, the aims of the present study were to identify the miR
expression specific to the esophageal mucosa of achalasia patients,
to determine potential target genes of these miRs and to assess the
alteration of miRs following POEM.
Materials and methods
Patients and clinical samples
A total of 29 achalasia patients who visited the
Showa University Northern Yokohama Hospital (Yokohama, Japan)
between October 2011 and June 2012 were enrolled into the current
study. Patients with any severe underlying illness, such as cancer,
or those who could not tolerate general anesthesia were excluded. A
total of 14 males and 15 females aged between 23 and 85 years
(mean=46) were enrolled. There were 15 smokers and 13 non-smokers
(1 unknown). They were all known to have achalasia. A total of 23
patients suffered from straight-type achalasia (the meandering of
the longitudinal axis of the esophagus appears weak on barium
esophagogram) and 6 patients suffered with sigmoid-type achalasia
(the meandering of the longitudinal axis of the esophagus appears
strong on barium esophagogram). The degree of esophageal dilatation
was grade I in 7 patients and grade II in 22 patients, as defined
by the Descriptive Rules for Achalasia of the Esophagus (24). A total of 4 healthy subjects with no
severe underlying illnesses were enrolled in the present study (2
male, 2 female; aged 63–68 years). Subsequent to obtaining informed
consent, 2 biopsy samples were collected from the middle esophageal
mucosa of each patient during esophagoscopy before POEM, and were
immediately placed into 1 ml RNAlater reagent (Ambion; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and stored at −80°C for
subsequent RNA isolation. All examinations were conducted according
to the 6th Good Clinical Practice guidelines and the Declaration of
Helsinki, and were approved by the Nagasaki and Showa University
Ethics Committees.
POEM
All patients underwent POEM, which was performed as
follows and the procedures were performed under general anesthesia
with positive pressure ventilation. A submucosal tunnel was formed
from the central esophagus to the esophago-gastric junction beyond,
using a technique similar to endoscopic mucosal dissection (ESD)
(25). An incision was subsequently
made of the circular muscle bundle from the entrance to the LES.
The incision mucosal invasion was closed with a hemostatic clip
(8).
RNA extraction
Total RNA, including small RNA, was extracted from
the tissue samples using the mirVana miRNA isolation kit (AM1560;
Ambion; Thermo Fisher Scientific, Inc.), and the total RNA was
quantified using a Nanodrop-1000 spectrophotometer (Nanodrop
Technologies, Wilmington, DE, USA). Next, the total RNA was
purified using the miRNeasy mini kit (cat. no. 217004; Qiagen,
Hilden, Germany), and the quality of the total RNA was determined
by UV absorption measurement and on Agilent 2100 Bioanalyzer
(Agilent Technologies, Santa Clara, CA, USA).
miR array hybridization and
analysis
In order to identify the miR(s) specific to the
esophageal mucosa of achalasia, total RNA was extracted from the
biopsy mucosal tissues of 8 representative cases of achalasia and
from those of 4 healthy volunteer controls. Following DNase
treatment, the isolated RNA samples were subjected to comprehensive
analysis of miR expression patterns using microarray-based
technology. These analyses were performed by Hokkaido System
Science Co., Ltd. (Sapporo, Japan) using the SurePrint G3 Human
8×60 K microarray version 2.0 (Agilent Technologies) and 50 ng
aliquots of each total RNA sample. The scan was performed using the
Agilent Technologies Microarray Scanner (Agilent Technologies) at 3
µm resolution, and each spot was digitized using Agilent Feature
Extraction version 10.7.3.1 software. To identify the miRs that
were differentially expressed in esophageal mucosa, data were
imported to GeneSpring GX version 10.7.3.1 (Agilent Technologies)
and analyzed; a feature is considered detected if the signal is
three-fold greater than the error. The differences in miR
expression were considered as statistically significant if the fold
change in expression values was >2.0 and P<0.05 using a
Student's t-test.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The expression levels of miRs that showed
significant differences based on the microarray results were
analyzed using RT-qPCR. Briefly, cDNA was prepared from total RNA
using the High Capacity cDNA Reverse Transcription kit (cat. no.
4374966; Applied Biosystems; Thermo Fisher Scientific, Inc.) and
the indicated TaqMan small RNA assay kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The RT reactions were performed in
a solution containing 10 ng total RNA, 1X RT primer, 1X RT buffer,
1 mM dNTP, 50 units MultiScribe Reverse Transcriptase and 3.8 units
of RNase inhibitor. Nuclease-free water was added to bring the
solution to a total volume of 15 µl. The reactions were run on the
TGradient thermocycler (Biometra GmbH, Göttingen, Germany) at 16°C
for 30 min, followed by 42°C for 30 min, and then 85°C for 5 min.
Subsequently, qPCR reactions were performed in a solution
containing 1.33 µl RT products with 1X TaqMan Universal Master Mix
II without Uracil-N glycosylase (UNG) (cat. no. 4440040; Applied
Biosystems; Thermo Fisher Scientific, Inc.) and 1 µl of each of the
TaqMan small RNA assay primers. Each Taqman small RNA primer set
contained primers for the analysis of has-miR-361-5p, has-miR-130a
and RNU6B (cat. nos. 000554, 000454 and 001093, respectively;
Applied Biosystems; Thermo Fisher Scientific, Inc.) Nuclease-free
water was added to obtain a solution with total volume of 20 µl.
All reactions were run in triplicate using the LightCycler 480 II
(Roche Diagnostics, Basel, Switzerland). The thermal cycling
reactions were initiated at 95°C for 10 min, followed by 45 cycles
of 95°C for 15 sec, and 60°C for 1 min. The cycle passing threshold
(Cq) was recorded for each candidate miR, and the 2−∆∆Cq
method was used with RNU6B as the endogenous control for data
normalization (12).
In order to determine potential target genes of
miR-130a, RT-qPCR was performed to determine changes in the mRNA
expression in the mucosa of achalasia patients compared with that
in healthy controls. cDNA was prepared from total RNA using the
High Capacity cDNA Reverse Transcription kit (cat. no. 4374966;
Applied Biosystems; Thermo Fisher Scientific, Inc.). The RT
reactions were performed in a solution containing 500 ng total RNA,
1X Random Primer, 1X RT Buffer, 4 mM dNTP, 50 units MultiScribe
Reverse Transcriptase and 20 units RNase Inhibitor. Nuclease-free
water was added to obtain a total volume of 20 µl. The reactions
were run on the TGradient thermocycler (Biometra) at 25°C for 10
min, followed by 37°C for 120 min, and 85°C for 5 min. Next, qPCR
reactions were performed in a solution containing 4 µl RT products
with 1X TaqMan Universal Master Mix II without UNG (cat. no.
4440040; Applied Biosystems; Thermo Fisher Scientific, Inc.) and 1
µl of each of the TaqMan mRNA assay primer sets. Nuclease-free
water was added to bring the total volume up to 20 µl. Of the
significantly altered genes, in silico Target Scan (version
6.2; Whitehead Institute for Biomedical Research, Cambridge, MA,
USA) prediction indicated that myotubularin related protein 10
(MTMR10) and WNK lysine deficient protein kinase 1 (WNK1) may be
candidate target genes of miR-130a, based on the seed sequence
matches in their 3′-UTRs. The TaqMan mRNA primer sets used were for
the amplification of the mRNAs for MTMR10, WNK1 and glyceraldehyde
3-phosphate dehydrogenase (GAPDH) (cat. nos. Hs01107504_m1,
Hs00219183_m1 and Hs99999905_m1, respectively; Applied Biosystems;
Thermo Fisher Scientific, Inc.). Reactions were run on the
LightCycler 480 II (Roche Diagnostics), and thermal cycling was
initiated at 95°C for 10 min, followed by 45 cycles of 95°C for 15
sec and 60°C for 1 min. Cq values were recorded for each candidate
mRNA, and the 2−∆∆Cq method was performed, using GAPDH
as the endogenous control for data normalization.
Statistical analysis
The differences between groups were analyzed using
the unpaired, one-tailed, Student's t-test. Data are expressed as
the mean ± standard error. Differences were considered to be
statistically significant at P<0.05. Multiple regression
analyses were also performed. All data were analysed using StatFlex
version 6 (Artech Information Systems LLC, Morristown, NJ,
USA).
Results
Patient characteristics
Of the patients enrolled in the present study, 23
suffered from straight-type achalasia and 6 with sigmoid-type
achalasia. The degree of esophageal dilatation was grade I in 7
patients and grade II in 22 patients (24). The mean disease duration was 60
months, ranging between 5 and 564 months.
Microarray analysis results
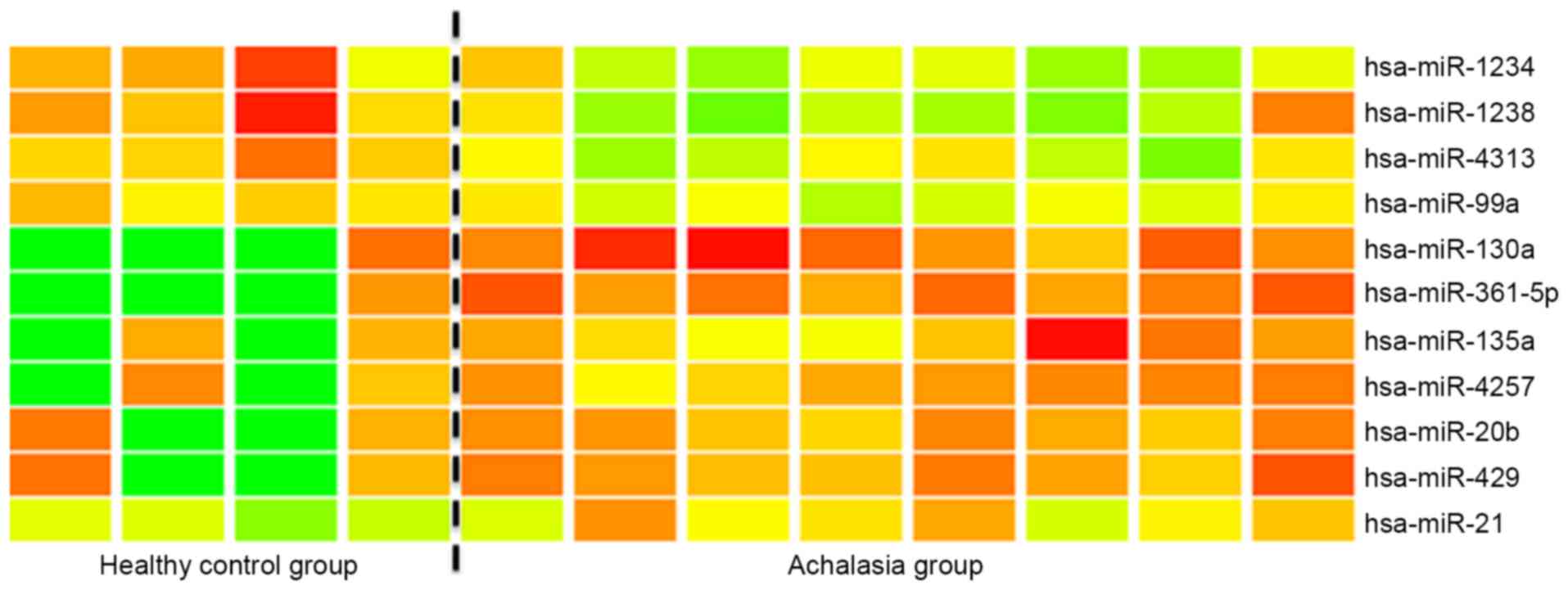
The microarray analysis of miR expression levels in
the esophageal mucosa of achalasia indicated that miR-361-5p and
miR-130a were significantly (>2-fold) overexpressed in the
esophageal mucosa of achalasia patients when compared with the
controls (Fig. 1). Subsequently,
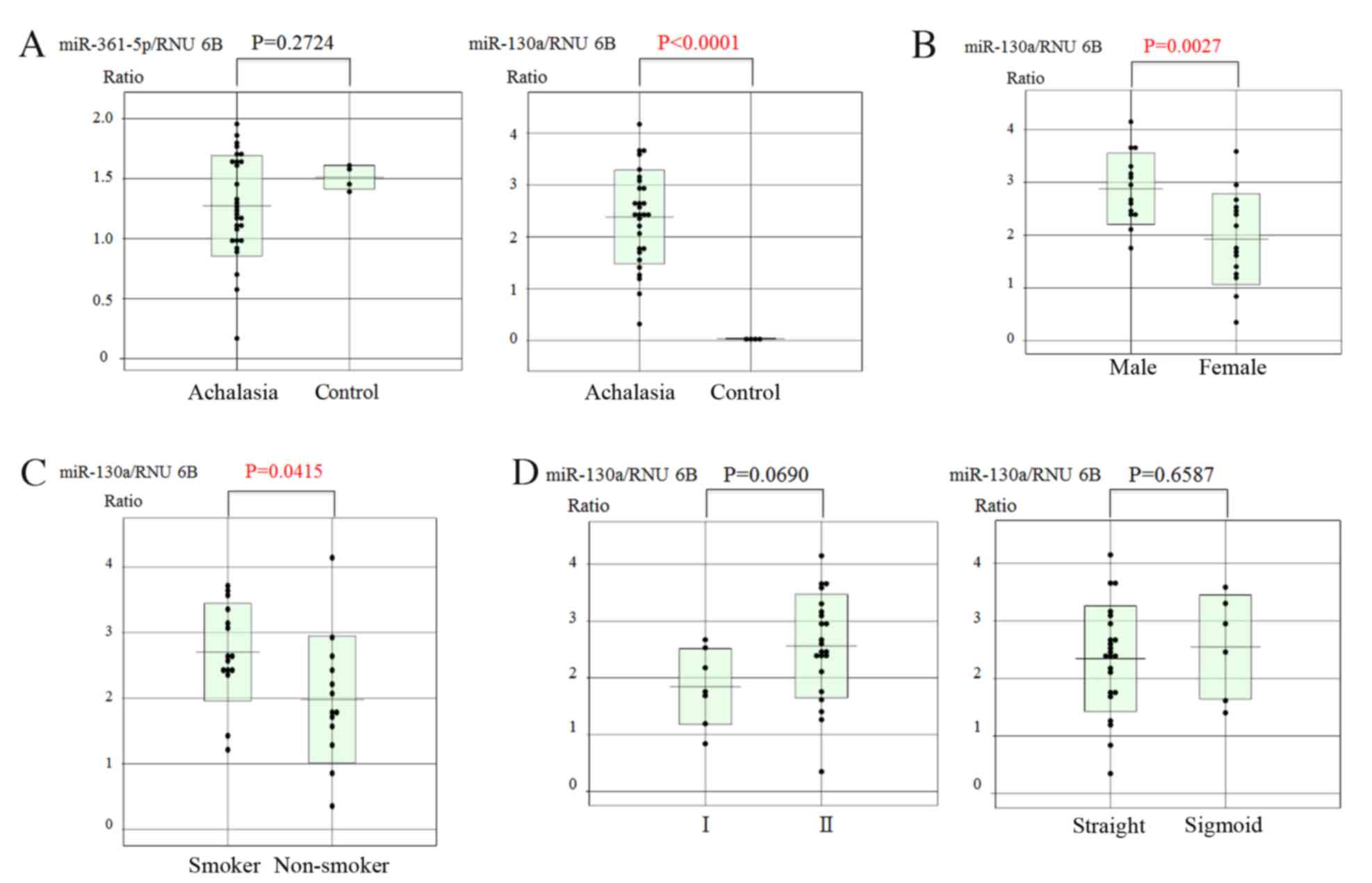
RT-qPCR was used to quantify the expression levels of these two
miRs in biopsy specimens of achalasia patients and controls. The
results revealed that only the expression of miR-130a was
significantly higher in achalasia patients compared with that in
healthy subjects (P<0.0001; Fig.
2A).
Correlation of miR-130a expression
with various parameters
The correlation between the miR-130a expression and
the background characteristics of the patients was then analyzed.
Significant correlations were observed between the expression
levels of miR-130a and sex, with males having significantly higher
levels than females (P=0.0027; Fig.
2B) and smoker status (elevated in smokers vs. non-smoker;
P=0.0415; Fig. 2C) in achalasia
patients. However, there were no correlations between miR-130a
expression and the degree of esophageal dilatation or the type of
achalasia (Fig. 2D). In addition,
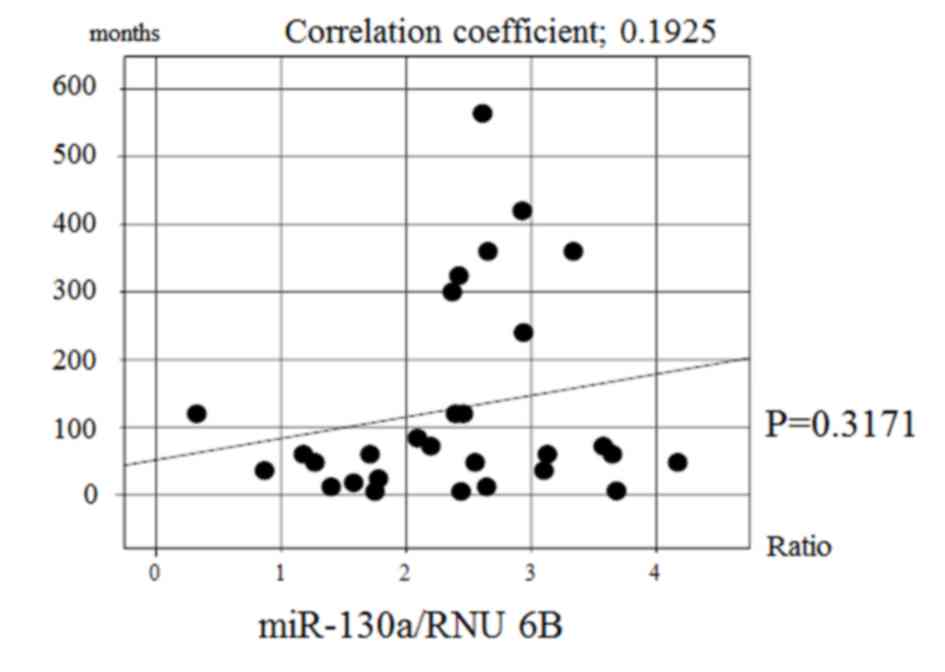
there was no significant correlation between the expression of
miR-130a and the disease duration in achalasia patients (Fig. 3). Multiple regression analysis
demonstrated that there was a significant correlation between
miR-130a expression and smoking (P=0.0084; data not shown).
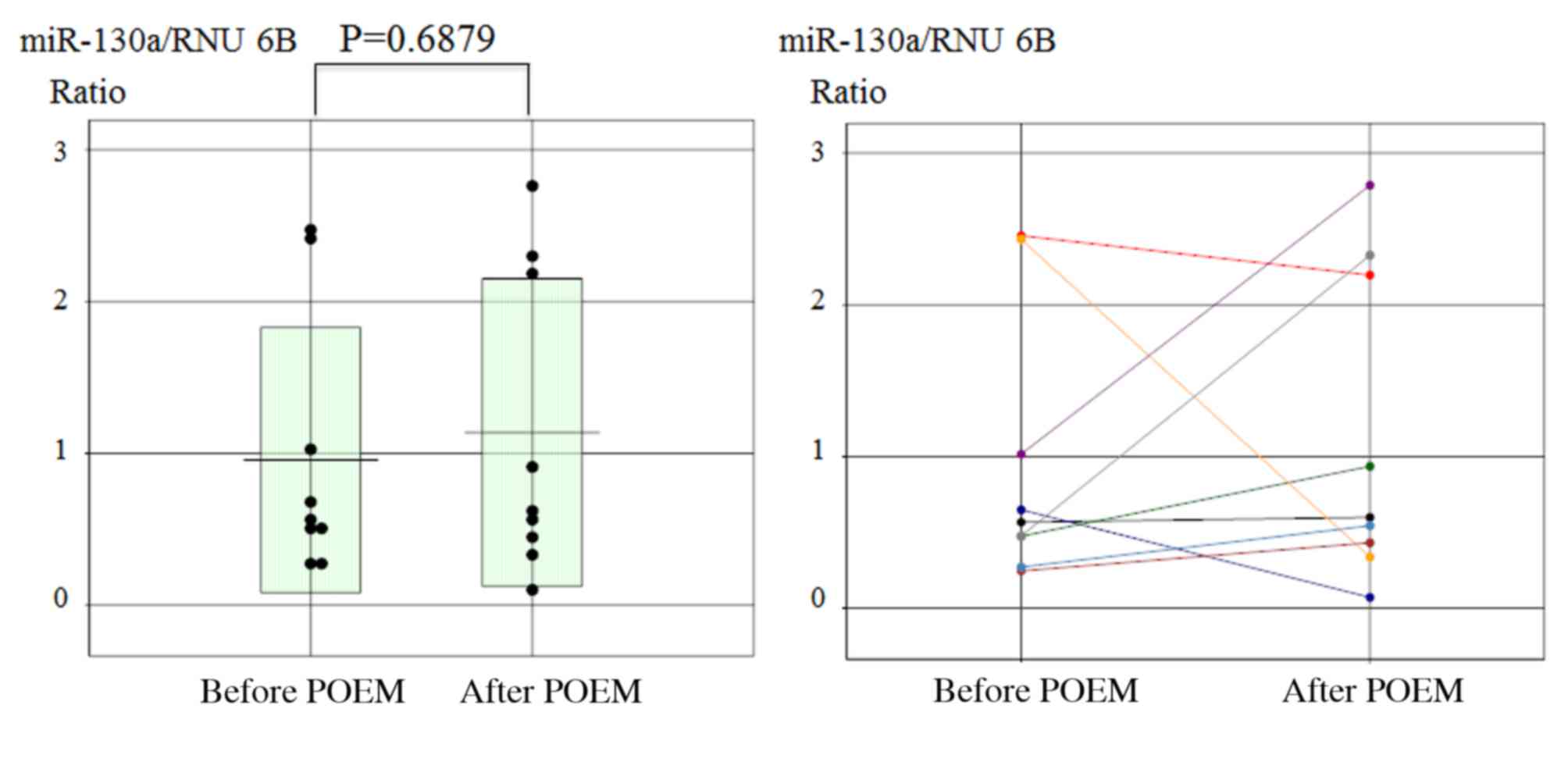
Furthermore, no significant change in miR-130a expression was
observed prior to and following the POEM procedure in achalasia
patients (Fig. 4).
cDNA array analysis results
In order to determine potential target genes of
miR-130a, a cDNA array analysis was performed to determine changes
in gene expression in the mucosa tissue of achalasia patients
compared with the healthy controls. This analysis indicated that
there were 845 genes that were upregulated 1.5-fold and 969 genes
that were downregulated by 1.5-fold in achalasia mucosa compared
with the control. The levels of these genes were substantially
decreased in achalasia patients according to the results of
comprehensive cDNA microarray.
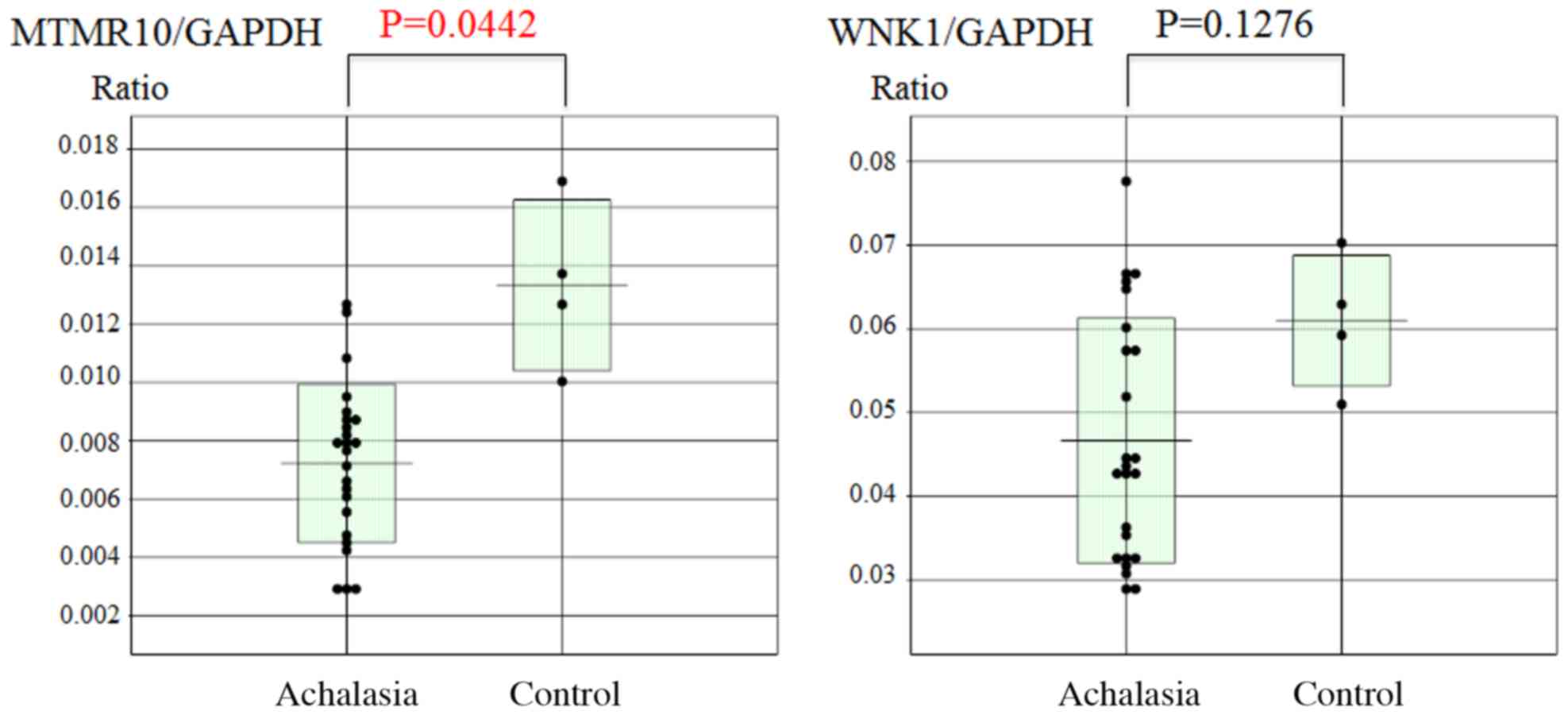
Subsequently, the esophageal mucosal mRNA expression
levels of MTMR10 and WNK1 were analyzed using RT-qPCR. The mucosal
MTMTR10 mRNA levels, but not WNK1, were significantly decreased in
achalasia patients compared with those in the controls (P=0.0442;
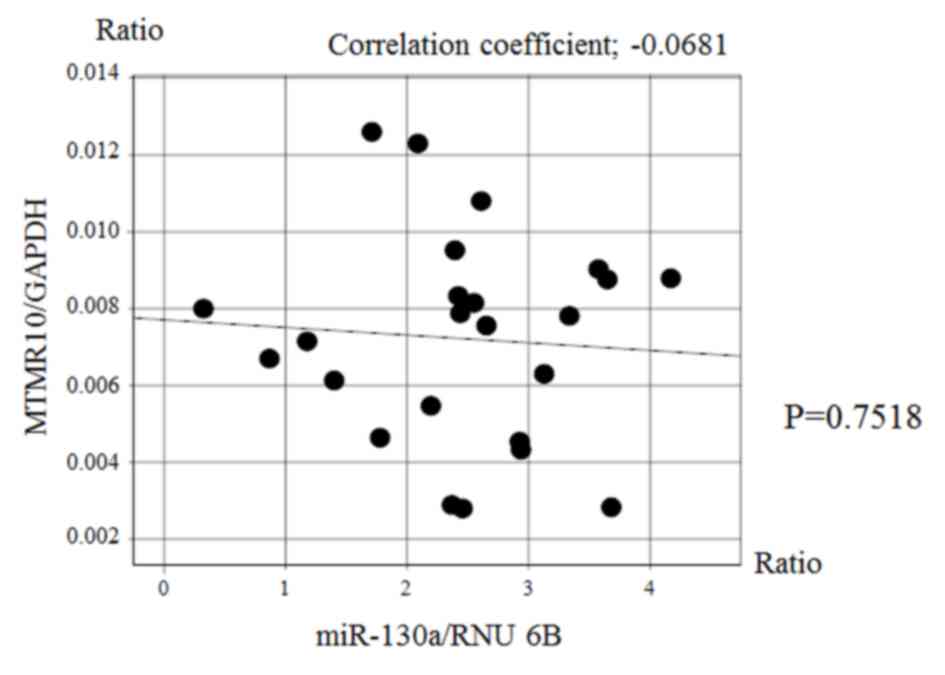
Fig. 5). However, there was no
significant correlation between the expression of miR-130a and that
of MTMR10 (Fig. 6).
Discussion
The present study demonstrated that miR-130a was
highly expressed in the esophageal mucosa of achalasia patients and
that smoking history was associated with a high expression of
miR-130a. The miR-130a gene is located at the chromosomal position
11q12, which is close to the region 11q13 that is frequently
amplified in cancer (26–29). In fact, miR-130a serves an important
role in multiple types of tumors. For instance, miR-130a has been
reported to be overexpressed in non-small-cell lung carcinoma
(30). Increased expression of
miR-130a is strongly associated with lymph node metastasis and poor
prognosis of this carcinoma (30).
By contrast, miR-130a is downregulated in prostate carcinomas and
jointly suppresses two major oncogenic pathways with miR-203 and
miR-205 (31). miR-130a also
increases drug resistance by regulating RUNX3 and Wnt signaling in
cisplatin-treated hepatocellular carcinoma cells (32), while upregulation of miR-130a has
been associated with MDR1/P- glycoprotein-mediated drug resistance
in ovarian cancer cells (33).
Acunzo et al (34) reported
that miR-130a was able to target Met and induce TNF-related
apoptosis-inducing ligand sensitivity in non-small-cell lung
carcinomas by downregulating miR-221 and miR-222.
In the current study, no significant change in
miR-130a expression was observed between before and after POEM.
Although POEM may be one option to reduce patient suffering and
decrease the risk of future carcinogenesis (8), it is unable to completely prevent
achalasia patients from developing esophageal cancer. Several
studies have reported the association of smoking with miRs, as well
as with lung diseases (35) and
various types of cancer (36),
including esophageal cancer (12).
In the present study, the results suggested that smoking history
may be associated with the expression level of miR-130a. However,
we were unable to compare the expression of miR-130a in healthy
non-smokers, healthy smokers, achalasia non-smokers and achalasia
smokers in the current study, and therefore further studies are
warranted.
The myotubularin gene (MTM1) was identified as a
gene mutated in X-linked myotubular myopathy (37). A subgroup of genes in the
myotubularin family encodes proteins that contain substitutions of
residues within the C(X)5R active site motif and are
catalytically inactive. Of the 14 known MTM related (MTMR) human
genes, 6 (MTMR5, MTMR9, MTMR10, MTMR11, MTMR12 and MTMR13) encode
inactive proteins, whereas the function of MTMR10 is largely
unknown (38). In the current study,
miR-130a and MTMR10 expression were not correlated at the mRNA
level; however, it is possible that they are associated at the
protein level. However, the association between eshophageal
achalasia and MTMR10 remains unclear, thus further studies are also
warranted in this regard.
In conclusion, miR-130a is highly expressed in the
esophageal mucosa of esophageal achalasia. Furthermore, smoking
history may be associated with the expression level of miR-130a.
Therefore, miR-130a may be a useful mucosal biomarker of esophageal
achalasia.
References
|
1
|
Leeuwenburgh I, Scholten P, Alderliesten
J, Tilanus HW, Looman CW, Steijerberg EW and Kuipers EJ: Long-term
esophageal cancer risk in patients with primary achalasia: A
prospective study. Am J Gastroenterol. 105:2144–2149. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gockel I, Muller M and Schumacher J:
Achalasia-a disease of unknown cause that is often diagnosed too
late. Dtsch Arztebl Int. 109:209–214. 2012.PubMed/NCBI
|
|
3
|
Zendehdel K, Nyrén O, Edberg A and Ye W:
Risk of esophageal adenocarcinoma in achalasia patients, a
retrospective cohort study in Sweden. Am J Gastroenterol.
106:57–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meijssen MA, Tilanus HW, van Blankenstein
M, Hop WC and Ong GL: Achalasia complicated by esophageal squamous
cell carcinoma: A prospective study in 195 patients. Gut.
33:155–158. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Porschen R, Molsberger G, Kühn A, Sarbia M
and Borchard F: Achalasia-associated squamous cell carcinoma of the
esophagus: Flow-cytometric and histological evaluation.
Gastroenterology. 108:5455491995. View Article : Google Scholar
|
|
6
|
Streitz JM Jr, Ellis FH Jr, Gibb SP and
Heatley GM: Achalasia and squamous cell carcinoma of the esophagus:
Analysis of 241 patients. Ann Thorac Surg. 59:1604–1609. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
West RL, Hirsch DP, Bartelsman JF, de
Borst J, Ferwerda G, Tytqat GN and Boeckxstaens GE: Long term
results of pneumatic dilation in achalasia followed for more than 5
years. Am J Gastroenterol. 97:1346–1351. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Minami H, Yamaguchi N, Matsushima K,
Akazawa Y, Ohnita K, Takeshima F, Nakayama T, Hayashi T, Inoue H,
Nakao K and Isomoto H: Improvement of endocytoscopic findings after
per oral endoscopic myotomy POEM) in esophageal achalasia; does
POEM reduce the risk of developing esophageal carcinoma? Per oral
endoscopic myotomy, endocytoscopy and carcinogenesis. BMC
Gastroenterol. 13:222013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Campos GM, Vittinqhoff E, Rabl C, Takata
M, Gadenstätter M, Lin F and Ciovica R: Endoscopic and surgical
treatments for achalasia: A systematic review and meta-analysis.
Ann Surg. 249:45–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Inoue H, Minami H, Kobayashi Y, Sato Y,
Kaga M, Suzuki M, Satodate H, Okada N, Itoh H and Kudo S: Peroral
endoscopic myotomy (POEM) for esophageal achalasia. Endoscopy.
42:265–271. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Minami H, Isomoto H, Yamaguchi N,
Matsushima K, Akazawa Y, Ohnita K, Takeshima F, Inoue H and Nakao
K: Peroral endoscopic myotomy for esophageal achalasia: Clinical
impact of 28 cases. Dig Endosc. 26:43–51. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsushima K, Isomoto H, Yamaguchi N,
Inoue N, Machida H, Nakayama T, Hayashi T, Kunizaki M, Hidaka S,
Nagayasu T, et al: MiRNA-205 modulates cellular invasion and
migration via regulating zinc finger E-box binding homeobox 2
expression in esophageal squamous cell carcinoma cells. J Transl
Med. 9:302011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmittgen TD: Regulation of microRNA
processing in development, differentiation and cancer. J Cell Mol
Med. 12:1811–1819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Y, Ridzon D, Wong L and Chen C:
Characterization of microRNA expression profiles in normal human
tissues. BMC Genomics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mathé EA, Nguyen GH, Bowman ED, Zhao Y,
Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: Associations with survival. Clin
Cancer Res. 15:6192–6200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lebanony D, Benjamin H, Gilad S, Ezagouri
M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, et
al: Diagnostic assay based on I-miR-205 expression distinguishes
squamous from nonsquamous non-small-cell lung carcinoma. J Clin
Oncol. 27:2030–2037. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kimura S, Naganuma S, Susuki D, Hirono Y,
Yamaguchi A, Fujieda S, Sano K and Itoh H: Expression of microRNAs
in squamous cell carcinoma of human head and neck and the
esophagus: miR-205 and miR-21 are specific markers for HNSCC and
ESCC. Oncol Rep. 23:1625–1633. 2010.PubMed/NCBI
|
|
22
|
Smith CM, Michael MZ, Watson DI, Tan G,
Astill DS, Hummel R and Hussey DJ: Impact of gastro-oesophageal
reflux on microRNA expression, location and function. BMC
Gastroenterol. 13:42013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leeuwenburgh I, Haringsma J, Van Dekken H,
Scholten P, Siersema PD and Kuipers EJ: Long-term risk of
oesophagitis, Barrett's oesophagus and oesophageal cancer in
achalasia patients. Scand J Gastroenterol. Suppl 7–10:2006.
View Article : Google Scholar
|
|
24
|
Descriptive Rules for Achalasia of the
Esophagus. 4th. Japan Society of Esophageal Diseases, Kanehara
& Co., Ltd.; Tokyo: 2012
|
|
25
|
Matsui N, Akahoshi K, Nakamura K, Ihara E
and Kita H: Endoscopic submucosal dissection for removal of
superficial gastrointestinal neoplasms: A technical review. World J
Gastrointest Endosc. 4:123–136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gibcus JH, Menkema L, Mastik MF, Hermsen
MA, de Bock GH, van Velthuysen ML, Takes RP, Kok K, Marcos CA
Alvarez, van der Laan BF, et al: Amplicon mapping and expression
profiling identify the Fas-associated death domain gene as a new
driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin
Cancer Res. 13:6257–6266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reshmi SC, Huang X, Schoppy DW, Black RC,
Saunders WS, Smith DI and Gollin SM: Relationship between FRA11F
and 11q13 gene amplification in oral cancer. Genes Chromosomes
Cancer. 46:143–54. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng CK, Chow LW, Loo WT, Chan TK and
Chan V: The cell cycle checkpoint gene Rad9 is a novel oncogene
activated by 11q13 amplification and DNA methylation in breast
cancer. Cancer Res. 65:8646–8654. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng SL, Stevens VL, Wiklund F, Isaacs
SD, Sun J, Smith S, Pruett K, Wiley KE, Kim ST, Zhu Y, et al: Two
independent prostate cancer risk-associated Loci at 11q13. Cancer
Epidemiol Biomarkers Prev. 18:1815–1820. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y and Gorski DH: Regulation of
angiogenesis through a microRNA (miR-130a) that down-regulates
antiangiogenic homeobox genes GAX and HOXA5. Blood. 111:1217–1226.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boll K, Reiche K, Kasack K, Mörbt N,
Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn
F and Hackermüller J: MiR-130a, miR-203 and miR-205 jointly repress
key oncogenic pathways and are downregulated in prostate carcinoma.
Oncogene. 32:277–285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q
and Zhang J: Upregulated miR-130a increases drug resistance by
regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell.
Biochem Biophys Res Commun. 425:468–272. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Li N, Wang H, Jia X, Wang X and
Luo J: Altered microRNA expression in cisplatin-resistant ovarian
cancer cells and upregulation of miR-130a associated with
MDR1/P-glycoprotein-mediated drug resistance. Oncol Rep.
28:592–600. 2012.PubMed/NCBI
|
|
34
|
Acunzo M, Visone R, Romano G, Veronese A,
Lovat F, Palmieri D, Bottoni A, Garofalo M, Gasparini P, Condorelli
G, et al: miR-130a targets MET and induces TRAIL-sensitivity in
NSCLC by downregulating miR-221 and 222. Oncogene. 31:634–642.
2012.PubMed/NCBI
|
|
35
|
Shi B, Gao H, Zhang T and Cui Q: Analysis
of plasma microRNA expression profiles revealed different cancer
susceptibility in healthy young adult smokers and middle-aged
smokers. Oncotarget. 7:21676–21685. 2016.PubMed/NCBI
|
|
36
|
Mullany LE, Herrick JS, Wolff RK, Stevens
JR and Slattery ML: Assosiation of cigarette smoking and microRNA
expression in rectal cancer: Insight into tumor phenotype. Cancer
Epidemiol. 45:98–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Laporte J, Hu LJ, Kretz C, Mandel JL,
Kioschis P, Coy JF, Kluack SM, Poustka A and Dahl N: A gene mutated
in X-linked myotubular myopathy defines a new putative tyrosine
phosphatase family conserved in yeast. Nat Genet. 13:175–182. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Senderek J, Bergmann C, Weber S, Ketelsen
UP, Schorle H, Rudnik-Shöneborn S, Büttner R, Buchheim E and Zerres
K: Mutation of the SBF2 gene, encoding a novel member of the
myotubularin family, in Charcot-Marie-Tooth neuropathy type
4B2/11p15. Hum Mol Genet. 12:349–356. 2003. View Article : Google Scholar : PubMed/NCBI
|