Introduction
Asthma is a common chronic airway inflammatory
disease characterized by T helper 2 (Th2) cell-mediated
eosinophilic inflammation and airway hyperresponsiveness (AHR)
(1). The World Health Organization
estimates that as many as 300 million people worldwide suffer from
asthma (2). Despite intense ongoing
research, the underlying molecular mechanisms leading to asthma
remain unclear. Although the majority of patients with asthma can
achieve a good level of control with existing treatments, asthma
still has a chronic and long disease course, and the effectiveness
of current treatments is not satisfactory for numerous
patients.
MicroRNAs (miRNAs/miRs) are a large family of
endogenous noncoding RNAs that post-transcriptionally modulate gene
expression by promoting mRNA degradation or inhibiting protein
translation (3). The alteration of
miRNA expression has been implicated in a range of human diseases
(4–6). Emerging evidence has identified that
miRNAs serve an essential role in allergic airway diseases.
Inhibition of miR-126 was demonstrated to suppress the function of
Th2 cells and the development of allergic airway inflammation
(7). In addition, antagonism of
miR-145 has been revealed to inhibit eosinophilic airway
inflammation, Th2 cytokine production and AHR (8). Furthermore, another previous study
identified that anti-let-7 treatment markedly inhibited the
production of allergic cytokines and asthmatic features (9). Based on the aforementioned evidence,
the manipulation of miRNA expression has a potential application
for the treatment of allergic airway diseases.
Attention has recently been focussed on to miR-155,
a multifunctional miRNA, due to its role in multiple physiological
processes in the immune system (10–13).
Data from clinical samples support the theory that miR-155 serves a
critical role in the pathogenesis of allergic diseases, including
allergic rhinitis (14,15) and atopic dermatitis (16). In regards to the lungs, mice
deficient in miR-155 exhibited enhanced airway remodelling
(12) and miR-155 was demonstrated
to contribute to the regulation of allergic airway inflammation by
modulating Th2 responses through the transcription factor PU.1
(17). In addition, miR-155
deficiency has been identified to alleviate allergic airway
inflammation in mice (18). Despite
limited evidence linking miR-155 to the etiology of asthma,
previous studies have evaluated the feasibility of using miR-155
antagonists to treat asthma (19,20).
The present study indicates that miR-155 serves an
important proinflammatory role in the development of AHR and
allergic airway inflammation. Decreasing the level of miR-155 with
small interfering (si)RNA delivered by a lentiviral vector
significantly reduced the severity of inflammatory lesions and AHR.
These findings suggest that a siRNA-expressing lentiviral vector
targeting miR-155 may be a novel approach for the therapy of
asthma.
Materials and methods
Mice
A total of 24 female BALB/c mice (weight, 18–20 g;
4–6 weeks old) were purchased from the Laboratory Animal Center of
Hubei Province (Wuhan, China). All mice were maintained in a vinyl
isolator in a room maintained at a constant temperature (22±2°C)
and humidity (55±5%) on a 12-h light/dark cycle. Mice were provided
with water and food ad libitum. The Institutional Animal
Care and Use Committee of Tongji Hospital (Wuhan, China) approved
the protocols used for animal experiments in the present study.
Construction of short hairpin (sh)RNA
and cloning of shRNAs into lentiviral vectors
The sequence of the miR-155-targeting complementary
shRNA was 5-TAC CCC TAT CAC AAT TAG CAT TAA-3. The negative control
shRNA used was part of a Lenti-KD Custom RNAi commercial kit
provided by Shanghai GeneChem Co., Ltd. (Shanghai, China).
Third-generation human immunodeficiency virus-1-derived lentiviral
vector stocks, pseudotyped with the vesicular stomatitis virus
envelope glycoprotein G, were produced as previously described
(21,22). shRNA lentiviral vectors were produced
by calcium phosphate-mediated transient transfection of HEK 293T
cells (Shanghai GeneChem Co., Ltd. Shanghai, China). Briefly, HEK
293T cells were cultured in DMEM supplemented with 10% FBS in 5%
CO2 at 37°C a humidified atmosphere and 5×106
cells seeded in 100-mm dishes were cotransfected with Lenti-Easy
Packaging Mix (25 µl, 1 µg/µl) and GFP Control Plasmid (20 µl, 0.8
µg/µl) (Shanghai GeneChem Co., Ltd. Shanghai, China). The viruses
were collected and concentrated 100-fold by ultracentrifugation
(4°C at 4,000 × g for 30 min). The concentrated virus stocks were
titered in 5×104 cells/ml HEK 293T cells at 37°C for 72
h based on green florescent protein (GFP) expression.
Generation of asthma model and
lentiviral vector transduction in vivo
To generate an asthmatic model (6 mice per group),
mice were sensitized and challenged with ovalbumin (OVA) as
previously reported (23). The mice
were immunized intraperitoneally on day 0 and 14 with 100 µg OVA
(S7951; Sigma-Aldrich; Merck KGaA; Darmstadt, Germany) and 1 mg
aluminium hydroxide (77161; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in 200 µl of 0.9% saline. The mice were then
challenged by intratracheal administration of 200 µg OVA in 20 µl
saline on days 22, 23 and 24. Solutions at 4×107 titer
units (TU)/ml containing 50 µl of the negative control lentivirus
or the miR-155 shRNA lentivirus (4×107 TU/ml,
~2×106 TU/mouse) were delivered intratracheally 3 days
prior to the first challenge with OVA. Scrambled miR-155 shRNA and
miR-155 shRNA lentiviral vectors contained a sequence encoding GFP.
Transduction efficiency was assessed via florescence microscopy
(magnification, ×100).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from lung tissue using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). cDNA was synthesized with a miRNA-specific primer using the
Fermentas First-Strand Synthesis kit (SuperScript III First-Strand
Synthesis SuperMix for qRT-PCR; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was performed to
determine miR-155 expression and U6 was used as the internal
control, as previously described (24). All primers used were provided by
Guangzhou RiboBio Co., Ltd. (mir-155, miRQ0000165-1-2; U6,
MQP-0201, Guangzhou, China). The primers sequences were not
released. Thermocycling was performed as follows: 60 min at 42°C
and 5 min at 70°C for reverse transcription and denaturation,
respectively, followed by 40 amplification cycles consisting of
95°C for 5 sec (denaturation) and 60°C for 30 sec (extension). The
2−ΔΔCq method (25) was
applied to calculate relative quantification of miRNA
expression.
Measurement of AHR
A total of 24 h after the last challenge, AHR was
measured using a computer-controlled small animal ventilator system
(flexiVent; SCIREQ Scientific Respiratory Equipment, Inc.,
Montreal, Canada) as described by Kramer et al (26). Mice were anesthetized by
intraperitoneal injection of pentobarbital sodium (70–90 mg/kg;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), tracheostomized
with an 18-gauge cannula and mechanically ventilated in a
quasi-sinusoidal fashion with a small animal ventilator at a
frequency of 2.5 Hz and a tidal volume of 12 ml/kg. The airway
resistance values (Raw; cm/H2O.sec/ml) were
recorded in response to increasing doses of nebulized methacholine
(0, 3, 6, 12, 25 mg/ml) (Sigma-Aldrich; Merck KGaA). Results were
expressed for each concentration of methacholine as a percentage of
the 0 mg/ml methacholine Raw value after exposure to
PBS.
Collection of bronchoalveolar lavage
fluid (BALF) and histological analysis
Mice were sacrificed 24 h after the last OVA or
saline challenge. The lungs were lavaged three times with 0.8 ml of
saline, and the collected cells were centrifuged (4°C at 300 × g
for 10 min) and subjected to Wright-Giemsa staining as previously
reported (27). A number of
differential cell counts, including eosinophils, macrophages,
lymphocytes and neutrophils, were performed on total of 200 cells
based on the staining of characteristics of morphology, as
previously described (28).
Supernatant samples were collected for cytokine assays. The left
lungs were isolated and fixed in 4% paraformaldehyde at room
temperature for 24 h, and then embedded in paraffin. Subsequently,
5-µm-thick sections were stained using hematoxylin and eosin
(H&E) and observed under a light microscope (magnification,
×200).
ELISA
Interleukin (IL)-4, IL-5 and IL-13 levels in BALF
were quantified by ELISA (EMC003.48, EMC108.48 and EMC124.48;
Neobioscience, Beijing, China) according to the manufacturer's
protocol.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Data were analyzed with GraphPad Prism software 5.1
(GraphPad Software, Inc., San Diego, CA, USA). Two-way analysis of
variance followed by Bonferroni's post hoc test was used for the
comparison of multiple groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
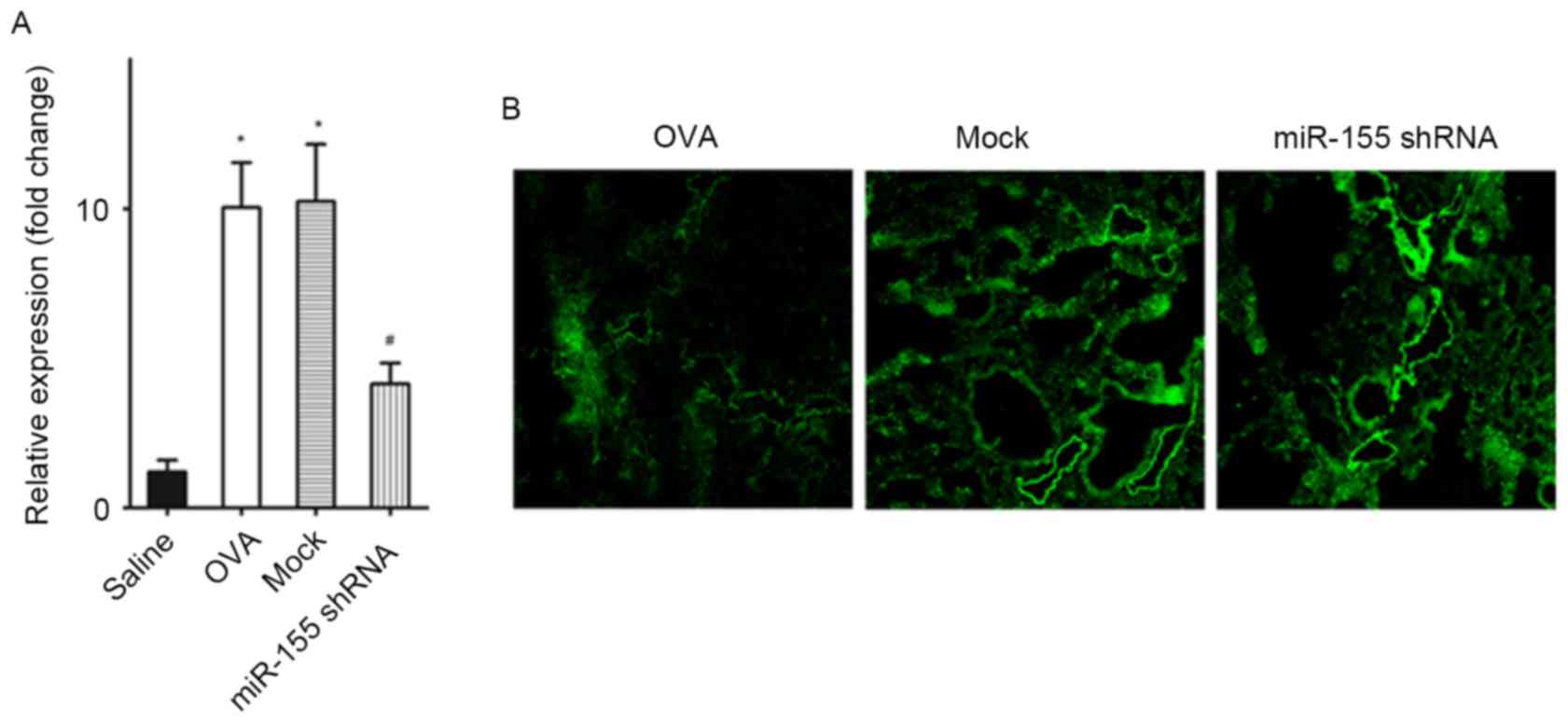
miRNA-155 expression is upregulated in
mice with OVA-induced asthma and miR-155 expression is successfully
silenced by intratracheal instillation of the anti-miR-155 shRNA
lentiviral vector in vivo
The miR-155 expression level in the lungs of the
OVA-sensitized and challenged mice was significantly increased
compared with that in the saline challenged mice (P<0.01;
Fig. 1A). After treatment with the
miR-155 shRNA lentiviral vector, miR-155 expression was
significantly decreased compared with the OVA group and the
negative control shRNA group (the mock group) (both P<0.01;
Fig. 1A). Transduction efficiency
was also assessed by detecting the expression of the marker gene,
GFP (Fig. 1B). The findings
indicated that the transfection efficiency of lentiviral vector was
good.
Delivery of the miR-155 shRNA
lentiviral vector before airway challenge prevents the development
of allergic airway inflammation
The miR-155 shRNA or the negative control shRNA
lentiviral vector were intratracheally administered 72 h before the
first OVA challenge in order to determine the effects of the
miR-155-taregting shRNA lentivirus on airway inflammation. Notably,
miR-155 shRNA lentiviral vector administration significantly
reduced the total cell count of eosinophils, macrophages and
lymphocytes count in the BALF after OVA sensitization and challenge
(P<0.01 vs. the OVA and mock groups), whereas the negative
control shRNA lentivirus treatment did not significantly decrease
the cell counts compared with the OVA group (Fig. 2A). Evidence of inflammatory cell
infiltration and the effects of the miR-155 shRNA lentiviral vector
treatment were further investigated by histologically examining
lung sections stained with H&E. When compared with the mock
group, miR-155 shRNA lentivirus treatment decreased inflammatory
cells infiltration in the lung tissue (Fig. 2B).
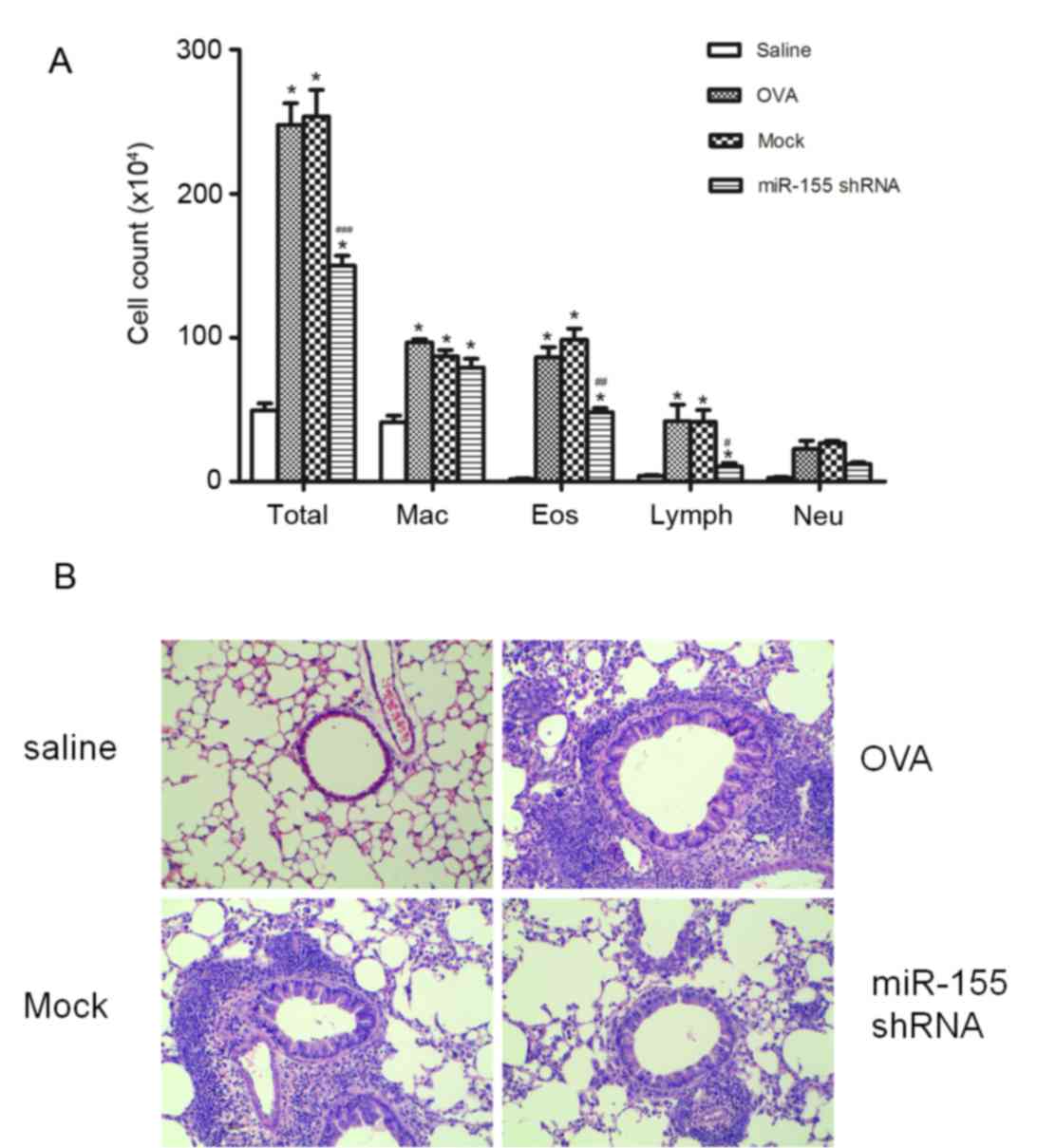 | Figure 2.miR-155 shRNA lentiviral vector
delivery attenuates airway inflammation after OVA challenge. (A)
Differential cell counts for Mac, Eos, Lymph and Neu were
calculated from cytospin preparations of bronchoalveolar lavage
fluid collected 24 h after the last OVA challenge. (B)
Histopathological sections of lungs collected 24 h after the last
OVA challenge, and stained with hematoxylin and eosin
(magnification, ×200). n=5 mice/group. *P<0.01 vs. the saline
group; #P<0.05, ##P<0.01 and
###P<0.001 vs. Mock group. Mac, macrophages; Eos,
eosinophils; Lymph, lymphocytes; Neu, neutrophils; OVA, ovalbumin;
miR-155, microRNA-155; shRNA, short hairpin RNA. |
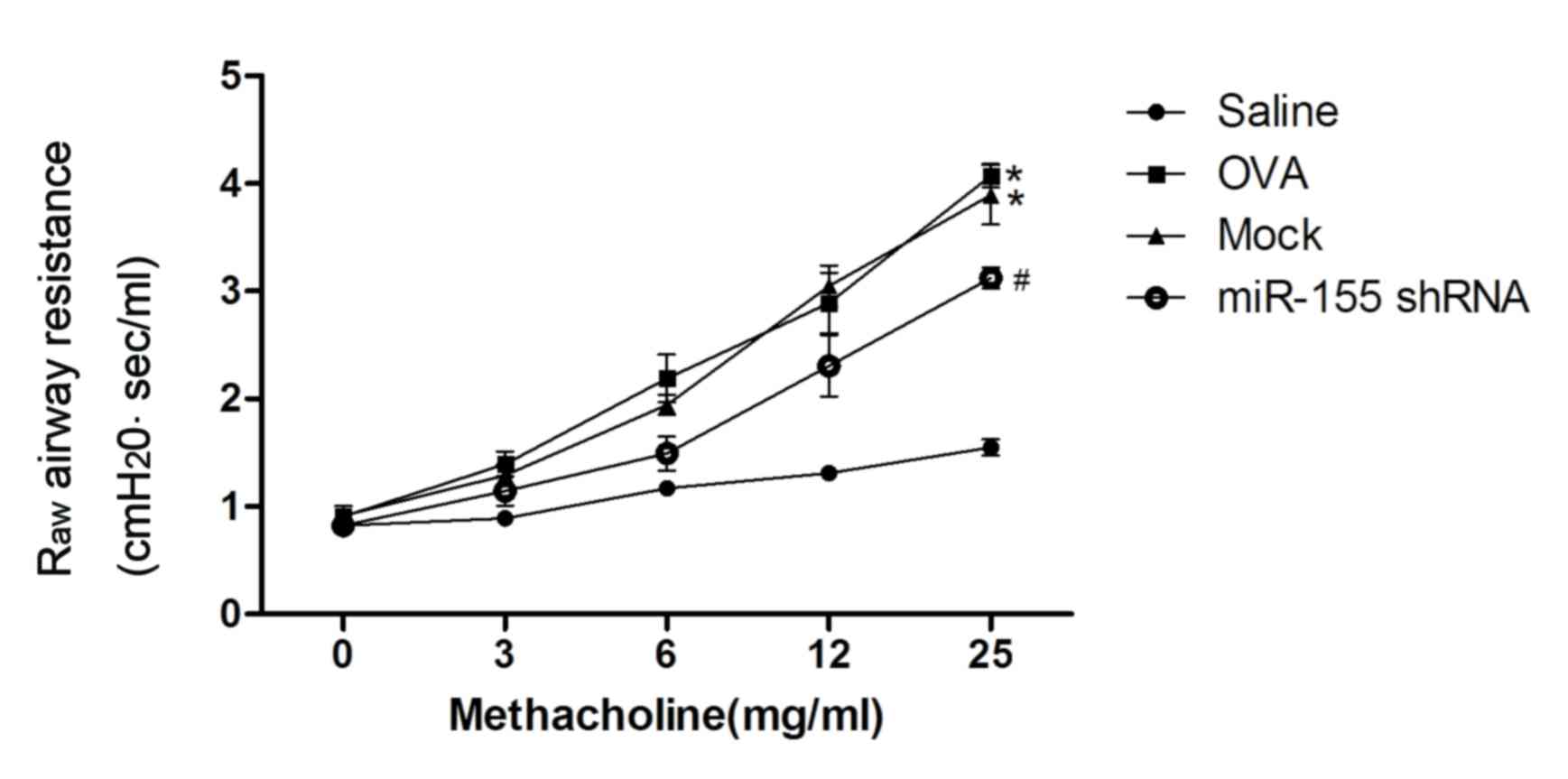
AHR is inhibited by miR-155 shRNA
lentivirus treatment
Administration of the miR-155 shRNA lentiviral
vector was identified to attenuate the development of airway
resistance as a reaction to increasing concentrations of inhaled
methacholine. Raw was significantly decreased in the
miR-155 shRNA group compared with the mock and OVA groups
(P<0.01; Fig. 3).
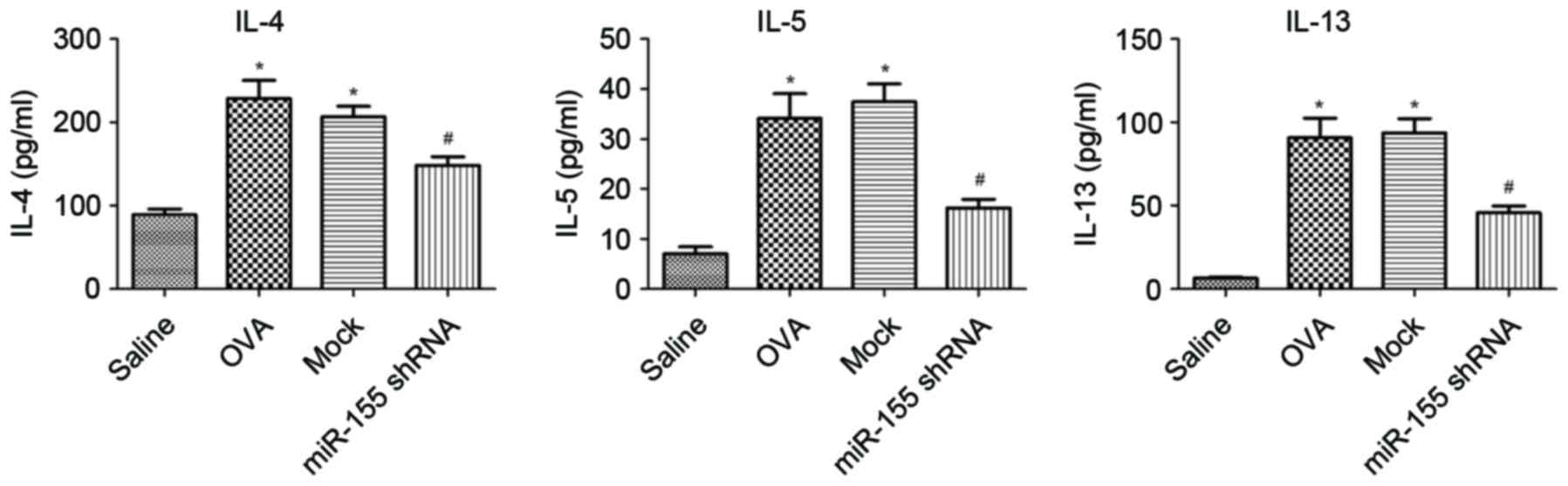
Th2 cytokine expression in BALF
decreases after intratracheal instillation of the miR-155 shRNA
lentiviral vector
The expression of IL-4, IL-5 and IL-13 in the BALF
was examined. The levels of IL-4, IL-5 and IL-13 were significantly
upregulated in the OVA group compared with the saline group, and
decreased significantly after treatment with miR-155 shRNA
lentivirus compared with that of the OVA and mock groups (all
P<0.01; Fig. 4).
Discussion
The role of miRNAs in the pathogenesis of allergic
asthma has been widely studied since 2007 (7–9,12,16,29) and
therapies that target miRNAs were considered to be important for
further study (30,31). The results of the present study
indicate that miR-155 is associated with the development of
allergic airway inflammation; miR-155 expression was significantly
higher in the lungs of mice with asthma compared with normal
controls. In addition, treatment with a miR-155-targeting shRNA
lentiviral vector in vivo substantially alleviated airway
inflammation and AHR due to miR-155 inhibition.
Previous microarray analysis has demonstrated that
miR-155 is upregulated in OVA-induced mouse models of asthma
(9,32). The present study confirmed that the
expression of miR-155 was significantly increased in the lungs of
mice with asthma by RT-qPCR. These data indicate that the
upregulation of miR-155 is associated with the pathogenesis of
asthma. Certain previous studies have revealed that higher miR-155
expression exists in the smooth muscle cells of asthmatic airways
(33) and bronchial epithelial cells
(34), which is partly consistent
with the results of the present study. However, further exploration
of the function of miR-155 in other cell types will be important to
understand the role of miR-155 in the development of the allergic
immune response.
Mice deficient in miR-155 have been reported to be
protected against allergen-induced eosinophilic airway inflammation
(17,18). In the present study, an shRNA was
applied as a therapeutic strategy to suppress miR-155 expression,
which successfully reduced inflammation, infiltration of
eosinophils, and the production of the Th2 cytokines IL-4, IL-5 and
IL-13. In addition, miR-155-targeting shRNA inhibited AHR, another
important feature of asthma (35).
These results support the suggested role of miR-155 in the
development of allergic airway inflammation. However, the precise
mechanism by which miR-155 regulates AHR remains to be clarified.
Interestingly, another group demonstrated that miR-155-deficient
mice spontaneously exhibited airway remodelling (12). A possible explanation for the
discrepancy between this study and the present study could be the
different mouse model used. An acute asthma mouse model was
employed in the present study, whereas in the study conducted by
Rodriguez et al (12), aging
mice were used. Airway remodeling may follow acute inflammation or
begin insidiously as a low-grade smoldering response (36). It is unknown whether airway
inflammation contributes to airway remodelling. Previous clinical
data has demonstrated that anti-inflammatory treatment may have no
effect on airway remodeling in patients with asthma (37,38).
These results suggest a paradoxical role of miR-155 in airway
inflammation and airway remodeling, and support the complex role of
miRNA in the development of asthma.
The knockdown of miRNA is an effective method to use
when characterizing the functions of miRNAs in vivo.
Antagomirs, chemically modified oligonucleotides complementary to
individual miRNAs, are widely used to transiently inhibit miRNA
function (39). However, this method
has several limitations. It is difficult to directly measure the
depletion of an miRNA, because the antisense oligonucleotide binds
to the miRNA and sequesters it from its target rather than inducing
its degradation. The lentiviral vector-mediated gene delivery
system also has some advantages over antagomirs, including a high
efficiency of gene transduction into a wide variety of cells,
including dividing and non-dividing cells, and long-term infection
(40,41). Lentiviral vectors were used to
knockdown miR-155, stably and specifically, in vivo. The
lentivirus encoding miR-155 shRNA used in the present study
successfully downregulated miR-155 expression. However, it was not
clear what the target cells for anti-miR-155 were in the present
study. A deficiency in miR-155 reduces cluster of differentiation
(CD)4+ T cell activation and transcription factor
expression in the lungs (17). In
addition, miR-155 deficient dendritic cells (DCs) have been
demonstrated to exhibit limited Th2 priming capacity and thus
failed to induce airway inflammation (18). These results indicate that
suppression of miR-155 expression, through anti-miR-155 treatment,
in CD4+ cells and/or DCs in vivo may contribute
to the attenuation of asthmatic features in mouse models of asthma.
Furthermore, other cell types, including bronchial epithelial cells
and airway smooth muscle cells, may be target cells for
anti-miR-155 treatment.
miRNAs are believed to function in vivo by
targeting multiple functionally related proteins or a key protein
target (42,43). Several target genes for miR-155 have
been identified, including transcription factor PU.1,
activation-induced cytidine deaminase, suppressor of cytokine
signaling 1 and inositol polyphosphate-5-phosphatase D (44,45).
Rodriguez et al (12)
confirmed that MAF bZIP transcription factor was a direct target of
miR-155 in T cells. However, in the present study, the proteins
that were targeted by miR-155 were not identified.
In conclusion, the present study identified that
miR-155 was significantly upregulated in an OVA-induced mouse model
of asthma, and that inhibition of miR-155 using a lentiviral vector
alleviated airway inflammation, AHR and Th2 cytokine release. These
results highlight the important role of miR-155 in the pathogenesis
of asthma and that miR-155 may serve as a novel target for the
treatment of allergic inflammatory diseases.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81570024).
References
|
1
|
Barnes PJ: Immunology of asthma and
chronic obstructive pulmonary disease. Nat Rev Immunol. 8:183–192.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bousquet J, Mantzouranis E, Cruz AA,
Aït-Khaled N, Baena-Cagnani CE, Bleecker ER, Brightling CE, Burney
P, Bush A, Busse WW, et al: Uniform definition of asthma severity,
control, and exacerbations: Document presented for the World Health
Organization consultation on severe asthma. J Allergy Clin Immunol.
126:926–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Du C, Liu C, Kang J, Zhao G, Ye Z, Huang
S, Li Z, Wu Z and Pei G: MicroRNA miR-326 regulates TH-17
differentiation and is associated with the pathogenesis of multiple
sclerosis. Nat Immunol. 10:1252–1259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thum T, Gross C, Fiedler J, Fischer T,
Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et
al: MicroRNA-21 contributes to myocardial disease by stimulating
MAP kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mattes J, Collison A, Plank M, Phipps S
and Foster PS: Antagonism of microRNA-126 suppresses the effector
function of TH2 cells and the development of allergic airways
disease. Proc Natl Acad Sci USA. 106:18704–18709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Collison A, Mattes J, Plank M and Foster
PS: Inhibition of house dust mite-induced allergic airways disease
by antagonism of microRNA-145 is comparable to glucocorticoid
treatment. J Allergy Clin Immunol. 128:160–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Polikepahad S, Knight JM, Naghavi AO, Oplt
T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, et
al: Proinflammatory role for let-7 microRNAS in experimental
asthma. J Biol Chem. 285:30139–30149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lind EF and Ohashi PS: Mir-155, a central
modulator of T-cell responses. Eur J Immunol. 44:11–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vigorito E, Kohlhaas S, Lu D and Leyland
R: miR-155: An ancient regulator of the immune system. Immunol Rev.
253:146–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rodriguez A, Vigorito E, Clare S, Warren
MV, Couttet P, Soond DR, van Dongen S, Grococ RJ, Das PP, Miska EA,
et al: Requirement of bic/microRNA-155 for normal immune function.
Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Seddiki N, Brezar V, Ruffin N, Lévy Y and
Swaminathan S: Role of miR-155 in the regulation of lymphocyte
immune function and disease. Immunology. 142:32–38. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suojalehto H, Lindström I, Majuri ML,
Mitts C, Karjalainen J, Wolff H and Alenius H: Altered microRNA
expression of nasal mucosa in long-term asthma and allergic
rhinitis. Int Arch Allergy Immunol. 163:168–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suojalehto H, Toskala E, Kilpeläinen M,
Majuri ML, Mitts C, Lindström I, Puustinen A, Plosila T, Sipilä J,
Wolff H and Alenius H: MicroRNA profiles in nasal mucosa of
patients with allergic and nonallergic rhinitis and asthma. Int
Forum Allergy Rhinol. 3:612–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sonkoly E, Janson P, Majuri ML, Savinko T,
Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al:
MiR-155 is overexpressed in patients with atopic dermatitis and
modulates T-cell proliferative responses by targeting cytotoxic T
lymphocyte-associated antigen 4. J Allergy Clin Immunol.
126(581–589): e1–e20. 2010.
|
|
17
|
Malmhäll C, Alawieh S, Lu Y, Sjöstrand M,
Bossios A, Eldh M and Rådinger M: MicroRNA-155 is essential for
T(H)2-mediated allergen-induced eosinophilic inflammation in the
lung. J Allergy Clin Immunol. 133(1429–1438): e1–e7. 2014.
|
|
18
|
Zech A, Ayata CK, Pankratz F, Meyer A,
Baudiß K, Cicko S, Yegutkin GG, Grundmann S and Idzko M:
MicroRNA-155 modulates P2R signaling and Th2 priming of dendritic
cells during allergic airway inflammation in mice. Allergy.
70:1121–1129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matsukura S, Osakabe Y, Sekiguchi A, Inoue
D, Kakiuchi Y, Funaki T, Yamazaki Y, Takayasu H, Tateno H, Kato E,
et al: Overexpression of microRNA-155 suppresses chemokine
expression induced by Interleukin-13 in BEAS-2B human bronchial
epithelial cells. Allergol Int. (65 Suppl):S17–S23. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Plank MW, Maltby S, Tay HL, Stewart J,
Eyers F, Hansbro PM and Foster PS: MicroRNA expression is altered
in an ovalbumin-induced asthma model and targeting miR-155 with
antagomirs reveals cellular specificity. PLoS One. 10:e1448102015.
View Article : Google Scholar
|
|
21
|
Naldini L, Blömer U, Gallay P, Ory D,
Mulligan R, Gage FH, Verma IM and Trono D: In vivo gene delivery
and stable transduction of nondividing cells by a lentiviral
vector. Science. 272:263–267. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee CC, Huang HY and Chiang BL:
Lentiviral-mediated GATA-3 RNAi decreases allergic airway
inflammation and hyperresponsiveness. Mol Ther. 16:60–65. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang A, Wang Z, Cao Y, Cheng S, Chen H,
Bunjhoo H, Xie J, Wang C, Xu Y and Xiong W: CCL2/CCR2-dependent
recruitment of Th17 cells but not Tc17 cells to the lung in a
murine asthma model. Int Arch Allergy Immunol. 166:52–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kramer EL, Mushaben EM, Pastura PA,
Acciani TH, Deutsch GH, Hershey Khurana GK, Korfhagen TR, Hardie
WD, Whitsett JA and Le Cras TD: Early growth response-1 suppresses
epidermal growth factor receptor-mediated airway
hyperresponsiveness and lung remodeling in mice. Am J Respir Cell
Mol Biol. 41:415–425. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen H, Xu X, Teng J, Cheng S, Bunjhoo H,
Cao Y, Liu J, Xie J, Wang C, Xu Y and Xiong W: CXCR4 inhibitor
attenuates allergen-induced lung inflammation by down-regulating
MMP-9 and ERK1/2. Int J Clin Exp Pathol. 8:6700–6707.
2015.PubMed/NCBI
|
|
28
|
Chen H, Cheng S, Wang A, Bunjhoo H, Cao Y,
Xie J, Wang C, Xu Y and Xiong W: IL-21 does not involve in
OVA-induced airway remodeling and chronic airway inflammation. Int
J Clin Exp Med. 8:10640–10645. 2015.PubMed/NCBI
|
|
29
|
Lu TX, Munitz A and Rothenberg ME:
MicroRNA-21 is up-regulated in allergic airway inflammation and
regulates IL-12p35 expression. J Immunol. 182:4994–5002. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharma A, Kumar M, Ahmad T, Mabalirajan U,
Aich J, Agrawal A and Ghosh B: Antagonism of mmu-mir-106a
attenuates asthma features in allergic murine model. J Appl Physiol
(1985). 113:459–464. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qin HB, Xu B, Mei JJ, Li D, Liu JJ, Zhao
DY and Liu F: Inhibition of miRNA-221 suppresses the airway
inflammation in asthma. Inflammation. 35:1595–1599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garbacki N, Di Valentin E, Huynh-Thu VA,
Geurts P, Irrthum A, Crahay C, Arnould T, Deroanne C, Piette J,
Cataldo D and Colige A: MicroRNAs profiling in murine models of
acute and chronic asthma: A relationship with mRNAs targets. PLoS
One. 6:e165092011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Comer BS, Camoretti-Mercado B, Kogut PC,
Halayko AJ, Solway J and Gerthoffer WT: Cyclooxygenase-2 and
microRNA-155 expression are elevated in asthmatic airway smooth
muscle cells. Am J Respir Cell Mol Biol. 52:438–447. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuo YC, Li YS, Zhou J, Shih YR, Miller M,
Broide D, Lee OK and Chien S: Human mesenchymal stem cells suppress
the stretch-induced inflammatory miR-155 and cytokines in bronchial
epithelial cells. PLoS One. 8:e713422013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Busse WW: The relationship of airway
hyperresponsiveness and airway inflammation: Airway
hyperresponsiveness in asthma: Its measurement and clinical
significance. Chest. 138 (2 Suppl):4S–10S. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vignola AM, Mirabella F, Costanzo G, Di
Giorgi R, Gjomarkaj M, Bellia V and Bonsignore G: Airway remodeling
in asthma. Chest. 123 (3 Suppl):417S–422S. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao JM, Cai F, Peng M, Ma Y and Wang B:
Montelukast improves air trapping, not airway remodeling, in
patients with moderate-to-severe asthma: A pilot study. Chin Med J
(Engl). 126:2229–2234. 2013.PubMed/NCBI
|
|
38
|
White SR and Dorscheid DR:
Corticosteroid-induced apoptosis of airway epithelium: A potential
mechanism for chronic airway epithelial damage in asthma. Chest.
122 (6 Suppl):278S–284S. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meister G, Landthaler M, Dorsett Y and
Tuschl T: Sequence-specific inhibition of microRNA- and
siRNA-induced RNA silencing. RNA. 10:544–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Abbas-Terki T, Blanco-Bose W, Déglon N,
Pralong W and Aebischer P: Lentiviral-mediated RNA interference.
Hum Gene Ther. 13:2197–2201. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matta H, Hozayev B, Tomar R, Chugh P and
Chaudhary PM: Use of lentiviral vectors for delivery of small
interfering RNA. Cancer Biol Ther. 2:206–210. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao C and Rajewsky K: MicroRNA control in
the immune system: Basic principles. Cell. 136:26–36. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hoefig KP and Heissmeyer V: MicroRNAs grow
up in the immune system. Curr Opin Immunol. 20:281–287. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
O'Connell RM, Rao DS, Chaudhuri AA and
Baltimore D: Physiological and pathological roles for microRNAs in
the immune system. Nat Rev Immunol. 10:111–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Teng G and Papavasiliou FN: Shhh!
silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci.
364:631–637. 2009. View Article : Google Scholar : PubMed/NCBI
|