Introduction
Despite considerable progresses in diagnostic and
therapeutic approaches in recent years, breast cancer remains the
most common malignant tumor in women worldwide (1). In the United States of America, breast
cancer accounts for ~29% of all new cancer cases in women annually
(2). Tumor invasion and metastasis
are multistep, dynamic and complex processes, which contribute to
the vast majority of breast cancer-associated mortalities (3). Previous studies have demonstrated that
B-cell lymphoma 6 (BCL6) serves an important role in breast cancer
invasion and metastasis (4–6). The oncogenic transcriptional modulator
BCL6 is a master regulator of B-lymphocyte development and growth,
and, in addition to facilitating the proliferation of B-lymphocytes
and inhibiting their differentiation into plasma and memory B cells
(7), BCL6 has been reported to
prevent the differentiation of mammary cells (8) and stimulate the oncogenicity of human
breast cancer cells (4).
The zinc finger E-box-binding homeobox (ZEB) family
includes ZEB1 and ZEB2, which are important nuclear transcription
factors and have been reported to be key factors in
epithelial-mesenchymal transition (EMT) (9,10). EMT
is recognized as an important process in the invasion and
metastasis of cancer cells (11),
and loss of the epithelial marker E-cadherin is considered to be a
hallmark of EMT. ZEB1 and ZEB2 are able to induce the initiation of
EMT by binding to the E-box sequence on the E-cadherin promoter,
thus repressing its expression (12). A recent study demonstrated that BCL6
facilitates EMT by enhancing the ZEB1-mediated transcriptional
repression of E-cadherin, promoting the invasion, migration and
growth of breast cancer cells (13).
The role of BCL6, ZEB1 and ZEB2 in different tumor types have been
widely studied; however, little is currently known about the
effects of and relationships between their expressions in breast
cancer.
In the present study, the mRNA and protein
expression of BCL6, ZEB1 and ZEB2 was assessed using in situ
hybridization and immunohistochemical (IHC) analysis, respectively,
in the breast tissue of 228 patients with breast cancer and 80
patients with benign breast disease. The association between BCL6,
ZEB1 and ZEB2 expression with the clinicopathological
characteristics and prognosis of patients with breast cancer was
also investigated. Finally, the association between the expression
of BCL6, ZEB1 and ZEB2 in breast cancer was assessed.
Materials and methods
Patients and specimens
A total of 228 consecutive female patients with
breast cancer (range, 30–81 years; mean, 51.0 years) and 80
consecutive female patients with benign breast disease (range,
18–57 years; mean, 37.5 years) were enrolled in the present study,
all of whom underwent radical mastectomy or modified radical
mastectomy at The Second People's Hospital of Hefei (Hefei, China)
or the First Affiliated Hospital of Anhui Medical University
(Anhui, China) between May 2002 and November 2014. Patients with
breast cancer who underwent chemotherapy or radiation therapy prior
to surgery, or who had rheumatic disease, acute infection, human
immunodeficiency virus infection or other types of cancer were
excluded. Complete follow-up data was available for 197 patients,
with a median follow-up time of 60 months. The histological grade
of cancerous tissues was based on the World Health Organization
classification of tumors (14), and
the pathological tumor stage was defined according to the sixth
edition of the tumor-node-metastasis classification of the
International Union Against Cancer (15). Estrogen receptor, progesterone
receptor and erb-b2 receptor tyrosine kinase 2 (c-erbB-2)
expression levels were obtained from patients' pathology records.
Levels were measured by immunohistochemistry scoring: For ER and PR
expression, a percentage of stained tumor cells >5% was
considered positive (16); for
c-erbB-2 expression, membrane staining intensity and pattern were
measured as follows: 0, completely negative or membrane positivity
in <10% of tumor cells; +, >10% of tumor cells had incomplete
faint membrane positivity; ++, >10% of tumor cells had complete
moderate membrane positivity; and +++, >10% of tumor cells had
complete strong circumferential membrane positivity (17). All tissue specimens collected during
surgery were formalin-fixed, paraffin-embedded and cut into
4-µm-thick sections. The protocol used in the present study was
approved by the Institutional Review Boards of the Second People's
Hospital of Hefei and the First Affiliated Hospital of Anhui
Medical University, and informed consent was provided by all
patients.
In situ hybridization
In situ hybridization analyses of BCL6, ZEB1
and ZEB2 mRNA expression was performed using human BCL6, ZEB1 and
ZEB2 ISH detection kit (MK1301-h, MK3730-h, and MK3731-h, Boshide
Biotech Co., Ltd., Wuhan, Hubei, China). Three digoxin-labeled
antisense oligonucleotide DNA probes for human BCL6, ZEB1 and ZEB2
were obtained from Boshide Biotech Co., Ltd. (Wuhan, China). The
probe sequences were as follows: BCL6,
5′-GACAGCTGTATCCAGTTCACCCGCCATGCCAGTGA-3′,
5′-TTCTATAGCATCTTTACAGACCAGTTGAAATGCAA-3′, and
5′-ATCCTGCAGATGGAGCATGTTGTGGACAGTTGCCG-3′; ZEB1,
5′-TGTAATCGTAAATTCAAATGCACTGAGTGTGGAAA-3′,
5′-TGGTTTGAAAAGATGCAAGCTGGACAGATTTCAGT-3′, and
5′-TATTCTCAACACATGAATCATCGCTACTCCTACTG-3′; and ZEB2,
5′-TACTATGCTATGAACATGGAGCCCAACTCCGATGA-3′,
5′-AAGGAATTTTCAAATTCAAATAATCTGGACAACAA-3′, and
5′-ATGAACCGGGCTTACTTGCAGAGCATTACCCCTCA-3′. All the materials used
for in situ hybridization were autoclaved and treated with
0.1% diethyl pyrocarbonate-double distilled water
(DEPC-ddH2O) for 24 h at room temperature, and all
solutions were prepared with 0.1% DEPC-ddH2O. Sections
were deparaffinized in xylene, rehydrated in a graded series of
ethanol solutions and incubated with 3% hydrogen peroxide for 10
min at room temperature. Sections were subsequently digested with
3% pepsin for 20 min at 37°C and rinsed with PBS three times (5
min/wash). A total of 50 µl of pre-hybridization solution (provided
with antibodies) was added to each section, and the sections were
incubated at 42°C for 4 h. The pre-hybridization solution was
removed and replaced with 50 µl of hybridization solution with
probes (or without probes for the negative control samples;
provided with antibodies), and the sections were incubated at 42°C
for 20 h. Slides were subsequently washed (5 min/wash) twice with
2X sodium chloride-sodium citrate (SSC), three times with 0.5X SSC
and three times with 0.02X SSC at 37°C. The washed sections were
blocked with serum-blocking solution for 30 min at 37°C and
incubated with mouse anti-digoxin antibodies against the following;
BCL6 (cat. no. MK1301-h), ZEB1 (cat. no. MK3730-h) and ZEB2: (cat.
no. MK3731-h; all 1:200; all Boshide Biotech Co., Ltd.) for 1 h at
37°C, following which they were washed with PBS three times (5
min/wash). Sections were subsequently incubated with
streptavidin-biotin-peroxidase complex solution (provided with
antibodies) for 20 min at 37°C, washed with PBS three times (5
min/wash) and further incubated with biotin-peroxidase solution for
20 min at 37°C (provided with antibodies). Sections were washed
three times with PBS (5 min/wash), stained with
3,3′-diaminobenzidine (DAB) solution for 5 min at room temperature,
and counterstained with hematoxylin solution for 3–5 min at room
temperature.
IHC analysis
IHC analyses of BCL6, ZEB1 and ZEB2 protein
expression was performed using a two-step immunohistochemical
staining kit (Shanghai Changdao Biotech Co., Ltd., Shanghai, China)
according to the manufacturer's protocol with rabbit anti-BCL6
polyclonal antibody (1:100; TA350798; Origene Technologies, Inc.,
Rockville, MD, USA), mouse anti-ZEB1 monoclonal antibody (1:200;
TA802313; Origene Technologies, Inc.), mouse anti-ZEB2 monoclonal
antibody (1:100; TA802113; Origene Technologies, Inc.). The
sections were deparaffinized in xylene and rehydrated in a graded
series of ethanol solutions. For antigen retrieval, slides were
heated in a microwave oven (800 W) in 0.01 M sodium citrate buffer
(pH 6.0) for 20 min. Slides were subsequently cooled in sodium
citrate buffer and immersed in 3% hydrogen peroxide in methanol for
10 min to block endogenous peroxidase activity. Slides were washed
with PBS three times (2 min/wash) and incubated with the primary
antibodies (as above) at 4°C overnight. Subsequently, the slides
were washed with PBS three times (2 min/wash), incubated for 20 min
with universal (anti-mouse/rabbit) horseradish peroxidase
conjugated detection reagent (D-3004; Shanghai Changdao Biotech
Co., Ltd.) at 37°C, washed again with PBS (three times; 2 min/wash)
and incubated with DAB solution (Shanghai Changdao Biotech Co.,
Ltd) for 5 min at room temperature. All slides were counterstained
with hematoxylin for 3–5 min at room temperature. Known positive
samples were used as the positive controls, and for the negative
controls, the primary antibody was replaced with 0.01 mol/l
PBS.
Scoring of stained sections
The mRNA and protein expression of BCL6, ZEB1, and
ZEB2 in breast tissue specimens were reviewed and scored
independently by two pathologists under double-blind conditions.
The stained sections were scored according to the staining
intensity and the amount of stained cells, as described previously
(18,19). Briefly, samples where <10% of the
breast cancer cells were stained with any intensity were considered
negative for expression of the mRNA/protein, whereas in samples
where ≥10% of the cancer cells were stained with any intensity were
considered as positive for expression of the mRNA/protein.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 13.0; SPSS, Inc., Chicago, IL, USA). The χ2 test
was used to examine the difference in the positive expression rate
of mRNA/protein between the groups. Kaplan-Meier survival curves
were constructed to determine patient overall survival (OS) and
relapse-free survival (RFS), and the variables associated with OS
and RFS rates were compared with a log-rank test. Spearman's rank
correlation coefficient analysis was used to determine the
correlation between the expression rate of mRNA/protein and
different clinicopathological parameters. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of BCL6, ZEB1 and ZEB2 mRNA
and protein is significantly increased in breast cancer tissue
compared with benign breast disease tissue
The mRNA expression of BCL6, ZEB1 and ZEB2 was
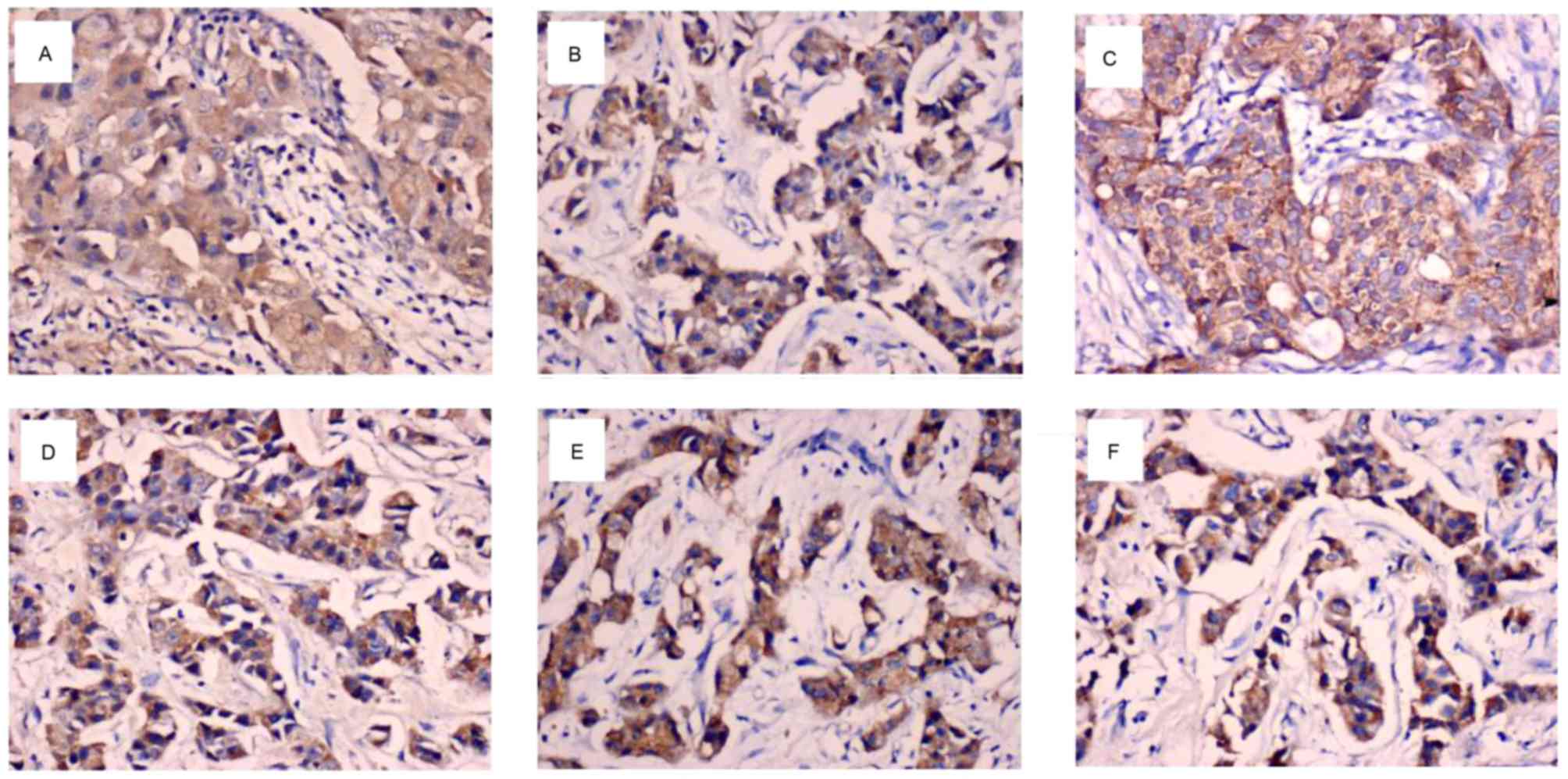
primarily localized to the cytoplasm and/or nucleus of breast
cancer cells (Fig. 1A-C). As
presented in Table I, the number of
patients with mRNA expression of BCL6, ZEB1 or ZEB2 was
significantly higher in the breast cancer group compared with the
benign breast disease group (P=0.001, 0.002 and 0.002,
respectively).
 | Table I.Expression of BCL6, ZEB1 and ZEB2 mRNA
in breast cancer and benign breast disease tissue. |
Table I.
Expression of BCL6, ZEB1 and ZEB2 mRNA
in breast cancer and benign breast disease tissue.
|
|
| BCL6 expression | ZEB1 expression | ZEB2 expression |
|---|
|
|
|
|
|
|
|---|
| Group P-value |
| Total no. of patients
n (%) |
| n (%) P-value |
| P-value | n (%) |
|---|
| Breast cancer | 228 | 123 (53.9) | 0.001 | 129 (56.6) | 0.002 | 119 (52.2) | 0.002 |
| Benign breast
disease | 80 | 25
(31.3) |
| 29
(36.3) |
| 26
(32.5) |
|
BCL6, ZEB1 and ZEB2 proteins were predominantly
expressed in the cytoplasm and/or the nucleus of the breast cancer
cells (Fig. 1D-F). As illustrated in
Table II, protein expression of
BCL6, ZEB1 or ZEB2 was significantly higher in breast cancer
tissues compared with benign breast disease tissues (P=0.002, 0.001
and 0.001, respectively).
 | Table II.Expression of BCL6, ZEB1 and ZEB2
protein in breast cancer and benign breast disease tissue. |
Table II.
Expression of BCL6, ZEB1 and ZEB2
protein in breast cancer and benign breast disease tissue.
|
|
| BCL6 expression | ZEB1 expression | ZEB2
expression |
|---|
|
|
|
|
|
|
|---|
| Group | Total no. of
patients | n (%) | P-value | n (%) | P-value | n (%) | P-value |
|---|
| Breast cancer | 228 | 125 (54.8) | 0.002 | 136 (59.6) | 0.001 | 126 (55.3) | 0.001 |
| Benign breast
disease | 80 | 28
(35.0) |
| 28
(35.0) |
| 26
(32.5) |
|
Clinical significance of BCL6, ZEB1
and ZEB2 expression in patients with breast cancer
The association between BCL6, ZEB1 and ZEB2 mRNA
expression and the clinicopathological characteristics of the
patients with breast cancer was investigated (Table III). The expression of BCL6 and
ZEB1 mRNA was significantly positively associated with tumor size
(P=0.02 and P=0.007, respectively), lymph node metastasis (P=0.038
and P=0.028, respectively), and a higher tumor stage (P=0.001).
ZEB2 mRNA levels were significantly positively associated with
tumor size (P=0.016), a higher tumor stage (P=0.002) and c-erbB-2
expression (P=0.022). No significant associations were found
between BCL6/ZEB1 mRNA expression and patient age, tumor grade, and
estrogen receptor, progesterone receptor and c-erbB-2 expression.
No significant associations were found between ZEB2 mRNA expression
and patient age, lymph node metastasis, tumor grade, or estrogen
receptor and progesterone receptor expression.
 | Table III.Association between BCL6, ZEB1 and
ZEB2 mRNA expression and the clinicopathological characteristics of
patients with breast cancer. |
Table III.
Association between BCL6, ZEB1 and
ZEB2 mRNA expression and the clinicopathological characteristics of
patients with breast cancer.
|
|
| BCL6
expression | ZEB1
expression | ZEB2
expression |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Total no. of
patients | n (%) | P-value | n (%) | P-value | n (%) | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
|
≤35 | 22 | 14 (63.6) | 0.56 | 16 (72.7) | 0.266 | 14 (63.6) | 0.341 |
|
35–55 | 131 | 71 (54.2) |
| 71 (54.2) |
| 70 (53.4) |
|
|
>55 | 75 | 38 (50.7) |
| 42 (56.0) |
| 35 (46.7) |
|
| Tumor size
(cm) |
|
|
|
|
|
|
|
| ≤2 | 19 | 5
(26.3) | 0.02 | 5
(26.3) | 0.007 | 4
(21.1) | 0.016 |
|
2–5 | 164 | 89
(54.3) |
| 93 (56.7) |
| 89 (54.3) |
|
|
>5 | 45 | 29 (64.4) |
| 31 (68.9) |
| 26 (57.8) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
| 0 | 82 | 36 (43.9) | 0.038 | 39 (47.6) | 0.028 | 36 (43.9) | 0.127 |
|
1–3 | 78 | 43 (55.1) |
| 43 (55.1) |
| 42 (53.8) |
|
|
>3 | 68 | 44 (64.7) |
| 47 (69.1) |
| 41 (60.3) |
|
| Tumor grade |
|
|
|
|
|
|
|
| I | 19 | 8
(42.1) | 0.142 | 8
(42.1) | 0.101 | 8
(42.1) | 0.561 |
| II | 141 | 72 (51.1) |
| 76 (53.9) |
| 73 (51.8) |
|
|
III | 68 | 43 (63.2) |
| 45 (66.2) |
| 38 (55.9) |
|
| Tumor stage |
|
|
|
|
|
|
|
| I |
9 | 1
(11.1) | 0.001 | 0 (0) | 0.001 | 1
(11.1) | 0.002 |
| II | 110 | 50 (45.5) |
| 53 (48.2) |
| 50 (45.5) |
|
|
III | 98 | 65 (66.3) |
| 68 (69.4) |
| 59 (60.2) |
|
| IV | 11 | 7
(63.6) |
| 8
(72.7) |
| 9
(81.8) |
|
| Estrogen
receptor |
|
|
|
|
|
|
|
| − | 136 | 73 (53.7) | 0.921 | 80 (58.8) | 0.406 | 78 (57.4) | 0.058 |
| + | 92 | 50 (54.3) |
| 49 (53.3) |
| 41 (44.6) |
|
| Progesterone
receptor |
|
|
|
|
|
|
|
| − | 136 | 73 (53.7) | 0.921 | 83 (61.0) | 0.099 | 76 (55.9) | 0.175 |
| + | 92 | 50 (54.3) |
| 46 (50.0) |
| 43 (46.7) |
|
| c-erbB-2 |
|
|
|
|
|
|
|
|
Low | 148 | 75 (50.7) | 0.178 | 79 (53.4) | 0.185 | 69 (46.6) | 0.022 |
|
High | 80 | 48 (60.0) |
| 50 (62.5) |
| 50 (62.5) |
|
The correlation between BCL6, ZEB1 and ZEB2 protein
expression and the clinicopathological features of breast cancer
were also investigated. As presented in Table IV, a significant positive
association was identified between BCL6 protein expression and
tumor size (P=0.019), lymph node metastasis (P=0.003), a higher
tumor grade (P=0.01), a higher tumor stage (P=0.001) and c-erbB-2
expression (P=0.023). ZEB1 and ZEB2 protein expression were
significantly positively associated with tumor size (P=0.02 and
P=0.007, respectively), lymph node metastasis (P=0.001 and P=0.008,
respectively) and a higher tumor stage (P=0.001; Table IV). Furthermore, a significant
positive association was observed between ZEB2 protein expression
and c-erbB-2 expression (P=0.03). No significant associations were
identified between BCL6 protein expression and patient age,
estrogen receptor expression or progesterone receptor expression.
No significant associations were observed between ZEB1 and ZEB2
protein expression and patient age, tumor grade, estrogen receptor
expression or and progesterone receptor expression, and there was
no significant association between ZEB1 protein expression and
c-erbB-2 expression.
 | Table IV.Association between BCL6, ZEB1 and
ZEB2 protein expression an the clinicopathological characteristics
of patients with breast cancer. |
Table IV.
Association between BCL6, ZEB1 and
ZEB2 protein expression an the clinicopathological characteristics
of patients with breast cancer.
|
|
| BCL6
expression | ZEB1
expression | ZEB2
expression |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Total no. of
patients | n (%) | P-value | n (%) | P-value | n (%) | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
|
≤35 | 22 | 15 (68.2) | 0.288 | 16 (72.7) | 0.334 | 13 (59.1) | 0.902 |
|
35–55 | 131 | 67 (51.1) |
| 74 (56.5) |
| 71 (54.2) |
|
|
>55 | 75 | 43 (57.3) |
| 46 (61.3) |
| 42 (56.0) |
|
| Tumor size
(cm) |
|
|
|
|
|
|
|
| ≤2 | 19 | 5
(26.3) | 0.019 | 6
(31.6) | 0.02 | 4
(21.1) | 0.007 |
|
2–5 | 164 | 91 (55.5) |
| 99 (60.4) |
| 95 (57.9) |
|
|
>5 | 45 | 29 (64.4) |
| 31 (68.9) |
| 27 (60.0) |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
| 0 | 82 | 34 (41.5) | 0.003 | 36 (43.9) | 0.001 | 36 (43.9) | 0.008 |
|
1–3 | 78 | 44 (56.4) |
| 49 (62.9) |
| 43 (55.1) |
|
|
>3 | 68 | 47 (69.1) |
| 51 (75.0) |
| 47 (69.1) |
|
| Tumor grade |
|
|
|
|
|
|
|
| I | 19 | 6
(31.6) | 0.01 | 10 (52.6) | 0.391 | 7
(36.8) | 0.15 |
| II | 141 | 73 (51.8) |
| 81 (57.4) |
| 77 (54.6) |
|
|
III | 68 | 46 (67.6) |
| 45 (66.2) |
| 42 (61.8) |
|
| Tumor stage |
|
|
|
|
|
|
|
| I |
9 | 1
(11.1) | 0.001 | 1
(11.1) | 0.001 | 1
(11.1) | 0.001 |
| II | 110 | 49 (44.5) |
| 51 (46.4) |
| 49 (44.5) |
|
|
III | 98 | 67 (68.4) |
| 75 (76.5) |
| 68 (69.4) |
|
| IV | 11 | 8
(72.7) |
| 9
(81.8) |
| 8
(72.7) |
|
| Estrogen
receptor |
|
|
|
|
|
|
|
| − | 136 | 78 (57.4) | 0.351 | 83 (61.0) | 0.605 | 81 (59.6) | 0.113 |
| + | 92 | 47 (51.1) |
| 53 (57.6) |
| 45 (48.9) |
|
| Progesterone
receptor |
|
|
|
|
|
|
|
| − | 136 | 80 (58.8) | 0.14 | 88 (64.7) | 0.058 | 80 (58.8) | 0.189 |
| + | 92 | 45 (48.9) |
| 48 (52.2) |
| 46 (50.0) |
|
| c-erbB-2 |
|
|
|
|
|
|
|
|
Low | 148 | 73 (49.3) | 0.023 | 82 (55.4) | 0.076 | 74 (50.0) | 0.03 |
|
High | 80 | 52 (65.0) |
| 54 (67.5) |
| 52 (65.0) |
|
BCL6, ZEB1 and ZEB2 protein expression
is associated with a significantly lower OS and RFS of patients
with breast cancer
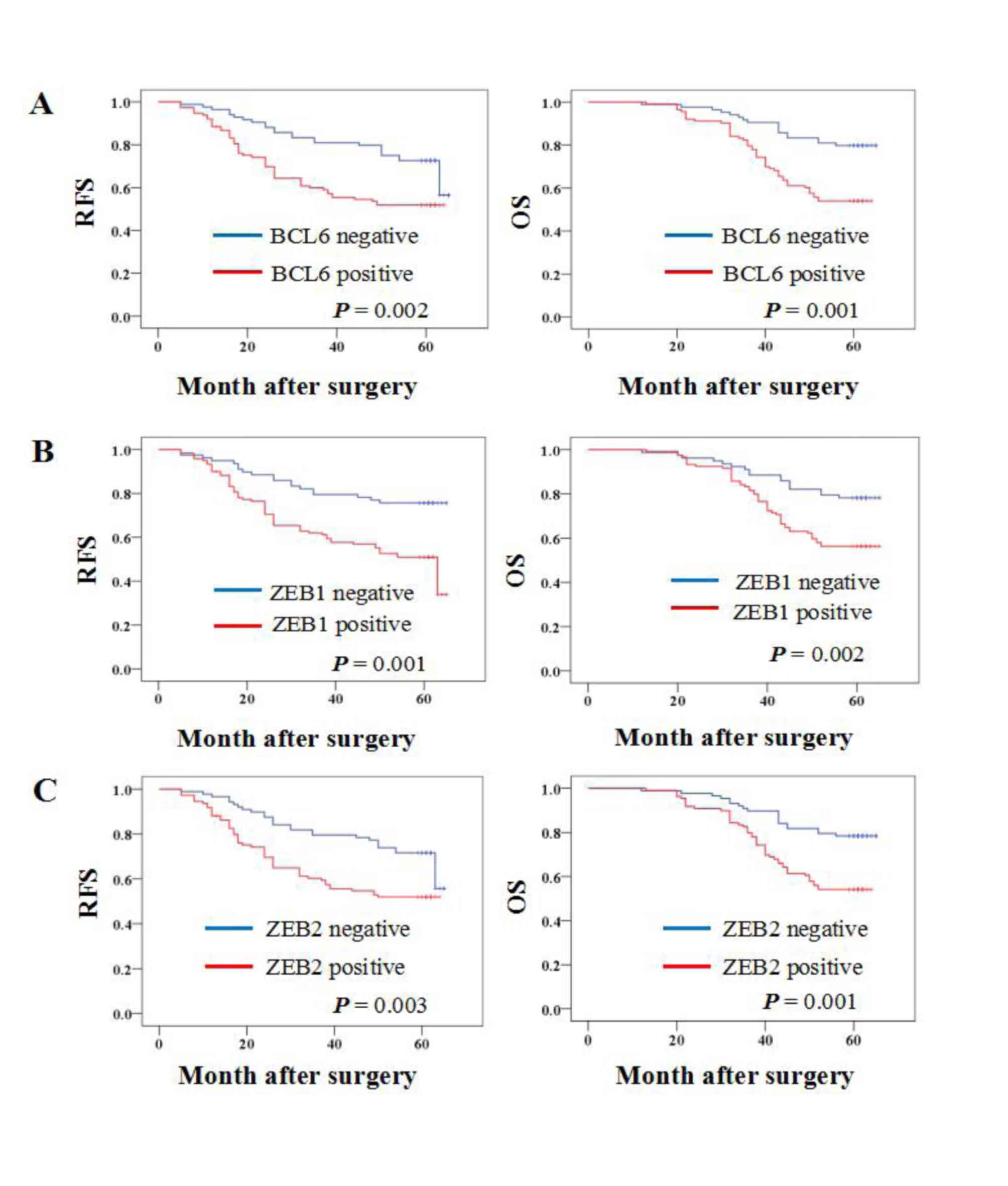
Kaplan-Meier survival analysis was performed to
determine whether BCL6, ZEB1 or ZEB2 protein expression was
associated with the OS or RFS of patients with breast cancer. The
results demonstrated that patients with BCL6, ZEB1 and ZEB2
protein-positive primary tumors had a significantly lower OS rate
(P=0.001, 0.002 and 0.001, respectively) and RFS rate (P=0.002,
0.001 and 0.003, respectively) (Fig.
2).
Correlation between BCL6, ZEB1 and
ZEB2 expression in breast cancer tissues
Spearman's rank correlation coefficient analysis
demonstrated that BCL6 mRNA expression was positively correlated
with ZEB1 (rs=0.326; P<0.001) and ZEB2
(rs=0.382; P<0.001) mRNA expression (data not
shown). A significant correlation was also identified between BCL6
protein expression and ZEB1 (rs=0.449;
P<0.001) and ZEB2 (rs=0.669; P<0.001)
protein expression (data not shown).
Discussion
BCL6 is a transcriptional repressor that serves an
oncogenic role in B cell lymphoma (20). Through binding to specific DNA
sequences, BCL6 regulates the transcription of a variety of genes
associated with B-cell development, differentiation and activation,
and, due to a functional mutation in the BCL6 promoter, BCL6 is
overexpressed in patients with diffuse large B cell lymphoma
(DLBCL) (21). It has been suggested
that BCL6 may serve a role in the progression of breast cancer.
BCL6 is expressed in the mammary epithelial cells of non-pregnant
animals, in addition to during early pregnancy (8). Although BCL6 protein is rarely
expressed in the normal mammary epithelium, it is overexpressed in
breast cancers, particularly high-grade ductal breast cancer
(22). ZEB1 and ZEB2 are members of
the ZEB family, and several lines of evidence have suggested that
ZEB1 and ZEB2 are inducers of EMT through regulating E-cadherin
expression, consequently promoting cancer progression (12,23–25). EMT
is a characteristic feature of aggressive metastatic cancers, and
is important for invasion and metastasis during cancer progression
(26,27).
In the present study, it was demonstrated that the
mRNA and protein expression of BCL6, ZEB1 and ZEB2 were
significantly higher in breast cancer tissues compared with benign
breast disease tissues. In addition, BCL6, ZEB1 and ZEB2 expression
was significantly positively associated with tumor size, lymph node
metastasis and a higher tumor stage. Furthermore, Kaplan-Meier
survival analyses revealed an association between BCL6, ZEB1 and
ZEB2 protein expression and poor OS and RFS in patients with breast
cancer. These data suggest that BCL6, ZEB1 and ZEB2 serve important
roles in breast cancer progression, and may therefore be useful
biomarkers for predicting patient survival.
Previous studies have reported altered expression of
BCL6, ZEB1 and ZEB2 in several types of tumor (28–30).
Overexpression of BCL6 has been observed in 40% of patients with
DLBCL (28), and Jia et al
(29) reported that overexpression
of ZEB1 may be associated with the occurrence and development, in
addition to the invasion and metastasis, of gastric carcinoma.
Prislei et al (30)
demonstrated that ZEB2 serves a role in ovarian cancer cell
migration and identified that high expression of ZEB2 mRNA was
significantly correlated with poor prognosis (OS and
progression-free survival) in a series of 143 patients with ovarian
cancer. These data are consistent with the results of the present
study. However, Pinto et al (18) reported that BCL6 protein expression
was significantly lower in metastatic lymph node tumors compared
with the corresponding primary breast cancer. The reason for this
is unclear and requires further research.
The results of the present study identified that the
mRNA and protein expression of BCL6 was significantly positively
associated with ZEB1 and ZEB2 mRNA and protein expression in breast
cancer tissues. This suggests that BCL6 is a potential regulator of
ZEB1 and ZEB2. Although the roles of BCL6, ZEB1 and ZEB2 in
different types of tumor have been widely studied, little is
currently known about their association with one another. Yu et
al (13) investigated the
effects of BCL6 in the regulation of EMT and the mechanisms
underlying this, and reported that BCL6 promoted EMT via enhancing
the ZEB1-mediated transcriptional repression of E-cadherin in
breast cancer cells. Brabletz et al (31) identified that ZEB1 and miR-200 family
members repressed the expression of one another in a reciprocal
feedback loop; this loop reportedly controls the Notch signaling
pathway in cancer cells. Nishijima et al (32) suggested that the miR200/ZEB axis
regulates sensitivity to nintedanib in non-small cell lung cancer
cells. Gregory et al (33)
reported a strong protein and mRNA expression correlation between
ZEB family members and transforming growth factor-β, and negative
mRNA expression correlations between miR-200 and TGF-β, and miR-200
and ZEB family members, in invasive ductal carcinomas. However,
further studies are required to determine whether there is a
correlation between BCL6 and miR-200.
In conclusion, the results of the present study
indicate that BCL6, ZEB1 and ZEB2 are potential biomarkers for
predicting the invasion, metastasis and prognosis of breast cancer,
and suggest that BCL6 may serve as a regulator of the ZEB family
(ZEB1 and ZEB2). Further studies are warranted to elucidate the
regulatory mechanisms underlying the association between BCL6 and
ZEB1/ZEB2 expression.
Acknowledgements
The present study was supported by the Fifth Cycle
Of Medical Key Specialist Construction Funds Of Hefei [grant no.
2016 (256)] and the Indigenous Innovation Policies Program of Hefei
[grant no. 2014 (71)-7)].
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nola S, Sin S, Bonin F, Lidereau R and
Driouch K: A methodological approach to unravel organ-specific
breast cancer metastasis. J Mammary Gland Biol Neoplasia.
17:135–145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu Q, Liu X, Yan H, He YH, Ye S, Cheng XW,
Zhu GL, Wu WY, Wang XN, Kong XJ, et al: B-cell lymphoma 6 protein
stimulates oncogenicity of human breast cancer cells. BMC Cancer.
14:4182014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan H, Zhao M, Huang S, Chen P, Wu WY,
Huang J, Wu ZS and Wu Q: Prolactin inhibits BCL6 expression in
breast cancer cells through a microRNA-339-5p-dependent pathway. J
Breast Cancer. 19:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walker SR, Liu S, Xiang M, Nicolais M,
Hatzi K, Giannopoulou E, Elemento O, Cerchietti L, Melnick A and
Frank DA: The transcriptional modulator BCL6 as a molecular target
for breast cancer therapy. Oncogene. 34:1073–1082. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shaffer AL, Yu X, He Y, Boldrick J, Chan
EP and Staudt LM: BCL-6 represses genes that function in lymphocyte
differentiation, inflammation, and cell cycle control. Immunity.
13:199–212. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Logarajah S, Hunter P, Kraman M, Steele D,
Lakhani S, Bobrow L, Venkitaraman A and Wagner S: BCL-6 is
expressed in breast cancer and prevents mammary epithelial
differentiation. Oncogene. 22:5572–5578. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eger A, Aigner K, Sonderegger S, Dampier
B, Oehler S, Schreiber M, Berx G, Cano A, Beug H and Foisner R:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Comijin J, Berx G, Vermassen P,
Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D
and van Roy F: The two-handed E box binding zinc finger protein
SIP1 downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guarino M, Rubino B and Ballabio G: The
role of epithelial-mesenchymal transition in cancer pathology.
Pathology. 39:305–318. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu JM, Sun W, Hua F, Xie J, Lin H, Zhou DD
and Hu ZW: BCL6 induces EMT by promoting the ZEB1-mediated
transcription repression of E-cadherin in breast cancer cells.
Cancer Lett. 365:190–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tavassoli FA and Devilee P: World Health
Organization Classification Of Tumors: Pathology And Genetics Of
Tumors Of The Breast And Female Genital Organs. Lyon: IARC Press;
pp. 13–59. 2003
|
|
15
|
Sobin LH and Wittekind C: TNM
classification of malignant tumours. 6th. New York: Wiley; 2002
|
|
16
|
Yoder BJ, Tso E, Skacel M, Pettay J, Tarr
S, Budd T, Tubbs RR, Adams JC and Hicks DG: The expression of
fascin, an actin-bundling motility protein, correlates with hormone
receptor-negative breast cancer and a more aggressive clinical
course. Clin Cancer Res. 11:186–192. 2005.PubMed/NCBI
|
|
17
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American Society of
Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pinto AE, André S, Silva G, Vieira S,
Santos AC, Dias S and Soares J: BCL-6 oncoprotein in breast cancer:
Loss of expression in disease progression. Pathobiology.
76:235–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kurahara H, Takao S, Maemura K, Mataki Y,
Kuwahata T, Maeda K, Ding Q, Sakoda M, Iino S, Ishigami S, et al:
Epithelial-mesenchymal transition and mesenchymal-epithelial
transition via regulation of ZEB-1 and ZEB-2 expression in
pancreatic cancer. J Surg Oncol. 105:655–661. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Basso K and Dalla-Favera R: Roles of BCL6
in normal and transformed germinal center B cells. Immunol Rev.
247:172–183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan S, Cermak L, Pagan JK, Rossi M,
Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R and Pagano
M: FBXO11 targets BCL6 for degradation and is inactivated in
diffuse large B-cell lymphomas. Nature. 481:90–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bos R, van Diest PJ, van der Groep P,
Greijer AE, Hermsen MA, Heijnen I, Meijer GA, Baak JP, Pinedo HM,
van der Wall E and Shvarts A: Protein expression of B-cell lymphoma
gene 6 (BCL-6) in invasive breast cancer is associated with cyclin
D1 and hypoxia-inducible factor-1alpha (HIF-1alpha). Oncogene.
22:8948–8951. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
You J, Li Y, Fang N, Liu B, Zu L, Chang R,
Li X and Zhou Q: MiR-132 suppresses the migration and invasion of
lung cancer cells via targeting the EMT regulator ZEB2. PLoS One.
9:e918272014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long non-coding RNA urothelial cancer-associated 1 promotes
bladder cancer cell migration and invasion by way of the
hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu DI, Liu L, Ren C, Kong D, Zhang P, Jin
X, Wang T and Zhang G: Epithelial-mesenchymal interconversions and
the regulatory function of the ZEB family during the development
and progression of ovarian cancer. Oncol Lett. 11:1463–1468.
2016.PubMed/NCBI
|
|
26
|
Bates RC and Mercurio AM: The
epithelial-mesenchymal transition (EMT) and colorectal cancer
progression. Cancer Biol Ther. 4:365–370. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xue C, Plieth D, Venkov C, Xu C and
Neilson EG: The gatekeeper effect of epithelial-mesenchymal
transition regulates the frequency of breast cancer metastasis.
Cancer Res. 63:3386–3394. 2003.PubMed/NCBI
|
|
28
|
Lossos IS, Jones CD, Warnke R, Natkunam Y,
Kaizer H, Zehnder JL, Tibshirani R and Levy R: Expression of a
single gene, BCL-6, strongly predicts survival in patients with
diffuse large B-cell lymphoma. Blood. 98:945–951. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jia B, Liu H, Kong Q and Li B:
Overexpression of ZEB1 associated with metastasis and invasion in
patients with gastric carcinoma. Mol Cell Biochem. 366:223–229.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prislei S, Martinelli E, Zannoni GF,
Petrillo M, Filippetti F, Mariani M, Mozzetti S, Raspaglio G,
Scambia G and Ferlini C: Role and prognostic significance of the
epithelial-mesenchymal transition factor ZEB2 in ovarian cancer.
Oncotarget. 6:18966–18979. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brabletz S, Bajdak K, Meidhof S, Burk U,
Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J
and Brabletz T: The ZEB1/miR-200 feedback loop controls Notch
signalling in cancer cells. EMBO J. 30:770–782. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
MiR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016.PubMed/NCBI
|
|
33
|
Gregory PA, Bracken CP, Smith E, Bert AG,
Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, et al:
An autocrine TGF-beta/ZEB/miR-200 signaling network regulates
establishment and maintenance of epithelial-mesenchymal transition.
Mol Biol Cell. 22:1686–1698. 2011. View Article : Google Scholar : PubMed/NCBI
|