Introduction
Staphylococcus aureus (S. aureus) is a
major causative agent of various infectious diseases worldwide,
ranging from furuncles to endocarditis, toxic shock syndrome and
sepsis (1,2). Of the pathogens most frequently
isolated from patients with bloodstream infections (BSIs), S.
aureus, which is common among neonates and elderly patients
(3,4), ranked third (11.4%) in 2012 in China
(5). Furthermore,
methicillin-resistant S. aureus (MRSA) strains accounted for
44.6 and 42.2% of BSIs in China in 2014 and 2015, respectively
(6,7). Invasive MRSA infection has been
identified as an independent risk factor for poor prognosis and is
associated with a significant increase in the duration of
hospitalization (8,9). Although MRSA-ST239-SCCmec III
represents the most common clone in China, others are also in
circulation, whose antimicrobial resistance profiles differ
(10). Thus, the provision of data
concerning antibiotic resistance is important to enable clinicians
to choose appropriate treatments.
S. aureus produces a wide variety of
virulence factors, including pan-valentine leucocidin (PVL), toxic
shock syndrome toxin-1, and staphylococcal enterotoxins (SEs),
which facilitate bacterial colonization of host tissues and evasion
of host immune responses, resulting in disease. MRSA appears to
harbour more virulence genes than other types. In a study by Yu
et al (1), the frequencies of
virulence genes, including the PVL and SEs genes, in S.
aureus isolated from BSIs in the MRSA group were higher than
those in the methicillin-susceptible S. aureus (MSSA) group.
The course of an infection may be affected by the genes encoding
these virulence factors; given their diversity and great
variability (11), toxin gene
profiling is of importance.
Conserved virulence factor B (CvfB) is a novel
virulence-associated protein that acts via activation of the
accessory gene regulator (AGR) locus, or through an AGR-independent
pathway, to control exoprotein production. Deletion of CvfB results
in decreased production of hemolysin, DNase and protease, and
reduced virulence (12,13). SEs are pyrogenic toxin superantigens
that are thought to be a leading cause of foodborne illness and
toxic shock (14). Staphylococcal
enterotoxin Q (SEQ) and staphylococcal enterotoxin K (SEK) are
newly described SEs with demonstrable superantigenic activity that
may cause severe gastroenteritis, nausea and vomiting (15–17). To
the best of our knowledge, carriage of CvfB by a clinical strain of
S. aureus and the prevalence of SEQ and SEK among S.
aureus isolates from paediatric patients with BSIs in China
have not yet been reported. Therefore, the present study examined
the prevalence of these three genes among isolates of this
bacterium from Chinese paediatric patients with BSIs.
Materials and methods
Bacterial strains
Between 2012 and 2015, a total of 53 clinical S.
aureus isolates were recovered from blood cultures derived from
paediatric patients with BSIs attending Guangzhou Women and
Children's Medical Center (Guangzhou, China). To avoid duplication,
strains consecutively isolated from the same patient were excluded.
All the isolates were stored at −70°C. The present study was
approved by the Ethics Committee of Guangzhou Women and Children's
Medical Center (Guangzhou, China; no. 2016081019). Informed consent
was obtained from the guardians of all participants.
Antibiotic susceptibility tests
S. aureus isolates were manually identified
by routine microbiological methods prior to final confirmation and
assessment of susceptibility to 12 antibiotics [penicillin,
erythromycin, clindamycin, trimethoprim/sulfamethoxazole (SXT),
tetracycline, ciprofloxacin, nitrofurantoin, rifampicin,
dalfopristin/quinupristin, gentamicin, linezolid and vancomycin] by
broth microdilution using an automated VITEK2 compact system
(bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibility
testing performance standards and minimum inhibitory concentration
(MIC) interpretive criteria followed the Clinical and Laboratory
Standards Institute guidelines. MRSA was determined based on
cefoxitin, a surrogate for oxacillin, for which the MIC value was
≥8 µg/ml (18). S. aureus
ATCC 29213 (American Type Culture Collection, Manassas, VA, USA)
was used as a reference strain.
Preparation of staphylococcal DNA
The boiling method (19,20) was
employed to extract crude DNA for use in PCR. S. aureus was
cultured at 37°C in lysogeny broth (Oxoid; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) for 16 h, 1 ml of which was
then centrifuged for 10 min at 4°C at 13,839 × g to collect the
cells, which were subsequently resuspended in 100 µl Tris-EDTA
buffer. This suspension was incubated in boiling water for 10 min
prior to being cooled on ice for 5 min. Following centrifugation at
4°C at 13,839 × g for 5 min, supernatants containing DNA were
recovered.
Primer design and polymerase chain
reaction (PCR) amplification
Primer design
Primer pairs specific for the CvfB, SEQ and SEK
genes were designed based on the S. aureus TW20 genome
(GenBank accession no. FN433596.1). Primer sequences, predicted PCR
product sizes, locations within the genome and GenBank accession
numbers are presented in Table I.
Primers were synthesized by the Beijing Genomics Institute
(Shenzhen, China).
 | Table I.Primer sequences, location and
anticipated sizes of polymerase chain reaction products for SEK,
CvfB and SEQ. |
Table I.
Primer sequences, location and
anticipated sizes of polymerase chain reaction products for SEK,
CvfB and SEQ.
| Primer | Sequence
(5′-3′) |
Locationa | Size (bp) |
|---|
| CvfB-F |
GCCGTCGACATGGCATTAGACAAAGATATAGTA |
1486946–1487848 | 903 |
| CvfB-R |
AAACTCGAGTTATTCTTTTGAGTCCATTCGACTC |
|
|
| SEQ-F |
GCAGTCGACATGCCTATATGGCGTTGTAATATA | 954332–955102 | 771 |
| SEQ-R |
CCGCTCGAGTTATTCAGTTTTCTCATATGAAATC |
|
|
| SEK-F |
GCCGTCGACATGAAAAAATTAATAAGCATCTTATTA | 953580–954308 | 729 |
| SEK-R |
CCGCTCGAGTTATATCGTTTCTTTATAAGAAATATCG |
|
|
PCR amplification
All PCRs were singleplex and performed using a
Takara taq™ Package (r001a) (Takara Bio, Inc., Otsu, Japan),
according to the manufacturer's instructions. Each reaction had a
final volume of 50 µl, comprising 1 µl DNA, 4 µl deoxynucleotide
triphosphates, 5 µl 10X PCR buffer, 1 µl of each primer, 0.5 µl
rTaq polymerase and 37.5 µl double-distilled H2O. For
each PCR run, a negative control was included using
ddH2O in place of the DNA template. Using a Biometra
personal PCR thermocycler (Biometra, Gottingen, Germany), the
following cycling conditions were applied: 94°C for 5 min, followed
by 30 cycles of amplification, consisting of denaturation at 94°C
for 30 sec, annealing at 52°C for 30 sec and extension at 72°C for
80 sec. PCR products and marker DL1,000 (Takara Bio, Inc.) were
electrophoresed using a DYY-8C instrument (Beijing Liuyi
Biotechnology Co., Beijing, China) on 2% (w/v) Biowest Agarose G10
gels (Gene Company Ltd., Hong Kong, China) and stained with
GoldView I dye (SBS Gene Tech, Shanghai, China), prior to
visualisation on an ultraviolet transilluminator GAS7508-T20
(Uvitec, Cambridge, UK).
Sequence analysis
The CVFB, SEQ and SEK PCR products from one isolate
were purified and subjected to Sanger sequencing using an ABI
3730xl instrument (Applied Biosystems; Thermo Fisher Scientific,
Inc.) to confirm the reliability of the PCR. The PCR products were
bi-directionally sequenced multiple times and the sequences were
assembled using ContigExpress software (ContigExpress LLC, New
York, NY, USA). The obtained DNA sequences and deduced protein
sequences were searched using the Basic Local Alignment Search Tool
(BLAST) (21) with the default
parameters of the National Center for Biotechnology Information
database (22) to assess homology
with TW20 and other S. aureus strains.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
Data Editor (SPSS, Inc., Chicago, IL, USA). Categorical variables
were described using frequencies and their proportion, and compared
using the Chi-square test. Correlations between the virulent genes,
SEK, SEQ, SEK + SEQ and MRSA, were determined using contingency
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical features of the patients with
S. aureus BSIs
The ages of the 53 patients with S. aureus
BSIs ranged from 5 days to 13 years, with a median age of 8 months.
Male patients accounted for 49.1% (26/53) of the cases, while the
remaining 50.9% (27/53) were female and one isolate strain was
obtained from each patient. The probable primary lesion was mostly
in the respiratory tract, being responsible for BSI in 39.6%
(21/53) of all cases, followed by skin and soft tissue (28.3%),
bone and joint (17.0%), and surgical site infection (1.9%)
(Table II). The distribution of
strains by clinical department was as follows: Neonatal and
paediatric intensive care units (n=16), emergency department
(n=14), internal medicine department (n=14) and surgery department
(n=9).
 | Table II.Clinical features of paediatric
patients with Staphylococcus aureus bloodstream
infections. |
Table II.
Clinical features of paediatric
patients with Staphylococcus aureus bloodstream
infections.
| Variable | n (%) |
|---|
| Year |
|
|
2012 | 6 (11.3) |
|
2013 | 10 (18.9) |
|
2014 | 24 (45.3) |
|
2015 | 13 (24.5) |
| Sex |
|
|
Male | 26 (49.1) |
|
Female | 27 (50.9) |
| Patient age
(years) |
|
| ≤1 | 32 (60.4) |
|
>1 | 21 (39.6) |
| Primary lesion |
|
|
Respiratory tract | 21 (39.6) |
| Skin
and soft tissue | 15 (28.3) |
| Bone
and joint | 9 (17.0) |
|
Surgical site | 1 (1.9) |
|
Unknown | 7 (13.2) |
| Clinical
department |
|
| NICU
and ICU | 16 (30.2) |
|
Emergency | 14 (26.4) |
|
Internal medicine | 14 (26.4) |
| Surgery
department | 9 (17.0) |
Antimicrobial susceptibility profiles
of S
aureus isolated from patients with BSIs. All
isolates were tested for susceptibility to the 12 antibiotics
mentioned above. In total, 43.4% (23/53) were MRSA, while the
remaining 56.6% (30/53) were MSSA. Antibiotic susceptibility
profiles for MRSA and MSSA isolates are listed in Table III. Among the 53 isolates
recovered, the highest rate of resistance was recorded in
association with penicillin (92.5%), followed by erythromycin
(66.0%), clindamycin (62.3%), tetracycline (20.8%), SXT (13.2%) and
ciprofloxacin (1.9%). None of the isolates was resistant to
nitrofurantoin, dalfopristin/quinupristin, rifampicin, gentamicin,
linezolid or vancomycin. All (23/23) MRSA strains and 33.3% (10/30)
of the MSSA strains were multidrug resistant (≥3 classes). Rates of
resistance to erythromycin, clindamycin and tetracycline were
significantly higher in the MRSA group than in the MSSA group.
Although resistance to penicillin and ciprofloxacin was more
prevalent among MRSA isolates and resistance to SXT was more common
among MSSA isolates, no significant differences were noted between
the two groups in this respect. The antimicrobial resistance
patterns between two age groups (≤1 year and >1 year) were also
not significantly different.
 | Table III.Antimicrobial resistance in MRSA,
MSSA and S. aureus isolates in n (%). |
Table III.
Antimicrobial resistance in MRSA,
MSSA and S. aureus isolates in n (%).
|
| S. aureus
strain |
| Age group
(years) |
|
|---|
|
|
|
|
|
|
|---|
| Antibiotic | MRSA + MSSA
(n=53) | MRSA (n=23) | MSSA (n=30) |
P-valuea | ≤1 (n=32) | >1 (n=21) |
P-valueb |
|---|
| PEN | 49 (92.5) | 23 (100) | 26 (86.7) | 0.12 | 30 (93.8) | 19 (90.5) | 1.00 |
| ERY | 35 (66.0) | 22 (95.7) | 13 (43.3) | <0.01 | 19 (59.4) | 16 (76.2) | 0.25 |
| CLI | 33 (62.3) | 22 (95.7) | 11 (36.6) | <0.01 | 17 (53.1) | 16 (76.2) | 0.15 |
| SXT | 7 (13.2) | 3 (13.0) | 4 (13.3) | 1.00 | 5 (15.6) | 2 (9.5) | 0.69 |
| GEN | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
| VAN | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
| CIP | 1 (1.9) | 1 (4.3) | 0 (0.0) |
| 1 (3.1) | 0 (0.0) |
|
| TCY | 11 (20.8) | 8 (34.8) | 3 (10.0) | 0.04 | 4 (12.5) | 7 (33.3) | 0.09 |
| NIT | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
| RFP | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
| LNZ | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
| QDA | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 0 (0.0) | 0 (0.0) |
|
Prevalence of CvfB, SEQ and SEK genes
among S. aureus isolates from paediatric patients with BSIs
As presented in Fig.
1, the amplified CvfB, SEQ and SEK fragments were ~903, 771 and
729 bp long, respectively. BLAST searches revealed that the
amplified sequences were highly similar to the corresponding genes
of strain TW20, as follows: CvfB, 99.0%; SEQ, 100%; and SEK, 99.0%.
High levels of similarity were also noted with sequences of other
S. aureus strains, such as those for AB297388.1 (99%
similarity in CvfB), KU574280.1 (99% similarity in SEK) and
U93688.2 (100% similarity in SEQ). The CvfB, SEQ and SEK sequences
generated in the present study have been deposited in GenBank under
the accession nos. KY705045, KY873303 and KY684176,
respectively.
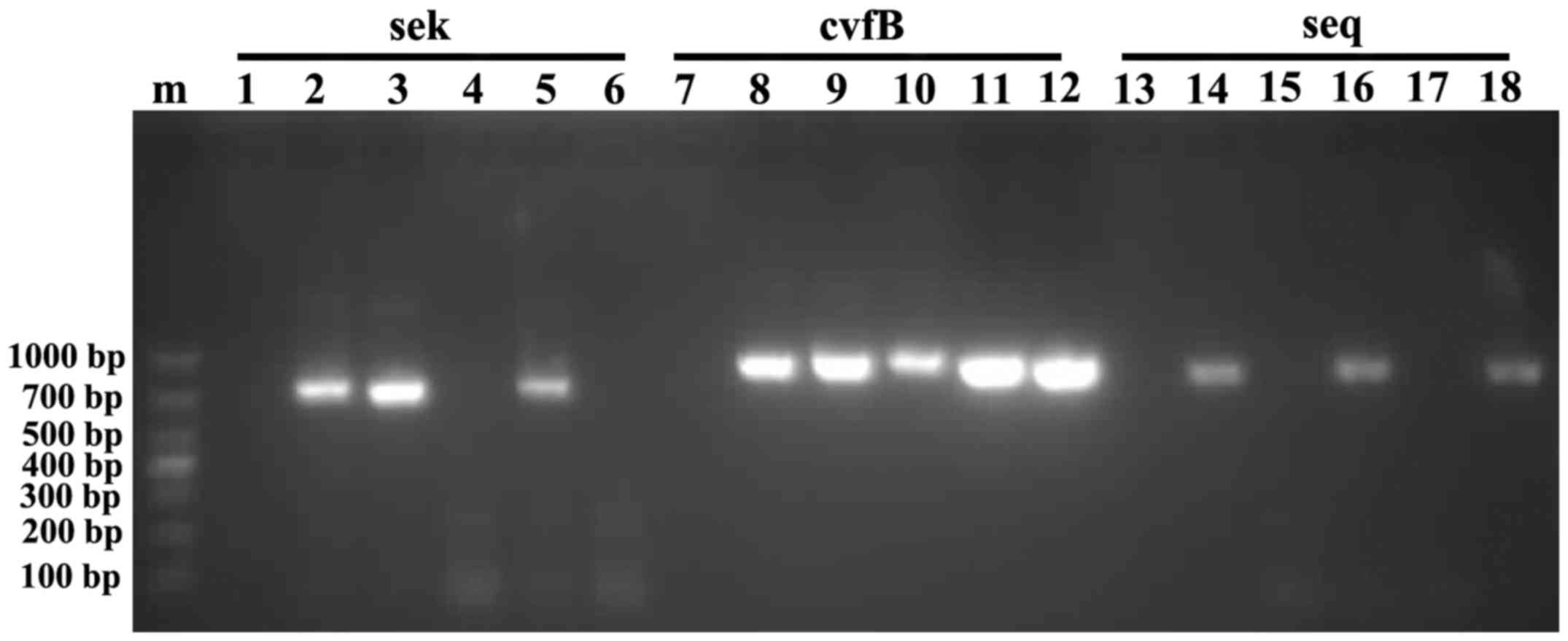 | Figure 1.Electrophoresis of SEK, CvfB and SEQ
PCR amplicons on a 2% agarose gel. Lanes: 1, negative control for
SEK; 2–6, SEK PCR products from five different isolates of MRSA; 7,
negative control for CvfB; 8–12, CvfB PCR products from the same
isolates as those in lanes 2–6; 13, negative control for SEQ;
14–18, SEQ PCR products from the same isolates as those in lanes
2–6. PCR, polymerase chain reaction; CvfB, conserved virulence
factor B; SEK, staphylococcal enterotoxin K; SEQ, staphylococcal
enterotoxin Q. |
Of the 53 strains isolated, all carried CvfB, 34.0%
(18/53) harboured SEQ and 35.8% (19/53) were positive for SEK.
Furthermore, 45.3% (24/53) possessed SEK or SEQ, while 24.5%
(13/53) carried both. The presence of SEK + SEQ was only identified
in MRSA isolates. The number of isolates harbouring SEK, SEQ or
both genes was significantly higher in the MRSA group than in the
MSSA group (P<0.01). In addition, correlations were found
between the virulent genes SEK, SEQ, SEK + SEQ and MRSA
[contingency coefficient 0.416, 0.500, 0.546 respectively;
P<0.01; Table IV).
 | Table IV.Occurrence of SEK, CvfB and SEQ genes
in 53 strains of S. aureus clinical isolates in n (%). |
Table IV.
Occurrence of SEK, CvfB and SEQ genes
in 53 strains of S. aureus clinical isolates in n (%).
| Gene | MRSA + MSSA
(n=53) | MRSA (n=23) | MSSA (n=30) |
P-valuea |
C-valueb |
|---|
| CvfB | 53 (100) | 23 (100) | 30 (100) |
|
|
| SEK | 19 (35.8) | 14 (60.9) | 5 (16.7) | <0.01 | 0.416 |
| SEQ | 18 (34.0) | 15 (65.2) | 3 (10.0) | <0.01 | 0.500 |
| SEK + SEQ | 13 (24.5) | 13 (56.5) | 0 (0.0) | <0.01 | 0.546 |
Discussion
In the present study, the antimicrobial resistance
profiles of S. aureus isolates recovered from paediatric
patients with BSIs in Guangzhou (China) were assessed and the
prevalence of the toxin-associated genes CvfB, SEQ and SEK was
estimated. In total, 43.4% were identified to be MRSA, and high
rates of resistance to penicillin (92.5%), erythromycin (66.0%) and
clindamycin (62.3%) were recorded. All isolates carried CvfB, while
34.0 and 35.8% harboured SEQ and SEK, respectively; 24.5% possessed
all three genes. Carriage of SEK, SEQ and SEK + SEQ, and resistance
to erythromycin, clindamycin and tetracycline, was significantly
higher among MRSA isolates than in those classified as MSSA.
The emergence of antimicrobial resistance and
presence of virulence genes among bacteraemia-associated S.
aureus strains result in limited therapeutic options and
challenging patient management, particularly for patients <1
year of age that involve MRSA. Among the 53 patients with S.
aureus BSIs, the age ranged from 5 days to 13 years, with a
median age of 8 months. The group aged ≤1 year accounted for 60.4%
(32/53). In a nationwide, multicentre surveillance study of
antimicrobial susceptibility among bacteria causing BSIs performed
in 2011, Yuan et al (5)
reported rates of S. aureus resistance to penicillin,
erythromycin and clindamycin of 90.2, 70.7 and 52.0%, respectively.
In the present study, high rates of resistance to penicillin
(92.5%), erythromycin (66.0%) and clindamycin (62.3%) were
identified among S. aureus isolates from paediatric patients
with BSIs in Guangzhou (China). These results indicated that these
drugs are not effective to treat invasive S. aureus
infections as empiric options.
In China, MRSA strains accounted for 44.6 and 42.2%
of BSIs on average in 2014 and 2015, respectively (6,7). In the
present study, 43.4% of isolates recovered from paediatric patients
with BSIs in Guangzhou were MRSA. Vancomycin, a glycopeptide
antibiotic, is the drug of choice for MRSA infections, as
recommended by the Infectious Diseases Society of America (23). The results of the present study imply
that vancomycin and linezolid may be effective treatments for MRSA
in paediatric BSIs, given that all isolates tested were susceptible
to these antimicrobial agents. Thus, for patients who are suspected
of having S. aureus bacteraemia and who are in a critical
condition, it is better to choose vancomycin as empiric option in
case of MRSA infections being present.
Casapao et al (24) reported that vancomycin treatment is
more likely to fail for patients with heterogeneous
vancomycin-intermediate S. aureus BSIs, and vancomycin
monotherapy may not be adequate for severe MRSA infections
(25). Linezolid or rifampicin may
be used as adjunctive therapies for severe, invasive MRSA
infections, as could gentamicin, which has been recommended by the
American Academy of Pediatrics for use with other drugs in the
treatment of endocarditis, persistent bacteraemia, meningitis and
ventriculitis (26). In the present
study, which included only paediatric patients, all strains were
susceptible to rifampicin and gentamicin.
According to a previous study, the invasiveness of
S. aureus largely depends on the carriage of a variety of
virulence factors (27). The
prevalence of SEQ and SEK among the isolates in the present study
was 34.0 and 35.8%, respectively, which represented similar
frequencies to those previously reported (31.1 and 34.4%,
respectively) for Korean patients with bacteraemia in intensive
care units in 2001 and 2008 (28).
Typically, genes of the SEA-SEK-SEQ cluster are present together in
a phage φ3 genomic island (29–31) and
are closely associated with the hospital-acquired MRSA
SCCmec III clone (32–35). In
the present study, the presence of SEK + SEQ was only identified in
MRSA isolates, which supported previous speculations. As carriage
of SEK, SEQ and SEK + SEQ was significantly more frequent among
MRSA strains than among MSSA strains, and correlations were
identified between the virulent genes SEK, SEQ, SEK + SEQ and MRSA.
MRSA may harbour more virulent genes than MSSA.
In the present study, CvfB was identified in all
isolates, and the obtained sequences exhibited high levels of
similarity to those of other strains, such as MS4 (CP009828.1; 99%
similarity), indicating that this gene is highly conserved in S.
aureus. In addition, a BLAST search revealed similarity values
of 78% with S. epidermidis 949 (CP010942.1) and 70% with
Enterococcus faecalis L12 (CP018102.1).
In conclusion, based on the observed antimicrobial
resistance rates, penicillin, erythromycin and clindamycin may not
be appropriate antibiotics for the treatment of paediatric patients
with BSIs, whereas vancomycin may be more effective as an empiric
option. Of the isolates recovered from paediatric patients with
BSIs in Guangzhou (China), 43.4% were MRSA, which demonstrated not
only higher rates of antimicrobial resistance, but also more
frequent carriage of SEK and SEQ genes compared with the MSSA
isolates obtained. The combination of the two aspects influenced
the dissemination of MRSA among children. The present study
clarified the characteristics of BSIs-associated S. aureus
and enhanced the current knowledge regarding the pathogenicity and
treatment of MRSA.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Guangdong (nos. 8451012001001570 and
9151012001000009), the Guangdong Science and Technology Department
(nos. 2014A020212013, 2015A030401007, 2016A020215013), the Medical
Health Science and Technology Foundation of Guangzhou (nos.
201102A212013 and 20171A010267) and the Guangzhou Science
Technology and Innovation Commission (no. 201707010010).
Glossary
Abbreviations
Abbreviations:
|
S. aureus
|
Staphylococcus aureus
|
|
BSIs
|
bloodstream infections
|
|
MRSA
|
methicillin-resistant S.
aureus
|
|
MSSA
|
methicillin-susceptible S.
aureus
|
|
SEs
|
staphylococcal enterotoxins
|
|
PVL
|
pan-valentine leucocidin
|
|
CvfB
|
conserved virulence factor B
|
|
SEQ
|
staphylococcal enterotoxin Q
|
|
SEK
|
staphylococcal enterotoxin K
|
References
|
1
|
Yu F, Li T, Huang X, Xie J, Xu Y, Tu J,
Qin Z, Parsons C, Wang J, Hu L and Wang L: Virulence gene profiling
and molecular characterization of hospital-acquired Staphylococcus
aureus isolates associated with bloodstream infection. Diagn
Microbiol Infect Dis. 74:363–368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cosgrove SE, Sakoulas G, Perencevich EN,
Schwaber MJ, Karchmer AW and Carmeli Y: Comparison of mortality
associated with methicillin-resistant and methicillin-susceptible
Staphylococcus aureus bacteremia: A meta-analysis. Clin Infect Dis.
36:53–59. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mejer N, Westh H, Schønheyder HC, Jensen
AG, Larsen AR, Skov R and Benfield T; Danish Staphylococcal
Bacteraemia Study Group, : Stable incidence and continued
improvement in short term mortality of Staphylococcus aureus
bacteraemia between 1995 and 2008. BMC Infect Dis. 12:2602012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou B, Liu X, Wu JB, Jin B and Zhang YY:
Clinical and microbiological profile of babies born with risk of
neonatal sepsis. Exp Ther Med. 12:3621–3625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan LV, Yu LI, Feng XUE, Xiu-Zhen Z,
Yun-Jian HU, Ting YU, Zhi-Dong HU, Jian-Hong ZHAO, Shi-Yang PAN,
Bi-Jie HU, et al: Mohnarin report of 2011–2012: Surveillance for
resistance of bacteria causing bloodstream infections. Chin J Clin
Pharmacol. 30:112014.
|
|
6
|
Fupin HU, Demei ZHU, Fu WANG, Xiaofei
JIANG, Yingchun XU, Xiaojiang ZHANG, Zhaoxia ZHANG, Ping JI, Yi
XIE, Mei KANG, et al: CHINET 2014 surveillance of bacterial
resistance in China. Chin J Infect Chemother. 15:401–410. 2015.
|
|
7
|
HU Fupin, ZHU Demei, WANG Fu, JIANG
Xiaofei, XU Yingchun and ZHANG Xiaojiang: Report of CHINET
antimicrobial resistance surveillance program in 2015. Chin J
Infect Chemother. 16:102016.
|
|
8
|
de Kraker ME, Wolkewitz M, Davey PG,
Koller W, Berger J, Nagler J, Icket C, Kalenic S, Horvatic J,
Seifert H, et al: Clinical impact of antimicrobial resistance in
European hospitals: Excess mortality and length of hospital stay
related to methicillin-resistant Staphylococcus aureus bloodstream
infections. Antimicrob Agents Chemother. 55:1598–1605. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park DA, Lee SM, Peck KR, Joo EJ and Oh
EG: Impact of methicillin-resistance on mortality in children and
neonates with Staphylococcus aureus bacteremia: A meta-analysis.
Infect Chemother. 45:202–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie X, Bao Y, Ouyang N, Dai X, Pan K, Chen
B, Deng Y, Wu X, Xu F, Li H and Huang S: Molecular epidemiology and
characteristic of virulence gene of community-acquired and
hospital-acquired methicillin-resistant Staphylococcus aureus
isolates in Sun Yat-sen Memorial hospital, Guangzhou, Southern
China. BMC Infect Dis. 16:3392016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dinges MM, Orwin PM and Schlievert PM:
Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 13:16–34.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsumoto Y, Xu Q, Miyazaki S, Kaito C,
Farr CL, Axelrod HL, Chiu HJ, Klock HE, Knuth MW, Miller MD, et al:
Structure of a virulence regulatory factor CvfB reveals a novel
winged helix RNA binding module. Structure. 18:537–547. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsumoto Y, Kaito C, Morishita D,
Kurokawa K and Sekimizu K: Regulation of exoprotein gene expression
by the Staphylococcus aureus cvfB gene. Infect Immun. 75:1964–1972.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aguilar JL, Varshney AK, Pechuan X, Dutta
K, Nosanchuk JD and Fries BC: Monoclonal antibodies protect from
Staphylococcal Enterotoxin K (SEK) induced toxic shock and sepsis
by USA300 Staphylococcus aureus. Virulence. 7:1–10. 2016.PubMed/NCBI
|
|
15
|
Orwin PM, Leung DY, Donahue HL, Novick RP
and Schlievert PM: Biochemical and biological properties of
Staphylococcal enterotoxin K. Infect Immun. 69:360–366. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yarwood JM, McCormick JK, Paustian ML,
Orwin PM, Kapur V and Schlievert PM: Characterization and
expression analysis of Staphylococcus aureus pathogenicity island
3. Implications for the evolution of staphylococcal pathogenicity
islands. J Biol Chem. 277:13138–13147. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Le Loir Y, Baron F and Gautier M:
Staphylococcus aureus and food poisoning. Genet Mol Res. 2:63–76.
2003.PubMed/NCBI
|
|
18
|
CLSI: Performance standards for
antimicrobial susceptibility testing: M100-S26. Clinical and
Laboratory Standards Institute; Wayne, PA, USA: 2016
|
|
19
|
Zhang QY, Zhou WW, Zhou Y, Wang XF and Xu
JF: Response surface methodology to design a selective
co-enrichment broth of Escherichia coli, Salmonella spp. and
Staphylococcus aureus for simultaneous detection by multiplex PCR.
Microbiol Res. 167:405–412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bai J, Shi X and Nagaraja TG: A multiplex
PCR procedure for the detection of six major virulence genes in
Escherichia coli O157:H7. J Microbiol Methods. 82:85–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
NCBI Resource Coordinators, . Database
resources of the national center for biotechnology information.
Nucleic Acids Res. 45:D12–D17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu C, Bayer A, Cosgrove SE, Daum RS,
Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray
BE, et al: Clinical practice guidelines by the infectious diseases
society of america for the treatment of methicillin-resistant
Staphylococcus aureus infections in adults and children. Clin
Infect Dis. 52:e18–e55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Casapao AM, Leonard SN, Davis SL, Lodise
TP, Patel N, Goff DA, LaPlante KL, Potoski BA and Rybak MJ:
Clinical outcomes in patients with heterogeneous
vancomycin-intermediate Staphylococcus aureus bloodstream
infection. Antimicrob Agents Chemother. 57:4252–4259. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Creel AM, Durham SH, Benner KW, Alten JA
and Winkler MK: Severe invasive community-associated
methicillin-resistant Staphylococcus aureus infections in
previously healthy children. Pediatr Crit Care Med. 10:323–327.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pickering LK, Baker CJ, Long SS and
McMillan JA: Red book: 2006 report of the committee on infectious
diseases. 27th. American Academy of Pediatrics, Elk Grove Village,
IL: American Academy of Pediatrics; 2006
|
|
27
|
Wardenburg J Bubeck, Patel RJ and
Schneewind O: Surface proteins and exotoxins are required for the
pathogenesis of Staphylococcus aureus pneumonia. Infect Immun.
75:1040–1044. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim T, Yi J, Hong KH, Park JS and Kim EC:
Distribution of virulence genes in spa types of
methicillin-resistant Staphylococcus aureus isolated from patients
in intensive care units. Korean J Lab Med. 31:30–36. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuroda M, Ohta T, Uchiyama I, Baba T,
Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al:
Whole genome sequencing of meticillin-resistant Staphylococcus
aureus. Lancet. 357:1225–1240. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baba T, Takeuchi F, Kuroda M, Yuzawa H,
Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, et al: Genome
and virulence determinants of high virulence community-acquired
MRSA. Lancet. 359:1819–1827. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Varshney AK, Mediavilla JR, Robiou N, Guh
A, Wang X, Gialanella P, Levi MH, Kreiswirth BN and Fries BC:
Diverse enterotoxin gene profiles among clonal complexes of
Staphylococcus aureus isolates from the Bronx, New York. Appl
Environ Microbiol. 75:6839–6849. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng J, Wang Y, Cao Y, Yan W, Niu X, Zhou
L, Chen J, Sun Y, Li C, Zhang X and Wu Y: The distribution of 18
enterotoxin and enterotoxin-like genes in Staphylococcus aureus
strains from different sources in East China. Foodborne Pathog Dis.
13:171–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chao G, Bao G, Cao Y, Yan W, Wang Y, Zhang
X, Zhou L and Wu Y: Prevalence and diversity of enterotoxin genes
with genetic background of Staphylococcus aureus isolates from
different origins in China. Int J Food Microbiol. 211:142–147.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie Y, He Y, Gehring A, Hu Y, Li Q, Tu SI
and Shi X: Genotypes and toxin gene profiles of Staphylococcus
aureus clinical isolates from China. PloS One. 6:e282762011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu D, Li X, Yang Y, Zheng Y, Wang C, Deng
L, Liu L, Li C, Shang Y, Zhao C, et al: Superantigen gene profiles
and presence of exfoliative toxin genes in community-acquired
meticillin-resistant Staphylococcus aureus isolated from Chinese
children. J Med Microbiol. 60:35–45. 2011. View Article : Google Scholar : PubMed/NCBI
|















