Introduction
Psoriasis is a chronic inflammatory disease of the
skin that affects 0.1–3% of the population worldwide (1,2).
Characteristics of the disease include altered differentiation of
keratinocytes and hyperplasia of the skin (1). Common treatments for psoriasis include
herbal therapy and systemic, topical, combination and phototherapy
treatments, which inhibit excessive keratinocyte proliferation
(3). Although there are a variety of
therapies available to treat psoriasis, no cure has been developed
and the relapsing-remitting nature of psoriasis continues to
adversely affect patient quality of life (4,5). The
pathophysiology of psoriasis is not well understood. Genetic and
auto-immunological mechanisms are suspected, but no single unified
theory exists to explain the nature of the disease.
MicroRNAs (miRNAs) are a group of small, non-coding
RNAs that are 22–25 nucleotides long and post-transcriptionally
regulate gene expression (6). miRNAs
control gene expression by pairing with incompletely matching
target sites of the 3′-untranslated regions (3′UTRs) of mRNAs,
causing translational repression and/or mRNA destabilization,
thereby downregulating the expression of the targeted gene. There
are an increasing number of studies on the vital function of miRNAs
in regulating expression at the post-transcriptional level. There
has been a focus on the role of miRNAs in numerous biological
progressions, including inflammatory disorders (7), and a variety of miRNAs are regarded as
a novel category of inflammatory mediators in human diseases
(8–11).
The transcription factor E2F transcription factor 2
is downregulated by miR-520a, which subsequently suppresses the
cell cycle progression and proliferation of hepatoma cells
(12). Glabridin (GLA) is a novel
antitumor medicine that suppresses inflammation, proliferation and
oxidization in cancer cells. miR-520a promotes the antitumor role
of GLA by inhibiting the nuclear factor (NF)-κB signalling pathway
and GLA upregulates the expression of miR-520a. miR-520a combines
with the 3′UTR of the NF-κB p65 subunit and inhibits its expression
(13). Wang et al (14) demonstrated that miR-520a may be a key
regulator of endoplasmic reticulum (ER) stress and the
proliferation of Raji cells (human B lymphoma cell line), and may
be associated with the PRKR-like ER kinase/eukaryotic translation
initiation factor 2 subunit α and protein kinase B (AKT1)/NF-κB
signalling pathways. miR-520a specifically binds to the 3′UTR of
AKT1 mRNA, and is a crucial mediator for inhibiting proliferation
of Raji cells. Therefore, the use of miR-520a may be a potential
therapeutic strategy for treating Burkitt's lymphoma.
In vitro studies on the differentiation of
keratinocytes are hindered due to the inherent properties of
keratinocytes and the limitations of the stringent requirements for
their culture (15). Primary
keratinocytes have a finite life span, while transformed cell lines
exhibit many phenotypic features not found in normal cells
(15). HaCaT cells are spontaneously
immortalized human keratinocytes, which have been widely used based
on their near normal phenotype and easy propagation (15). They have been used previously as a
model for epidermal keratinocytes in the study of multiple
diseases, including psoriasis (16).
The present study demonstrated that miR-520a suppresses HaCaT cell
proliferation.
Treatment strategies for psoriasis remain elusive.
The phosphatidylinositol 3-kinase (PI3K)/AKT pathway has recently
been identified to be associated with psoriasis (17). AKT was predicted as the target gene
of miR-520a in the present study and AKT may serve a crucial role
in the proliferation of HaCaT cells. Therefore, the present study
highlights a potential method to suppress the proliferation of
HaCaT cells with miR-520a through the AKT pathway.
Materials and methods
Cell line
The HaCaT cell line was purchased from NTCC (cat.
no. 340383; Biovector Science Lab, Inc., Beijing, China). HaCaT
cells were used for an in vitro model of psoriasis and
cultured in Dulbecco's modified Eagle's medium (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.),
penicillin and streptomycin. The cells were cultured at 37°C with
5% CO2.
Reagents
Lipofectamine 2000, the mirVana miRNA Isolation kit
and the TaqMan miRNA assay kit were obtained from Thermo Fisher
Scientific, Inc. The Luc-Pair™ miR Luciferase Assay kit (cat. no.
S900042) for detecting the 3′-UTR luciferase reporter gene of AKT
was obtained from SwitchGear Genomics (Menlo Park, CA, USA). The
assay kits for cell proliferation (BrdU Assay kits) were purchased
from Promega Corporation (Madison, WI, USA). The control small
interfering (si)RNA (cat. no. sc-37077) and anti-AKT siRNA (cat no.
sc-38910) were both obtained from Santa Cruz Biotechnology Inc.
(Dallas, TX, USA). The mimics of human miR-520a were purchased from
Applied Biosystems (cat. no. KIT 001168; Thermo Fisher Scientific,
Inc.), and the scramble miRNA as negative control (cat. no.
SI03650325) and inhibitors (cat. no. GS574467) were purchased from
Qiagen China Co., Ltd. (Shanghai, China). The miR-520a mimics and
inhibitors were transfected based on the manufacturer's
instructions for Lipofectamine 2000.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from cultured HaCaT cells
with TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions, and RT was then
performed using the Access Quick RT-PCR system (Promega
Corporation, Madison, WI, USA) at 44°C for 1 h followed by 92°C for
10 min. RT-qPCR analysis was conducted using the ABI 7,500
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Thermocycling conditions were as follows: 2 min at 95°C
followed by 30 to 32 cycles of 30 sec at 94°C, 30 sec at 58°C and
30 sec at 72°C, with a final extension for 10 min at 72°C. GAPDH
was used as an internal control and results were quantified using
the 2−∆∆Cq method (18).
GAPDH was used as the control. The primer sequences were as
follows: AKT forward, 5′-TGGACTACCTGCACTCGGAGAA-3′ and reverse,
5′-GTGCCGCAAAAGGTCTTCATGG-3′; GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Western blot analysis
Total protein was extracted from lysed cells with
radioimmunoprecipitation assay buffer (Cell Signaling Technology,
Inc., Danvers, MA, USA) and the protein concentrations were
measured using a BCA Protein Assay (Pierce; Thermo Fisher
Scientific, Inc.). Equivalent amounts of protein (20 µg/well) were
separated by 10% SDS-PAGE. Following electrophoretic transfer of
the proteins to a nitrocellulose membrane, the membrane was blocked
for 1 h at 22°C with 5% dried milk. The membrane was incubated with
primary antibodies (dilution 1:2,000) overnight at 4°C followed by
secondary antibodies in 5% dried milk for 1 h at 22°C. Primary
antibodies were as follows: β-actin (sc-58673) and anti-AKT
(sc-24500) (both from Santa Cruz Biotechnology, Inc.). The
secondary antibodies, rabbit anti-mouse IgG (Light Chain Specific;
D3V2A) and mAb (no. 58802; both 1:5,000) were obtained from Cell
Signaling Technology, Inc. Protein bands were detected using an
enhanced chemiluminescence detection kit (GE Healthcare Life
Sciences, Little Chalfont, UK).
Luciferase reporter assays
miRNA-target prediction software DIANA microT
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microtv4/index;
version 4.0) was used to predict the 3′UTR targets of miR-520a,
5′-AAUAAAAGUGACUUGAGCACUUA-3′, which were then amplified by PCR
with the Geneamp XL PCR kit (Thermo Fisher Scientific, Inc.) using
DNA extracted from HaCaT cells and subsequently cloned into pmirGLO
plasmids. The thermocycling conditions of PCR were as follows: 5
min at 95°C followed by 25 cycles of 30 sec at 94°C, 30 sec at 55°C
and 1 min at 72°C, with a final extension for 10 min at 72°C. The
following primers were used: forward,
5′-CUCAGGCUGUGACCCUCCAGAGGGAAGUACUUUCUGUUGUCUG-3′ and reverse,
5′-GAGUUUGGCUUUGUCAGGUUUCCCUUCGUGAAAGAAAAGAGAG-3′ (Invitrogen;
Thermo Fisher Scientific, Inc.). The pmirGLO-3′-UTR of AKT
construct plasmids and the control were transfected into cells
using Lipofectamine 2000 according to the manufacturer's
instructions. The activity of luciferase was then analyzed with a
dual-luciferase reporter system (cat. no. E1910, Dual-luciferase
assay system; Promega Corporation) following transfection for 48 h.
Signals from reporter genes were normalized to Renilla
luciferase.
Bromodoexyuridine (BrdU) assays and
mitotic index analysis
Cells were synchronized for 16 h using the block
method with double thymidine (19).
Cells were then released and collected, or fixed for assays, 1 ml
of Carnoy's fixative (3 parts methanol, 1 part glacial acetic acid)
at −20°C for 20 min. DNA synthesis or mitotic entry analysis was
performed using the BrdU-fluorescein isothiocyanate (FITC) labeling
method (20). Cells were counted
using an immunofluorescence microscope and then the number of
BrdU-FITC-positive cells was quantified. The fluorescence
excitation and emission wavelengths were set to 488 and 525 nm,
respectively.
Mitotic events were determined using Hoechst 33258
DNA staining for 5 min at 37°C and time-lapse video microscopy.
Following synchronization, real-time images of the cells were
obtained at 10 min intervals. Mitotic events in the cells were
detected by changes in morphology. Mitotic cells were counted based
on DNA condensation and nuclear morphology.
Detection of apoptosis
HaCaT cells fluorescently stained for 15 min at room
temperature with Annexin V-FITC/propidium iodide (PI) from
MitoCapture Apoptosis Detection kit (BioVision, San Francisco, CA,
USA) were analyzed using a flow cytometer at 12, 24, 36 and 48 h
following treatment with miR-520a mimics or inhibitors. Annexin
V-FITC and PI were excited by the 488 or 532 nm laser emitted at
wavelengths >610 nm. The flow cytometry (FCS) filter was used to
filter the data files. The program has been designed to gate out
microbead populations and remove debris and other unnecessary
events from the FCS data files that may interfere with the
clustering procedure of the FCAP Array software. The filtering
process is based on a gate defined by the user. The gate should be
defined on a single FCS data file and the FCS Filter program will
automatically apply the same gate for all FCS files in a selected
folder. The software saves the filtered events into new FCS 2.0
data files and keeps the original files. Data analysis was
performed using FlowJo software version 7.6.3 (FlowJo LLC, Ashland,
OR, USA).
Statistical analysis
Continuous variables were determined to be normally
distributed using SPSS software (version 18.0; SPSS, Inc., Chicago,
IL, USA). Data are expressed as the mean ± standard deviation from
three independent experiments. A unpaired Student's t-test or
one-way analysis of variance and post hoc pairwise multiple
comparisons tests were used for the comparisons of multiple groups.
Data with a non-normal distribution were analyzed using the
Kruskal-Wallis test. P<0.05 was determined to indicate
statistically significant difference.
Results
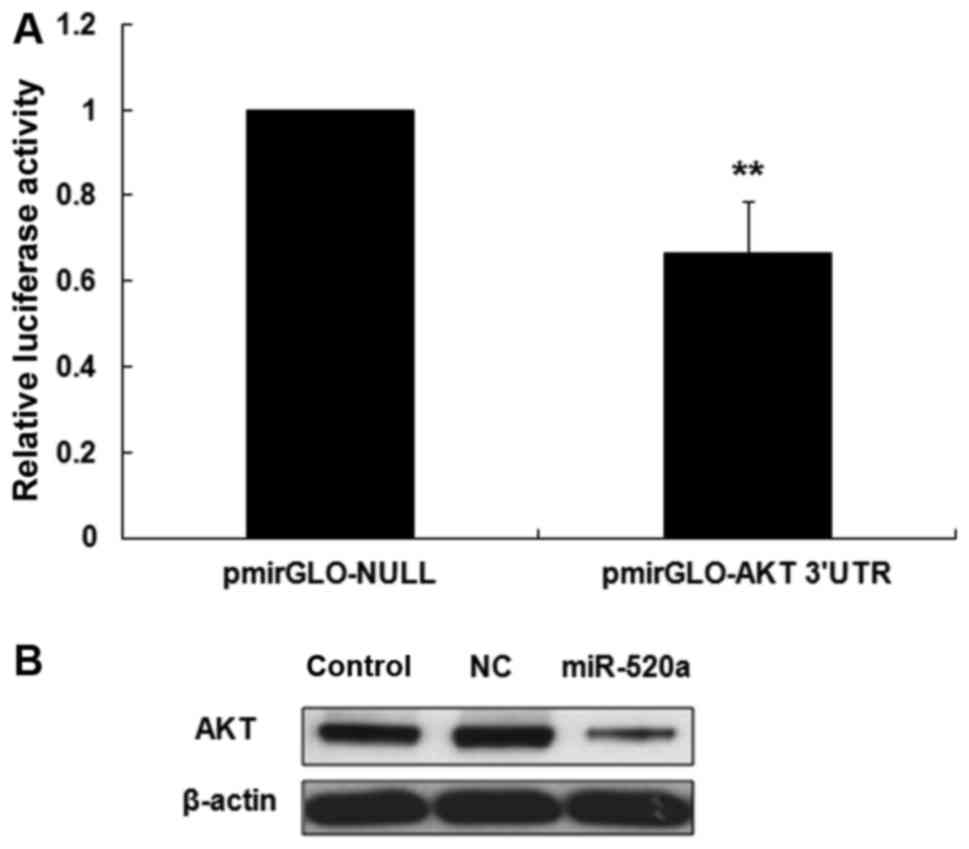
miR-520a inhibits the expression of
AKT in HaCaT cells
DIANA microT software was used to identify the
targets of miR-520a in HaCaT cells. The 3′UTR of AKT mRNA was
identified as a potential target of miR-520a (data not shown). It
was then demonstrated that the luciferase activity of the AKT 3′UTR
was significantly reduced in miR-520a treated HaCaT cells compared
with the control group (P<0.001; Fig.
1A). Western blotting then revealed that miR-520a notably
inhibited the expression of AKT in HaCaT cells (Fig. 1B).
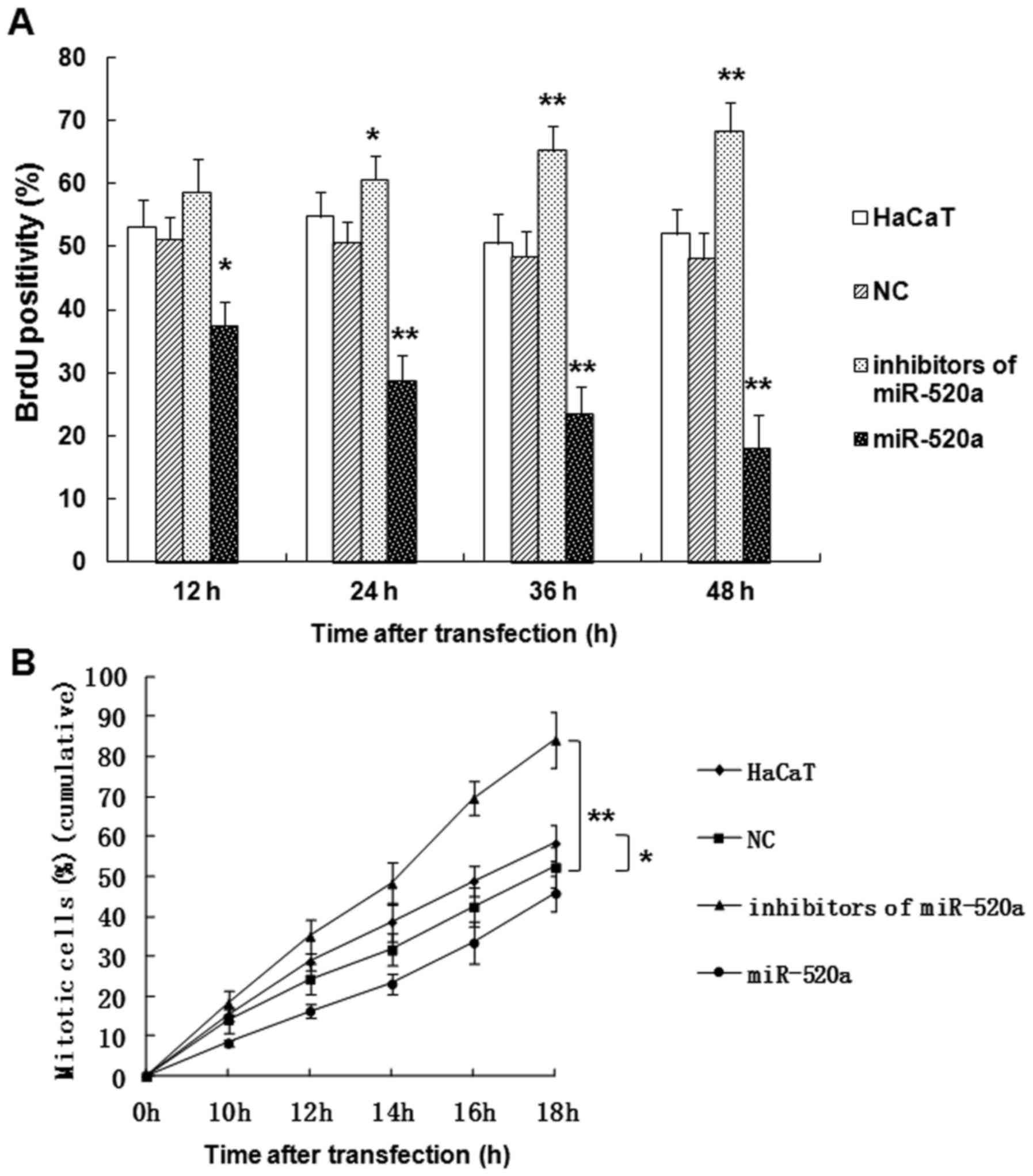
miR-520a suppresses the mitotic entry
and proliferation of HaCaT cells
The role served by miR-520a in HaCaT cells was
further investigated using a BrdU assay (Fig. 2). The results indicated that miR-520a
significantly suppressed the proliferation (P<0.05; Fig. 2A) and mitotic entry (P<0.05;
Fig. 2B) of HaCaT cells in a
time-dependent manner, while the inhibitors of miR-520a promoted
the proliferation (P<0.05; Fig.
2A) and mitotic entry (P<0.01; Fig. 2B) of HaCaT cells compared with the
control group.
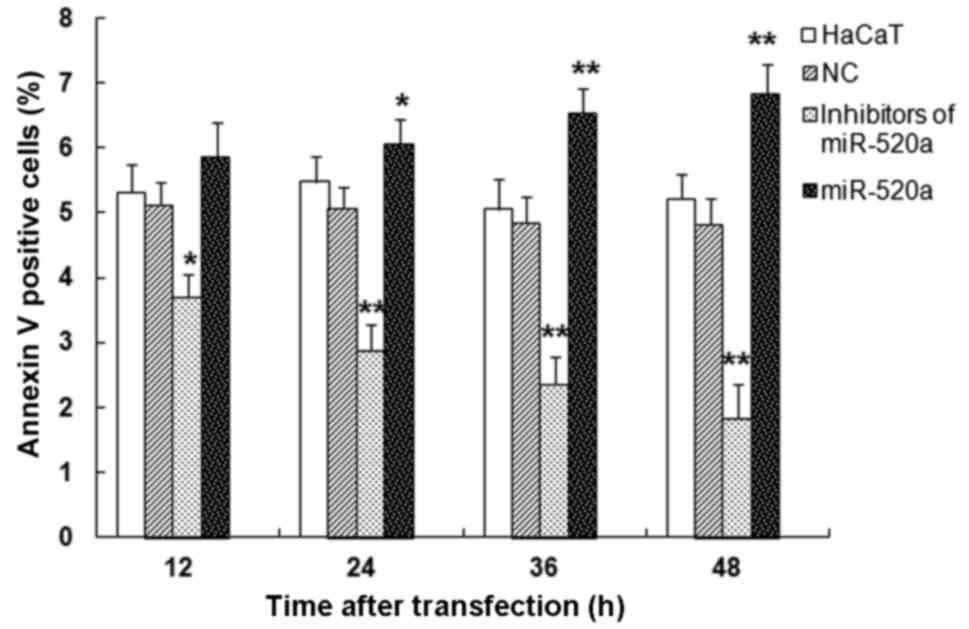
miR-520a induces HaCaT cell
apoptosis
Flow cytometry was used to measure the apoptosis
induced by miR-520a mimics and inhibitors (Fig. 3). The mimics of miR-520a
significantly increased HaCaT cell apoptosis in a time-dependent
manner, while the inhibitors of miR-520a repressed HaCaT cell
apoptosis (all P<0.05 vs. the control group). These results
indicate that miR-520a suppresses the survival of HaCaT cells.
The function of AKT in HaCaT cell
proliferation and mitosis
AKT is a key regulatory factor of cell proliferation
(21). In the present study, the
expression of AKT was knocked down in a HaCaT cell model via siRNA
silencing. The expression of AKT was examined using western blot
analysis and RT-qPCR. The proliferation and mitosis of HaCaT cells
were then detected. The results confirmed that specific siRNAs for
AKT induced significant downregulation of AKT mRNA expression
compared with the untransfected HaCaT cell group or the siRNA
control group, respectively (Fig.
4A; P<0.01). Furthermore, the protein expression of AKT was
downregulated by siRNA (Fig. 4B). In
addition, the decreased expression of AKT in HaCaT cells
significantly suppressed the proliferation and mitosis of HaCaT
cells in a time-dependent manner (Fig.
4C and D; all P<0.05 vs. the siRNA control group).
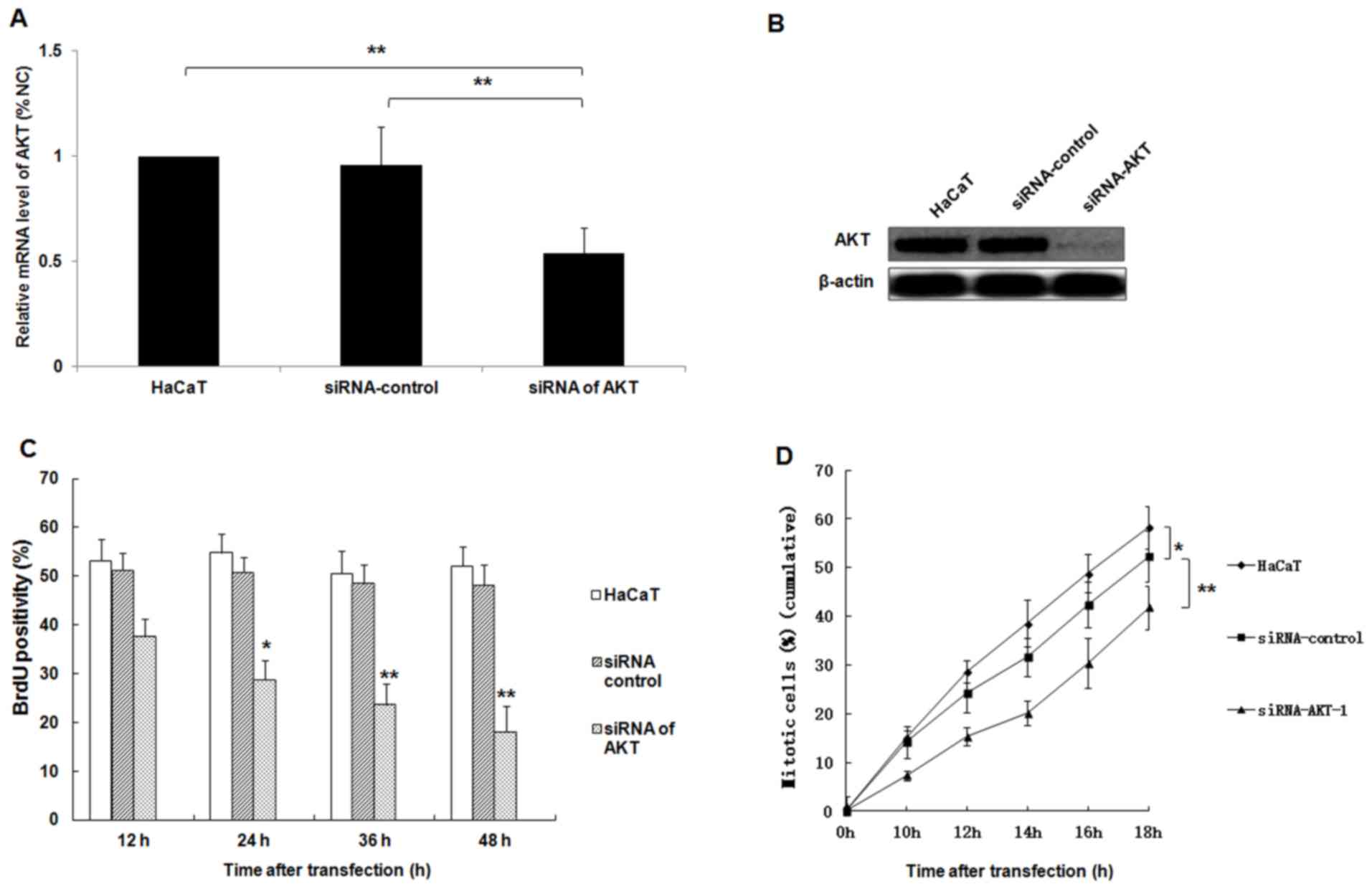 | Figure 4.siRNA silencing of AKT represses the
proliferation and mitosis of HaCaT cells. Following the
transfection of HaCaT cells with AKT-specific siRNA, the expression
of AKT mRNA and protein was analyzed using (A) reverse
transcription-quantitative polymerase chain reaction and (B)
western blotting analyses, respectively. (C) The proliferation of
HaCaT cells was determined at 12, 24, 36 and 48 h following
transfection using a BrdU assay. (D) The percentage of mitotic
HaCaT cells were determined at 10, 12, 14, 16 and 18 h following
transfection. *P<0.05, **P<0.01 vs. the siRNA control group;
miR, microRNA; AKT, protein kinase B; siRNA, small interfering
RNA. |
Discussion
Psoriasis is a long-lasting autoimmune disease. Its
primary clinical feature is skin injury, where the skin appears
scaly, red and itchy. The severity of psoriasis varies from its
presence in a small area to coverage of the entire body (22). The Koebner phenomenon is when
Psoriatic changes occur depending on the location of skin lesions
(23). Skin lesions may be pustular,
guttate, erythrodermic, inverse or plaque (22). They are commonly located on the
scalp, around the navel, shin and the back of the forearms
(24,25). Genetic factors associated with the
immune reaction of skin cells serve a role in the pathogenesis of
psoriasis, which may be affected by the environment, psychological
stress or infections (22–24). However, the mechanisms of the
pathogenesis of psoriasis remain unclear; therefore, further study
is required. Furthermore, cardiovascular disease, lymphomas,
psoriatic arthritis, depression and Crohn's disease may increase
the risk of psoriasis (24).
There are no effective treatments for psoriasis;
however, various therapies may be used to improve its symptoms
(24), including ultraviolet light,
vitamin D3 cream and immune system suppressing medications, such as
methotrexate and steroid creams (22,26).
Genetic susceptibility serves a crucial role in the development of
psoriasis (27,28). The majority of genes associated with
psoriasis have been identified as serving a role in the immune
system, particularly in T cells and major histocompatibility
complexes (29). Genomic research
may potentially be used to identify the molecular mechanisms and
pathways involved in psoriasis, as well as potential novel drug
targets.
Classic genome-wide linkage analysis has indicated
that there are nine loci on different chromosomes associated with
psoriasis, including those containing genes whose products serve a
role in inflammation, such as AKT (29–32).
These loci are associated with the NF-κB and tumor necrosis factor
(TNF)-α signaling pathways (29). In
addition, T lymphocytes induce the abnormal proliferation of
keratinocytes, which may trigger psoriasis (29).
During inflammation, TNF-α regulates its downstream
molecules, including the transcription factors AKT or NF-κB, and
controls several cellular pathophysiological progressions (30). The crucial role served by AKT in cell
proliferation and mitosis has been demonstrated previously
(33). AKT is activated by a range
stimuli and regulates multiple steps in angiogenesis, including
invasion, migration and cell survival. The phosphorylation of AKT
and the inhibition of the PI3K-AKT signaling pathway act upstream
of NF-κB, leading to its activation in acute inflammatory responses
(34). However, the endogenous
miRNA-dependent mechanisms of silencing AKT expression in HaCaT
cells remain unknown. Thus, the present study investigated the
upstream mechanisms of AKT activation associated with miRNA
regulation.
The present study demonstrated that miR-520a
inhibits the proliferation and mitotic entry of the human
keratinocyte cell line HaCaT. Furthermore, AKT was identified as a
predicted target gene of miRNA-520a. A luciferase reporter assay
was then used to confirm that miR-520a directly binds with the
3′UTR of AKT. The results indicated that in the 3′UTR of AKT, the
relative luciferase activities were significantly decreased in
HaCaT cells transfected with miR-520a. This confirmed that miR-520a
directly regulates the expression of its target gene, AKT, at the
mRNA and protein level.
miRNAs are non-coding RNAs that influence gene
expression. They do this through mediating post-transcriptional
gene silencing by suppressing the translation of mRNAs or degrading
mRNAs. miRNAs serve a role in a variety of cellular processes,
including differentiation, development, proliferation and
tumorigenesis (35).
The present study aimed to investigate the role
served by miR-520a in regulating the proliferation and mitosis of
the human keratinocyte cell line HaCaT. The results indicated that
miR-520a directly regulates the expression of AKT, and
significantly inhibits the mitotic entry and proliferation of HaCaT
cells, while promoting apoptosis. Furthermore, the proliferation
and mitotic entry of HaCaT cells was increased in a time-dependent
manner by inhibiting miR-520a, and HaCaT apoptosis was inhibited.
These results demonstrate that miR-520a suppresses HaCaT cell
survival.
Knockdown of AKT was established in a HaCaT cell
model via transfection with siRNAs specific for AKT. The results
indicated that the expression of AKT was significantly
downregulated, which significantly inhibited the mitotic entry and
proliferation of HaCaT cells.
The present study only investigated one member of
the PI3K-AKT-mammalian target of rapamycin kinase system;
therefore, the regulation of the entirety of this cascade system by
miRNA-520a remains unclear. Additionally, further investigation
into the other effects of miRNA-520a in psoriasis, including its
effect on NF-κB, TNF-α and ER-stress is required. The challenges of
using miRNA-520a as a treatment for patients with psoriasis may be
solved by using mimics of miRNA-520a or siRNA specific for AKT.
In conclusion, the results of the present study
indicate that miRNA-520a induces the downregulation of AKT
expression, and suppresses the proliferation and mitotic entry of
the human keratinocyte cell line HaCaT via the AKT signalling
pathway. Thus, miRNA-520a and AKT may be a novel target for the
treatment of psoriasis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30600524; Beijing,
China).
References
|
1
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gudjonsson JE and Elder JT: Psoriasis:
Epidemiology. Clin Dermatol. 25:535–546. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahman M, Alam K, Ahmad MZ, Gupta G, Afzal
M, Akhter S, Kazmi I, Jyoti, Ahmad FJ and Anwar F: Classical to
current approach for treatment of psoriasis: A review. Endocr Metab
Immune Disord Drug Targets. 12:287–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Menter A, Gottlieb A, Feldman SR, Van
Voorhees AS, Leonardi CL, Gordon KB, Lebwohl M, Koo JY, Elmets CA,
Korman NJ, et al: Guidelines of care for the management of
psoriasis and psoriatic arthritis: Section 1. Overview of psoriasis
and guidelines of care for the treatment of psoriasis with
biologics. J Am Acad Dermatol. 58:826–850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garshick MK and Kimball AB: Psoriasis and
the life cycle of persistent life effects. Dermatol Clin. 33:25–39.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong Y, Zou J, Su S, Huang H, Deng Y, Wang
B and Li W: MicroRNA-218 and microRNA-520a inhibit cell
proliferation by downregulating E2F2 in hepatocellular carcinoma.
Mol Med Rep. 12:1016–1022. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Juan M, Shilong N, Xingxing W, Lu S, Fei
J, Yuan L and Zhong L: The repressive effect of miR-520a on
NF-κB/IL-6/STAT-3 signal involved in the glabridin-induced
anti-angiogenesis in human breast cancer cells. RSC Advances.
5:34257–34264. 2014.
|
|
14
|
Wang X, Wang P, Zhu Y, Zhang Z, Zhang J
and Wang H: MicroRNA-520a attenuates proliferation of Raji cells
through inhibition of AKT1/NF-κB and PERK/eIF2α signaling pathway.
Oncol Rep. 36:1702–1708. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deyrieux AF and Wilson VG: In vitro
culture conditions to study keratinocyte differentiation using the
HaCaT cell line. Cytotechnology. 54:77–83. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Breitkreutz D, Schoop VM, Mirancea N, Baur
M, Stark HJ and Fusenig NE: Epidermal differentiation and basement
membrane formation by HaCaT cells in surfacetransplants. Eur J Cell
Biol. 75:273–286. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coppo R, Camilla R, Alfarano A, Balegno S,
Mancuso D, Peruzzi L, Amore A, Canton Dal A, Sepe V and Tovo P:
Upregulation of the immunoproteasome in peripheral blood
mononuclear cells of patients with IgA nephropathy. Kidney Int.
75:536–541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T). Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumagai-Sano F, Hayashi T, Sano T and
Hasezawa S: Cell cycle synchronizationn of tobacco BY-2 cells. Nat
Protoc. 1:2621–2627. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lei Y, Liu H, Yang Y, Wang X, Ren N, Li B,
Liu S, Cheng J, Fu X and Zhang J: Interaction of LHBs with C53
promotes hepatocyte mitotic entry: A novel mechanism for
HBV-induced hepatocellular carcinoma. Oncol Rep. 27:151–159.
2012.PubMed/NCBI
|
|
21
|
Fu X, Wen H, Jing L, Yang Y, Wang W, Liang
X, Nan K, Yao Y and Tian T: MicroRNA-155-5p promotes hepatocellular
carcinoma progression by suppressing PTEN through the PI3K/Akt
pathway. Cancer Sci. 108:620–631. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meier M and Sheth PB: Clinical spectrum
and severity of psoriasis. Curr Probl Dermatol. 38:1–20. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khurrum H, AlGhamdi KM, Bedaiwi KM and
AlBalahi NM: Multivariate analysis of factors associated with the
koebner phenomenon in vitiligo: An observational study of 381
patients. Ann Dermatol. 29:302–306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boehncke WH and Schön MP: Psoriasis.
Lancet. 386:983–994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sima J: Dermatology: Illustrated Study
Guide and Comprehensive Board Review. Springer; New York: pp.
83–87. 2012, ISBN: 978-3-319-47395-6.
|
|
26
|
Parisi R, Symmons DP, Griffiths CE and
Ashcroft DM: Identification and Management of Psoriasis and
Associated ComorbidiTy (IMPACT) project team: Global epidemiology
of psoriasis: A systematic review of incidence and prevalence. J
Invest Dermatol. 133:377–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tamparo CD and Lewis MA: Diseases of the
Human Body. 5th edition. F.A. Davis Company; Philadelphia, PA: pp.
1802011, ISBN-13: 978-0-8036-2505-1.
|
|
28
|
Krueger G and Ellis CN: Psoriasis-recent
advances in understanding its pathogenesis and treatment. J Am Acad
Dermatol. 53 Suppl 1:S94–S100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamamoto S, Yamane M, Yoshida O, Okazaki
M, Waki N, Toyooka S, Oto T and Miyoshi S: Activations of
mitogen-activated protein kinases and regulation of their
downstream moleculesafter rat lung transplantation from donors
after cardiac death. Transplant Proc. 43:3628–3633. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smith CH and Barker JN: Psoriasis and its
management. BMJ. 333:380–384. 2015. View Article : Google Scholar
|
|
32
|
Prieto-Pérez R, Cabaleiro T, Daudén E,
Ochoa D, Roman M and Abad-Santos F: Genetics of Psoriasis and
Pharmacogenetics of Biological Drugs. Autoimmune Dis.
2013:6130862013.PubMed/NCBI
|
|
33
|
Beaudoin GM: Mosaic cellular patterning in
the nose: Adhesion molecules give their two scents. J Cell Biol.
212:495–497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi EK, Jang HC, Kim JH, Kim HJ, Kang HC,
Paek YW, Lee HC, Lee SH, Oh WM and Kang IC: Enhancement of
cytokine-mediated NF-kappaB activation by phosphatidylinositol
3-kinase inhibitors inmonocytic cells. Int Immunopharmacol.
6:908–915. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|