Introduction
Cervical disc disease is a common disease, of which
the prevalence is rising and the age of onset is decreasing
(1,2). Cervical disc disease is characterized
by degeneration in the neck, bones, cartilage and ligaments, which
can stimulate the peripheral spinal cord, nerve roots, vascular,
soft tissues and the sympathetic nerve. This degeneration causes
pain in different areas of the body, leading to serious health
problems (3). It has been suggested
that cervical disc disease is primarily caused by the breakdown of
intervertebral discs (4).
Intervertebral discs consist of a nucleus, annulus and endplate.
Pathological changes in the intervertebral disc primarily include
calcification of cervical cartilage endplate (CEP), abnormal
annulus fibrosis and proteoglycan, and loss of water in the nucleus
pulposus (5).
Vertebral CEP, a thin layer of hyaline cartilage
located between the upper and lower surfaces of the vertebra body,
is an important part of the intervertebral disc, which is composed
of cartilage cells and extracellular matrix. Its degeneration is
accompanied by a loss of nutrients in the extracellular matrix,
which is considered to contribute to the initiation and development
of degenerative disc disease (6). As
the largest avascular structure of the body, CEP functions
mechanically as a shock absorber and is also an important gateway
allowing nutrients and metabolites to pass freely between the
avascular nucleus pulposus and vertebral body (7). CEP also serves an important role in the
growth of the vertebral body, and is important in facilitating the
normal physiological function of the spine (5,8). It also
serves as a natural barrier to prevent damaging substances, such as
matrix metalloproteinases, inflammatory cytokines and immune
molecules, from entering the nucleus (8,9). A
number of studies have indicated that the initiation of disc
degeneration may be due to the degeneration, dysfunction and
calcification of endplates (10,11). CEP
is almost totally reliant on the orderly expression of genes to
maintain its normal development and function; therefore, any minor
errors in genetic pathways can lead to severe endplate degeneration
(12–14).
In recent years, microRNAs (miRs), which are small
RNAs that regulate gene expression at the post-transcriptional
level, have been an important focus of various studies. miRs are a
class of highly conserved, non-coding RNA molecules that are 21–25
bases long and participate in gene regulation by pairing with a
non-coding region of the target mRNA molecule. This leads to
changes in mRNA stability and translation performance. miRs also
serve crucial roles in cell proliferation, apoptosis and
differentiation (15).
Bioinformatics research predicts that around a third of mRNA is
regulated by miRs in humans (16).
Furthermore, a number of studies have demonstrated that miR is not
only involved in the regulation of cartilage development, but also
serves pivotal roles in the pathogenesis of the nucleus pulposus
degeneration and arthritis (17,18).
However, it remains unknown whether miR is associated with the
molecular mechanism of CEP degeneration.
Fas, also known as apoptosis antigen 1 or cluster of
differentiation 95, is located on the cell membrane and is a type I
membrane receptor protein consisting of extracellular,
transmembrane and cytoplasmic domains (19). An amino acid sequence of the
cytoplasmic domain, Fas-associated death domain (FADD), is highly
homologous to the tumor necrosis factor receptor, which in turn
interacts with the death domain (20). The Fas system has been extensively
studied in apoptosis. The extracellular portion of the Fas can
specifically bind to the anti-Fas monoclonal antibody or Fas ligand
(FasL), mediating apoptosis through the death domain or death
signal transduction pathway (21).
Most research so far has focused on endplate apoptosis in
intervertebral disc degeneration with aging. Ariga et al
(10) demonstrated that CEP
degenerative mice have apoptotic cells that increase with age and
external pressures, including excessive exercise and overload,
leading to a decline of cartilaginous endplate cells (CEC). The
higher the rate of apoptosis, the more quickly CEP disappeared
(22). Although there are a few
studies that reveal abnormal expression profiles of miRNAs in
degenerative CEP, the specific molecular mechanisms involved in the
disease process remain unclear (23–25).
In the present study, the association between
miR-625 and Fas expression in human cervical degenerative disc
endplates was investigated, with the aim of identifying the
mechanism of miRNAs in the development of cervical disease.
Materials and methods
Patients
A total of 30 patients were enrolled from Quzhou
People's Hospital (Quzhou, China) and divided into two groups as
described below. The case group consisted of 15 patients with
cervical disease, including 8 males and 7 females (mean age, 52.6
years; age range, 46–62 years). All patients were diagnosed with
cervical disease by X-ray, computed tomography and magnetic
resonance imaging (MRI) tests. The control group was composed of 15
patients with cervical burst fracture and vertebral reconstruction
surgery due to trauma, and included 9 males and 6 females (mean
age, 47.2 years; age range, 38–58 years). Patients with disc
degeneration detected by MRI test, tuberculosis, cancer, diabetes,
genetic and metabolic diseases and congenital malformation were
excluded from the current study. Tissue samples were isolated
during surgery and were immediately frozen with liquid nitrogen and
stored at −80°C. Use of the stored human specimens in the present
study was approved by the Ethical Committee of Quzhou People's
Hospital and informed consent was obtained from all patients.
Reagents
TRIzol® reagent, Lipofectamine™ 2000
reagent, radioimmunoprecipitation assay buffer (RIPA), lentiviral
vector encoding pre-miR-625 and a scrambled sequence for control
were all purchased from Invitrogen (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Rabbit anti-human Fas/B-cell lymphoma 2 (Bcl-2)
polyclonal antibody was obtained from Abcam (Cambridge, MA, USA),
PrimeScript RT Reagent kit and SYBR® PrimeScript™ miRNA
RT-PCR kit were purchased from Takara Biotechnology Co., Ltd.
(Dalian, China). mRNA SYBR Green RT-PCR reagent was purchased from
Kapa Biosystems, Inc., (Wilmington, MA, USA), enhanced
chemiluminescence (ECL) kit was purchased from Pierce Biotechnology
(Thermo Fisher Scientific, Inc.) and QuikChange XL site-directed
mutagenesis kit was from Stratagene (Agilent Technologies, Inc.,
Santa Clara, CA, USA).
CEP cell culture
Degenerative cervical CEP samples were obtained from
clinical specimens [Modic type 1 or 2 (26), isolated in lumbar fusion surgery] and
were used for cell culture. Tissues were separated by a dissecting
microscope to remove cross-contamination between the tissues and
the surrounding ligaments, thus obtaining CEP tissue alone.
Following removal of the endplate tissue, tissues were washed
repeatedly with 0.1 M phosphate-buffered saline containing 100 U/ml
penicillin and streptomycin to remove the blood on the surface of
the endplate. Subsequently, tissues were digested with 0.2% type II
collagenase (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
and Dulbecco's modified Eagle medium (DMEM)/F12 (HyClone; GE
Healthcare Life Sciences) for 6 h and then filtered through a mesh
with a pore size of 100 µm. The filtrate was centrifuged at 350 × g
for 10 min at room temperature. Supernatant was removed and
DMEM/F12 medium containing 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences) was added to re-suspend the cells. In
total, ~2×106 cells could be harvested from each
specimen.
The total number of viable and nonviable cells was
counted under an inverted microscope (Olympus BX50; Olympus Corp.,
Tokyo, Japan) with the help of a haemocytometer and the trypan blue
method (Beijing Solarbio Science and Technology Co., Ltd., Beijing,
China) was used to determine the number of viable cells present in
the cell suspension. Cells were cultured in an incubator containing
5% CO2 at 37°C and observed under an inverted light
microscope. Media were changed every 2–3 days until the culture
flask bottom was completely covered.
RNA extraction and reverse
transcription
CEP tissues (100 mg) were weighed, ground to a
powder in liquid nitrogen and then placed in a centrifuge tube with
1 ml TRIzol reagent for lysis. Total RNA was isolated using the
phenol-chloroform method (27), and
RNA quality was assessed by electrophoresis and the UV absorption
ratio 260/280 using a UV spectrophotometer. A total of 1 µg total
RNA was used in the reverse transcription reaction to obtain cDNA
and stored at −20°C. Reverse transcription of miRNA in the samples
was performed using the PolyA tailing method. Briefly, 6 µl miRNA
was added in ice-cold RNase-free eppendorf (EP) tubes and the
following were gently mixed in the EP tubes: 10 µl 2X miRNA
reaction buffer mix, 2 µl 0.1% bovine serum albumin (Beyotime
Institute of Biotechnology, Haimen, China) and 2 µl miRNA
PrimeScript RT Enzyme mix (Takara Biotechnology Co., Ltd.). Total
final volume was made up to 20 µl with double distilled
H2O. The miRNA was treated with cDNAse and the reverse
transcription step was performed as follows: PolyA tailing and
reverse transcription reaction were performed at 37°C for 60 min
and 80 µl RNAase-free H2O was then added to the RT
reaction. Finally, quantitative polymerase chain reaction (qPCR)
was performed with 2 µl product.
Bioinformatics analysis
The miRNA binding sites on the 3′-untranslated (UTR)
region of Fas mRNA were predicted using TargetScan (www.targetscan.org).
pFL-Fas 3′-UTR expression vector
construction
The potential target sequences for miR-625 in the
3′-UTR of Fas mRNA (Gen-Bank Accession No. NM_000043) were
amplified and cloned into pGL3-control reporter plasmids (Promega
Corporation, Madison, WI, USA) between the EcoRI and
HindIII restriction sites. The stop codon of firefly
luciferase was inserted using the same method.
Luciferase reporter gene analysis
Transient co-transfections of 0.8 µg pFL-Fas 3′-UTR
plasmid, 100 nM miR-625 mimics/miR-control (Ambion; Thermo Fisher
Scientific, Inc.) and 0.04 µg thymidine kinase promoter-Renilla
luciferase reporter plasmid (pRL-TK) were conducted in HEK293 cells
(American Type Culture Collection, Manassas, VA, USA) using
Lipofectamine 2000 following the manufacturer's instructions.
Firefly and Renilla luciferase activities were measured using the
Dual-Luciferase Reporter Assay kit (cat. no. E1910; Promega
Corporation) according to manufacturer's instructions 48 h
following transfection. pRL-TK is commonly used as a normalization
control for transfection efficiency to confirm the inhibition of
Fas 3′UTR expression by miR-625. The assay was repeated at least 3
times independently. In order to further explore the role of
miR-625 on Fas 3′-UTRs, the QuikChange XL site-directed mutagenesis
kit (Stratagene; Agilent Technologies, Inc.) was employed to
generate point mutations in the 3′-UTR of the Fas gene of miR-625
binding sites and each mutation contained four sequential
bases.
Overexpression or inhibition of
miR-625 in CECs
Green fluorescent protein, lentiviral expression
vector of pre-miR-625 and the control scrambled oligonucleotides
were purchased from Invitrogen (Thermo Fisher Scientific, Inc.).
CECs were cultured in each well (1.5×105/well) of a
24-well culture plate in 250 µl DMEM/F12 supplemented with 10% FBS.
Transfected cells were subsequently infected with virus at a
multiplicity of infection of 10, incubated at 37°C for 5 h, and
observed under an inverted microscope (Olympus Corp.) to confirm
successful transfection. To further determine the regulation of
miR-625 to Fas in CECs, transient transfection of antigomiR-625 and
negative control were preformed in CECs according to the
aforementioned method.
qPCR of miR-625 and Fas mRNA
The SYBR® PrimeScript miRNA RT-PCR kit
was employed using small nuclear RNAs (U6) as an internal
reference. The primer sequence for miR-625 was
5′-CCAGGGGGAAAGTTCTATAGTCC-3′. The PCR system (20 µl) was as
follows: 12.5 µl SYBR EX Taq-Mix, 2 µl cDNA, 1 µl PCR forward
primer and 1 µl Uni-miR RT-qPCR primer with 8.5 µl of 0.1% diethyl
pyrocarbonate (DEPC)-H2O added to 25 µl. Each sample was
performed in triplicate. The relative expression levels of miR-625
were calculated using the 2−ΔΔCq method (28).
To determine the mRNA expression level of Fas in
tissues and CECs, the SYBR Green qRT-PCR reagent was employed and
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an
internal reference. The mRNA was reverse transcribed into cDNA
using a PrimeScript RT reagent Kit with gDNA Eraser (cat. no.
RR047Q; Takara Biotechnology Co., Ltd). The primers for Fas mRNA
were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
Primer sequences were as follows: FAS, forward
5′-GAGCTCGTCTCTGATCTCGC-3′, and reverse 5′-AAAGAGCTTCCCCAACTCCG-3′;
GAPDH, forward 5′AAG GTG AAG GTC GGA GTCA3′ and reverse: 5′GGA AGA
TGG TGA TGG GAT TT3′. The PCR system (20 µl) was as follows: 10 µl
SYBR EX Taq-Mix, 1 µl cDNA, 0.5 µl primer 1, 0.5 µl primer 2, with
8 µl of 0.1% DEPC-H2O added to 20 µl. Each sample was
performed in triplicate. The PCR conditions consisted of
denaturation at 95°C for 10 min, 95°C for 1 min, 60°C for 40 sec
and 72°C for 30 sec, for a total of 40 cycles. A final extension
was performed at 72°C for 1 min. Finally, the relative expression
levels of Fas were calculated using the 2−ΔΔCq method
(19).
Western blot analysis
A total of 100 µg cervical tissue was weighed,
ground to a power in liquid nitrogen and then placed in a
centrifuge tube with 600 µl ice-cold RIPA reagent (50 mM Tris-base,
1 mM EDTA, 150 mM NaCl, 0.1% SDS, 1% TritonX-100 and 1% sodium
deoxycholate) for 50 min. Subsequently, the sample was centrifuged
at 12,000 × g for 5 min at 4°C to remove precipitates and the
supernatant was used as the total protein. Protein concentration
was measured using the bicinchoninic acid protein assay reagent kit
(cat. no. 23225; Thermo Fisher Scientific, Inc.).
To obtain the total protein from CEC cells, the
cells were collected and lysed with the ice-cold RIPA lysis buffer
and the protein extraction steps using the aforementioned
procedure. A total of 50 µg total protein were mixed with 2X
loading buffer and were boiled for 5 min. In total, 5 µg protein
samples from CEC cells and cervical tissues were separated by 12%
SDS-PAGE at 100 V, and then transferred onto a polyvinylidene
difluoride membrane at 300 mA, and 4°C for 1.5 h. The membrane was
then blocked with 50 g/l fat-free milk at room temperature for 1 h,
and incubated with anti-Fas primary antibody (1:800; Abcam,
Cambridge, UK; cat. no. ab82419), anti-Bcl-2 primary antibody
(1:1,000; Abcam; cat. no. ab32124) and anti-GAPDH primary antibody
(1:1,000; Abcam; cat. no. ab9485), respectively, at 4°C overnight.
The membrane was washed 3 times for 15 min each time with
phosphate-buffered saline and Tween-20 (PBST) and the membrane was
then incubated with goat anti-rabbit secondary antibody (1:1,000;
Abcam; cat. no. ab6721) at room temperature for 1 h. The membrane
was then washed 3 times for 15 min each time with PBST. Finally,
the membrane was developed using an ECL kit and visualized by
exposure on films (Merck KGaA, Darmstadt, Germany). Images were
captured and quantified by Image lab software 4.0 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The relative content of the
objective protein was represented by the ratio of the objective
protein gray value and the gray value of GAPDH.
Statistical analysis
Data were recorded and statistically analyzed using
Microsoft® ExcelXP® and SPSS for
Windows® version 13.0.1 (SPSS, Inc., Chicago, IL, USA).
Statistical differences were detected using the Student's t test.
Results with a two-sided P≤0.05 were considered to indicate a
statistically significant difference.
Results
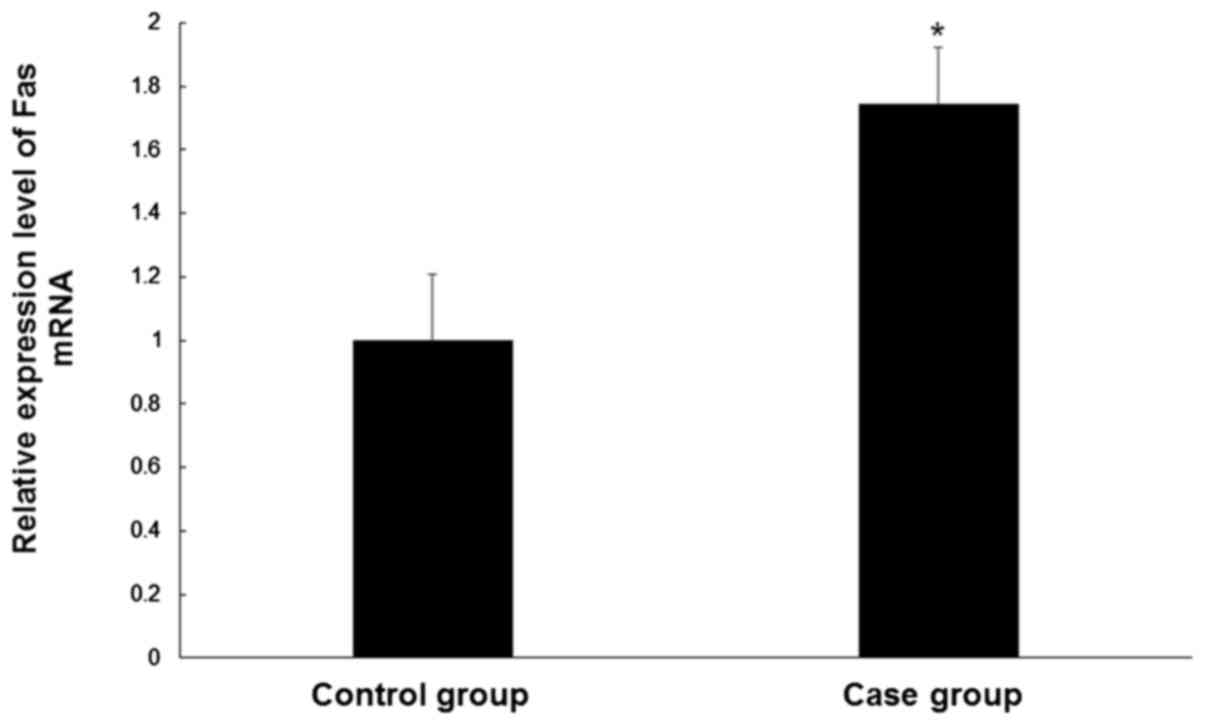
Fas mRNA levels are increased in CEP
tissue
In order to determine the Fas mRNA levels in the CEP
tissue samples of controls and patients, qPCR was performed. Levels
of Fas mRNA were significantly higher (~1.7 fold) in degenerative
cervical CEP tissues compared with normal CEP tissues (P<0.05;
Fig. 1). These results suggest a
role for Fas in CEP degeneration.
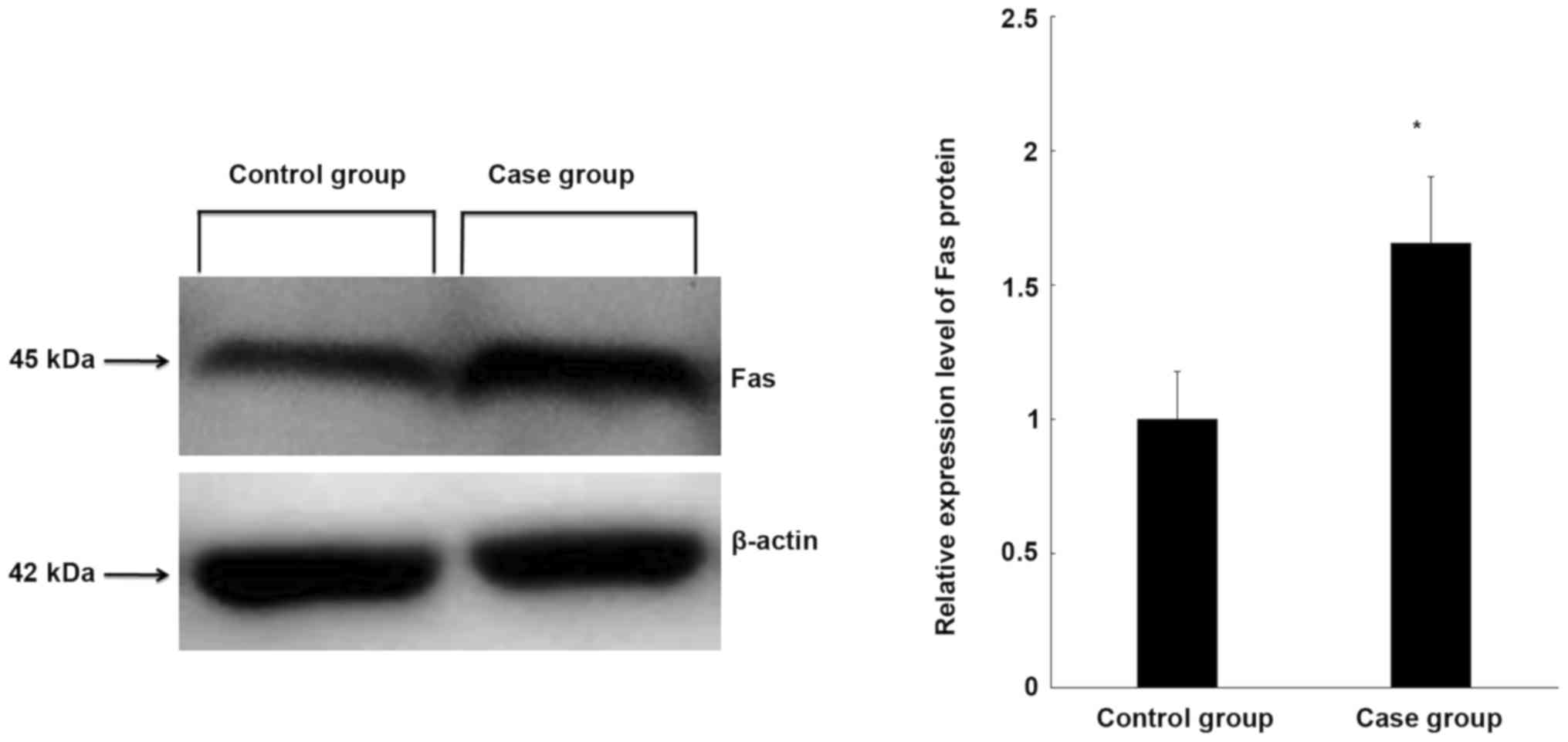
Fas protein expression is increased in
CEP tissue
To further clarify the expression of Fas in the CEP
tissue of controls and patients, Fas protein expression was
measured using western blot analysis. Fas protein expression was
significantly higher in degenerative cervical CEP tissues than in
normal CEP tissues (P<0.05; Fig.
2), which is consistent with the expression of Fas at the mRNA
level.
Fas is a potential target of
miR-625
In order to identify the effect of miR-625 on gene
regulation in CEP tissue, TargetScan was used to predict the target
gene of miR-625. Fas was selected as the candidate target gene of
miR-625 and the binding site was shown to be on the sequential area
of 8-base pairs of the Fas 3′UTR (Fig.
3).
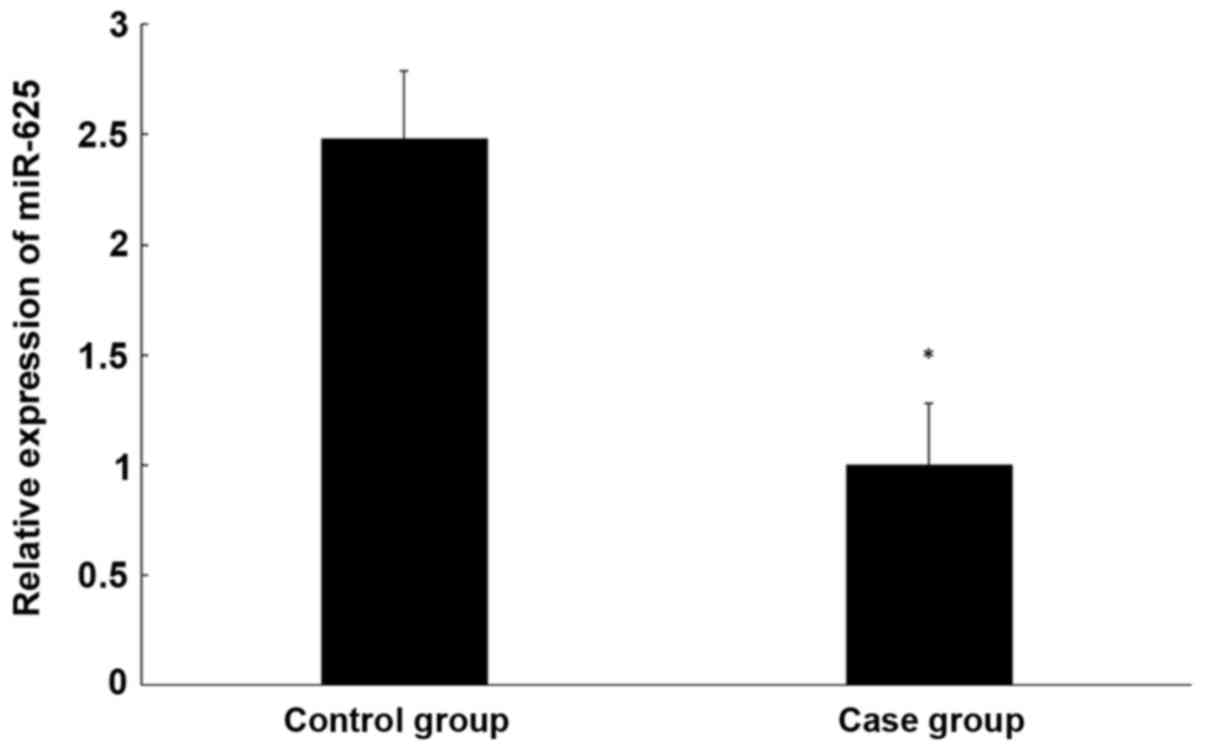
miR-625 expression in CEP tissue is
lower than in healthy tissue
To determine miR-625 expression in the CEP tissue of
patients and controls, qPCR was performed. It was determined that
miR-625 expression is significantly lower in degenerative CEP than
normal CEP tissue (P<0.05; Fig.
4).
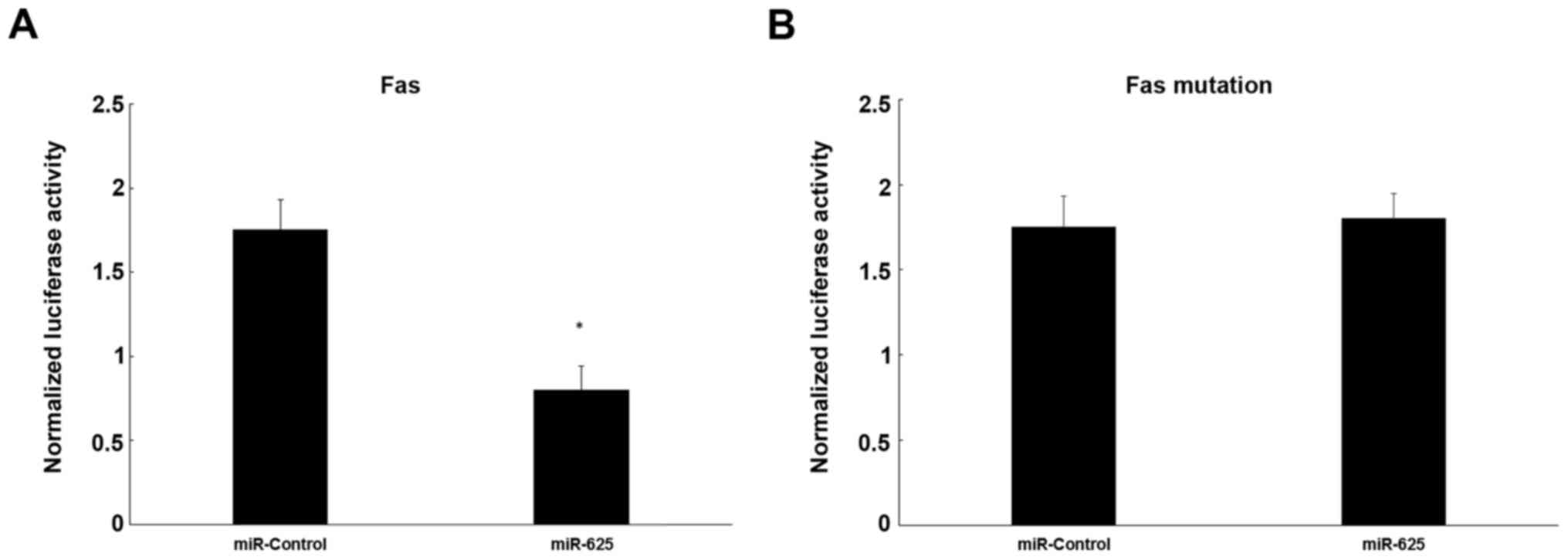
Dual-luciferase reporter assay
To evaluate the influence of miR-625 on target gene
activation, a dual-luciferase reporter assay was used, showing that
miR-625 decreased luciferase activity significantly by
co-transfection of a mixture of pRL-TK vector and miR-625 mimics
together with pFL-Fas 3′-UTR plasmid in cells, compared with the
negative control (P<0.05; Fig.
5A). However, mutation of Fas mRNA did not decrease luciferase
activity significantly in cells co-transfected with the mutation
pFL-Fas 3′-UTR and pRL-TK together with mimics, in comparison to
cells transfected with the negative control (P>0.05; Fig. 5B).
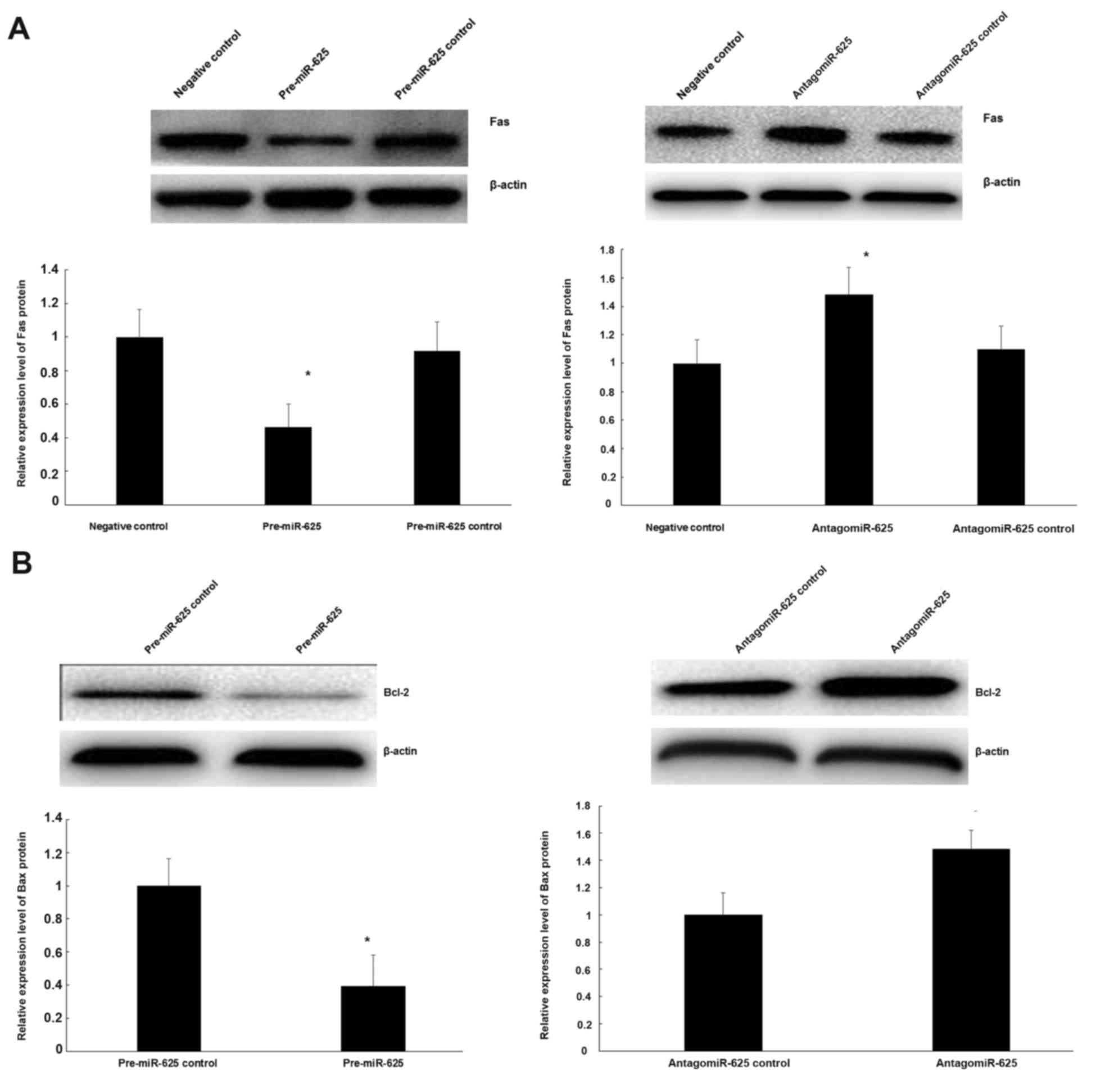
Role of miR-625 on Fas protein
expression and its inhibition of the Bcl-2 gene
To ascertain if miR-625 regulates Fas protein, CECs
were transfected with pre-miR-625. Upregulation of miR-625
expression significantly reduced the expression of Fas compared
with the negative control (P<0.05; Fig. 6A). To further analyze the association
between miR-625 and Fas expression in CECs, antigomiR-625 were
transfected into CECs. It was demonstrated that downregulation of
miR-625 significantly increased Fas expression compared with the
negative control (P<0.05; Fig.
6A). Meanwhile, to examine the functional role of miR-625 on
the Bcl-2 gene, pre-miR-625 and antigomiR-625 were transfected into
CECs. As indicated in Fig. 6B,
upregulation of miR-625 increased Bcl-2 protein expression, while
reducing the expression of miR-625 significantly reduced Bcl-2
protein expression (P<0.05).
Discussion
CEP degeneration is directly associated with
intervertebral disc degeneration. However, the exact molecular
mechanism of CEP degeneration remains to be elucidated. In the
present study, the role of miR-625 in CEP degeneration was
examined. Fas is a transmembrane protein of the death receptor
family, which can interact with FasL and induce apoptosis by
activating a hierarchy of caspases (21). The Fas and FasL system constitute an
important pathway mediating apoptosis in immune and tumor cells,
and serve pivotal roles in numerous physiological and pathological
processes, including immune homeostasis, inflammation, tumor
monitoring, organ transplantation, autoimmune diseases and T- and
B-lymphocyte maturation (21).
Previous reports have demonstrated that there are
two major Fas-mediated mechanisms of apoptosis: One is the cell
surface death receptor signaling (extracellular pathway); the other
is the mitochondrial pathway (intracellular pathway) (29,30). The
cell surface death receptor signaling pathway is initiated on the
ligand-receptor interactions at the cell surface, including the
FAS-FASL system, followed by binding between receptors of the death
domain and signaling molecules, and interaction with pro caspase 8
to form a death-inducing signaling complex. Subsequently caspase 3,
6 and 7 are activated, inducing cell death (31).
The Bcl-2 gene was first identified in follicular B
cell lymphomas in 1985, and it was determined that it was closely
associated with apoptosis (32) and
was subsequently determined to be an anti-apoptotic gene. Recently,
it was found that cell-death regulation proteins, including Bcl-2,
Bcl-xL, Bcl-2-associated X protein and the Bcl-2 gene are capable
of inhibiting apoptosis in disc cells, which is the primary
mechanism of action of the Bcl-2 protein (33). In the present study, the expression
of Fas was determined in degenerative and normal CEP using western
blot analysis and qPCR at the protein and mRNA levels,
respectively. It was observed that expression of Fas mRNA and
protein in degenerative cervical CEP tissue was higher than in the
normal CEP either. A previous study by Ariga et al (10) confirmed that apoptosis of the
cartilage endplate is an important pathological basis for lumbar
degeneration. The results of the present study suggest that
Fas-induced apoptosis of the cartilage endplate may be involved in
cervical degeneration. Using a bioinformatics approach, it was
predicted that miR-625 would bind to the 3′UTR of Fas through a
binding domain. Therefore, the expression of miR-625 in
degenerative CEP tissues was examined further and it was revealed
that miR-625 expression was significantly lower in degenerative CEP
compared with normal CEP, suggesting that Fas gene expression may
be directly regulated by miR-625. In order to verify the regulation
of Fas expression by miR-625, dual luciferase reporter gene assay
was performed and this found that miR-625 can bind directly to the
3′-UTR of the Fas gene, thus inhibiting Fas protein expression. The
role of abnormal expression of miRNAs in the cartilage endplate and
cervical or lumbar degeneration has been studied previously; for
example, Chen et al (23)
reported that miR-34a regulated the apoptosis of cartilage endplate
cells by targeting Bcl-2. In addition, Wang et al (34) reported that miR-155 regulated
Fas-mediated apoptosis of lumbar nucleus pulposus through FADD and
caspase-3. These results suggest that miRNA-mediated apoptosis may
serve an important role in cervical/lumbar degeneration.
Lentiviral vector transfection experiments showed
that downregulation of miR-625 in CECs can induce upregulation of
Fas expression. Therefore, Bcl-2 expression was detected further
and it was revealed that the upregulation of miR-625 may increase
anti-apoptotic Bcl-2 expression in CECs. By contrast,
downregulation of miR-625 expression can reduce Bcl-2
expression.
In conclusion, the results of the current study
demonstrated that during degeneration of CEP, miR-625 is important
in inhibiting apoptosis. This suggests that in the pathogenesis of
cervical disease, downregulation of miR-625 promotes Fas-mediated
cell endplate apoptosis. Therefore, miR-625 may be a novel target
for the treatment of cervical disease using lentivirus-mediated
pre-miR-625 expression.
Acknowledgements
The present study was supported by the Projects of
Nonprofit Technology & Research of Zhejiang Province (grant no.
2015C33294).
References
|
1
|
Irvine DH, Foster JB, Newell DJ and
Klukvin BN: Prevalence of cervical spondylosis in a general
practice. Lancet. 1:1089–1092. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Al-Ryalat NT, Saleh SA, Mahafza WS, Samara
OA, Ryalat AT and Al-Hadidy AM: Myelopathy associated with
age-related cervical disc herniation: A retrospective review of
magnetic resonance images. Ann Saudi Med. 37:130–137. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rao RD, Currier BL, Albert TJ, Bono CM,
Marawar SV, Poelstra KA and Eck JC: Degenerative cervical
spondylosis: Clinical syndromes, pathogenesis, and management. J
Bone Joint Surg Am. 89:1360–1378. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hughes SP, Freemont AJ, Hukins DW,
McGregor AH and Roberts S: The pathogenesis of degeneration of the
intervertebral disc and emerging therapies in the management of
back pain. J Bone Joint Surg Br. 94:1298–1304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Humzah MD and Soames RW: Human
intervertebral disc: Structure and function. Anat Rec. 220:337–356.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grunhagen T, Wilde G, Soukane DM,
Shirazi-Adl SA and Urban JP: Nutrient supply and intervertebral
disc metabolism. J Bone Joint Surg Am. 88 Suppl 2:S30–S35. 2006.
View Article : Google Scholar
|
|
7
|
Huang YC, Urban JP and Luk KD:
Intervertebral disc regeneration: Do nutrients lead the way? Nat
Rev Rheumatol. 10:561–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Urban JP, Smith S and Fairbank JC:
Nutrition of the intervertebral disc. Spine (Phila Pa 1976).
29:2700–2709. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moon SM, Yoder JH, Wright AC, Smith LJ,
Vresilovic EJ and Elliott DM: Evaluation of intervertebral disc
cartilaginous endplate structure using magnetic resonance imaging.
Eur Spine J. 22:1820–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ariga K, Miyamoto S, Nakase T, Okuda S,
Meng W, Yonenobu K and Yoshikawa H: The relationship between
apoptosis of endplate chondrocytes and aging and degeneration of
the intervertebral disc. Spine (Phila Pa 1976). 26:2414–2420. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Holm S, Holm AK, Ekström L, Karladani A
and Hansson T: Experimental disc degeneration due to endplate
injury. J Spinal Disord Tech. 17:64–71. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuss P, Kraft K, Stumm J, Ibrahim D,
Vallecillo-Garcia P, Mundlos S and Stricker S: Regulation of cell
polarity in the cartilage growth plate and perichondrium of
metacarpal elements by HOXD13 and WNT5A. Dev Biol. 385:83–93. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Macsai CE, Georgiou KR, Foster BK,
Zannettino AC and Xian CJ: Microarray expression analysis of genes
and pathways involved in growth plate cartilage injury responses
and bony repair. Bone. 50:1081–1091. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pattappa G, Li Z, Peroglio M, Wismer N,
Alini M and Grad S: Diversity of intervertebral disc cells:
Phenotype and function. J Anat. 221:480–496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miyaki S, Sato T, Inoue A, Otsuki S, Ito
Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al:
MicroRNA-140 plays dual roles in both cartilage development and
homeostasis. Genes Dev. 24:1173–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sumiyoshi K, Kubota S, Ohgawara T, Kawata
K, Nishida T, Shimo T, Yamashiro T and Takigawa M: Identification
of miR-1 as a micro RNA that supports late-stage differentiation of
growth cartilage cells. Biochem Biophys Res Commun. 402:286–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang BS, Minn AJ, Muchmore SW, Fesik SW
and Thompson CB: Identification of a novel regulatory domain in
Bcl-X (L) and Bcl-2. EMBO J. 16:968–977. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yabuki S, Onda A, Kikuchi S and Myers RR:
Prevention of compartment syndrome in dorsal root ganglia caused by
exposure to nucleus pulposus. Spine (Phila Pa 1976). 26:870–875.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagata S and Golstein P: The Fas death
factor. Science. 267:1449–1456. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blanco FJ, Guitian R, Vazquez-Martul E, de
Toro FJ and Galdo F: Osteoarthritis chondrocytes die by apoptosis.
A possible pathway for osteoarthritis pathology. Arthritis Rheum.
41:284–289. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen H, Wang J, Hu B, Wu X, Chen Y, Li R
and Yuan W: MiR-34a promotes Fas-mediated cartilage endplate
chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem.
406:21–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of type II collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E717–E724. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Wang WJ, Yan YG, Xiang YX, Zhang
J, Tang ZH and Jiang ZS: MicroRNAs: New players in intervertebral
disc degeneration. Clin Chim Acta. 450:333–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuisma M, Karppinen J, Haapea M,
Lammentausta E, Niinimäki J and Tervonen O: Modic changes in
vertebral endplates: A comparison of MR imaging and multislice CT.
Skeletal Radiol. 38:141–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nagata S: Apoptosis by death factor. Cell.
88:355–365. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X: The expanding role of mitochondria
in apoptosis. Genes Dev. 15:2922–2933. 2001.PubMed/NCBI
|
|
31
|
Nagata S: Fas ligand-induced apoptosis.
Annu Rev Genet. 33:29–55. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsujimoto Y, Cossman J, Jaffe E and Croce
CM: Involvement of the bcl-2 gene in human follicular lymphoma.
Science. 228:1440–1443. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scaffidi C, Fulda S, Srinivasan A, Friesen
C, Li F, Tomaselli KJ, Debatin KM, Krammer PH and Peter ME: Two
CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675–1687. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis
D, Jia LT, Wu SX, Huang J, Chen J and Luo ZJ: Deregulated miR-155
promotes Fas-mediated apoptosis in human intervertebral disc
degeneration by targeting FADD and caspase-3. J Patho. 225:232–242.
2011. View Article : Google Scholar
|