Introduction
Glioma originates from glial cells and is the most
aggressive and lethal primary brain tumor of the adult central
nervous system (1,2). Patients with glioma are routinely
treated with surgical resection combined with radiotherapy,
chemotherapy and other comprehensive treatments (3,4). Severe
symptoms of glioblastoma, including seizures and cerebral
hemorrhage, make the effective treatment of this tumor a challenge
(5). It has been demonstrated that
that glioblastomas account for ~75% of all malignant brain tumors
(6). The World Health Organization
categorizes glioblastoma into four grades based on the pathological
characteristics of malignancy and indicates that the five-year
survival is lower than other types of cancer (7,8). Due to
variations in infiltrative growth, the malignant grades of
glioblastoma are diverse in appearance (9). Therefore, whilst glioblastoma has
accrued clinical interest, developing an effective treatment for
patients remains a challenge.
Oncolytic virotherapy may be a promising form of
gene therapy for the treatment of various types of cancer. It
utilizes a combination of viral oncolytic properties and functional
genes to destroy malignant cells (10). Cassel and Murray (11) demonstrated that Newcastle disease
virus (NDV) oncolysates may be a promising anticancer agent in
patients with stage III malignant melanoma. The potential
therapeutic effects of NDV have prompted research into the
underlying mechanism of oncolytic viruses (12). Previous studies have demonstrated
that NDV exhibits cancer cell selectively by inducing interferon
(IFN) and inhibiting NDV replication via its sialic acid receptor.
This is unusual, as the majority of tumor cells impair the IFN
signaling pathway (13,14). NDV therefore replicates in tumor
cells and induces a potent IFN immune response to inhibit tumor
cell growth. However, previous studies have also demonstrated that
NDV treatment alone is insufficient to fully inhibit tumor growth
(15,16).
The tumor suppressor protein p53 is encoded by the
tumor protein 53 gene and serves a primary role in a number of
pathways, including cell cycle, cell growth, differentiation,
apoptosis and cell death (17,18). It
has been demonstrated that p53 regulates the variation and repair
of cells when exposed to DNA-damaging agents, including ultraviolet
radiation, toxins and certain drugs (19). Similar to various other cancers, the
occurrence of glioma is associated with genetic mutations in p53
(20–24). The alteration or inactivation of p53
induced by mutations or its interaction with oncogene products of
DNA tumor viruses may lead to cancer (25). Low p53 expression occurs in glioma,
which leads to dysfunction of the p53-associated regulatory
pathways (26). Therefore,
dysregulation of p53 may contribute to the regression of glioma
therapy.
In the present study, the therapeutic effects of
recombinant (r)NDV-p53 in glioma cell lines and tumor models were
assessed. The role of p53 as a molecular marker of glioma remains
controversial as previous studies have demonstrated that no
association exists between p53 and prognosis (27,28).
However, the results of the present study demonstrate that rAd-p53
exhibits an anticancer effect in tumor-bearing mice. Intravenous
injections of rNDV-p53 in pre-clinical examinations demonstrated
that gene-targeted oncolytic virotherapy exhibited marked effects
on glioma growth. The present study aimed to assess the efficacy
and impact of p53 on the growth, aggressiveness, apoptosis,
prognosis and immunoregulatory function of NDV in the treatment of
glioma, as well as its effects on T lymphocyte infiltration,
immunologic memory and specific toxicity in vivo. The
results of the present study provide an insight into the
pathophysiology of glioma and suggest that rNDV-p53 may serve as a
potential anti-cancer oncolytic drug for the treatment of
glioma.
Materials and methods
Ethics statement
The present study was performed in strict accordance
with the Guide for the Care and Use of Laboratory Animals of
Qianfoshan Hospital (Shandong, China). All experimental protocols
involving animals were performed in accordance with the National
Institutes of Health and approved by the Ethics Committee of
Qianfoshan Hospital of Shandong Province (Shandong, China). All
surgery and euthanasia were performed to minimize suffering. The
use of human tissue samples was also approved by the Qianfoshan
Hospital of Shandong Province.
Patient tissue samples
A total of 4 patients with glioma (2 females, 2
females, aged 46–62 years old) were recruited from the Qianfoshan
Hospital of Shandong Province between January 2015 and May 2016.
Glioma samples and adjacent non-tumor tissues were obtained from
the same individuals. All patients signed a study-specific written
informed consent prior to inclusion.
Construction of rNDV
The expression system of NDV was used to construct
the rNDV virion. The 585 base pair DNA sequence encoding human p53
(forward primer, 5′-TGGAGGAGCCGCAGTCAGAT-3′, reverse primer,
5′-ATATCGTCCGGGGACAGC-3′) and the enhanced green fluorescent
protein (EGFP) were amplified from plasmid expression p53 (pMD-p53)
using polymerase chain reaction [PCR; 25 µl volume: Primers, 1 µl,
DNA polymerase (Takara Bio, Inc., Otsu, Japan), 1 µl dNTP, 2 µl
Buffer, 2 µl water] and subcloned into rNDV plasmids using TA
Cloning™ kit (cat. no. K200001; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) as described previously and named rNDV-P53
(29). The thermocycling conditions
were as follows: 30 cycles of 95°C for 9 sec, 54.5°C for 5 sec and
72°C for 20 sec. The PCR products were analyzed by 1% agarose gel
electrophoresis and photographed by Image Master VDS Gel Imaging
system (Pharmacia Biotech, Uppsala, Sweden). rNDV-p53 and rNDV-EGFP
plasmids were acquired. PCR and gene sequencing were used to select
the correct clone and translated into E. coli. rNDV,
rNDV-EGFP and rNDV-p53 (10 µg, constructed by the Department of
Neurosurgery, Qianfoshan Hospital of Shandong Province) were
generated through transfection into CEF1 cells (BeNa Biotechnology,
Shanghai, China) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Following 72 h transfection, rNDVs were propagated in specific
pathogen free (SPF) embryonated chickens (Microbiology Laboratory,
Shangdong University, Jinan, China). rNDV viruses were purified
following a previously described protocol (30). NDV titers were determined using a
TCID50 assay using the Reed-Muench method and recorded
as plaque-forming units (pfu)/ml, following the protocol of a
previous study (31). MOI was
calculated by measuring TCID50.
MTT cytotoxicity
Glioblastoma cell lines G422 (cat. no. BNCC340295)
and U251 (cat. no. BNCC337874) were purchased from Beijing BeiNa
Institute of Biotechnology (Beijing, China) and cultured in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified
atmosphere containing 5% CO2. G422 and U251 cells were
then incubated with p53 (10 mg/ml; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), rNDV-EGFP, rNDV-p53 or PBS in a 96-well plate
for 96 h at 37°C in triplicate for each condition for 48 h. A total
of 20 µl MTT (5 mg/ml) in PBS solution was added to each well and
cells were incubated for a further 4 h at 37°C. Medium was removed
and 100 µl dimethylsulfoxide was added to dissolve the formazan
crystals. Optical density was measured using an ELISA reader at a
wavelength of 450 nm.
Animal analysis
A total of 45 SPF female BALB/c nude mice (6 weeks
old; body weight, 26–32 g) were purchased from the Harbin
Veterinary Research Institute (Harbin, China). All animals were
housed in a temperature-controlled facility at 23±1°C and a
relative humidity of 50±5%. Animals were subjected to a 12 h
light/dark cycle and had ad libitum access to food and
water. A total of 100 µl U251 cells at a density of
5×105 were injected into the right flank of mice.
Treatment for tumor-bearing mice, rNDV-EGFP or rNDV-p53 was
initiated when tumor diameters reached 6–8 mm at 7 days following
inoculation. Mice with glioma were randomly divided into 3 groups
(n=15) and injected intratumorally with 2×107 pfu
rNDV-p53, rAd-EGFP or PBS. Treatment was performed once every other
day for a total of 10 days. Tumor diameters were recorded once
every 2 days and tumor volume was calculated by using the following
formula: 0.52 × smallest diameter2 × largest diameter.
Tumor volume was recorded over a 30 day period of observation
following the 10 day treatment period. The survival rate of
experimental mice was calculated in a long-term experiment
conducted over 180 days using Kaplan-Meier method (32).
Cell culture and flow cytometric
analysis (FACS)
Cell suspensions (5×106) from the tumors
of treated mice were prepared for FACS on day 30. Tumor cell
suspensions from experimental mice were filtered through a 100 µm
nylon strainer. Tumor cells were then labeled with cluster of
differentiation (CD)31 (1:500; cat. no. ab28364; Abcam, Cambridge,
UK) and CD69 (1:500; cat. no. ab202909; Abcam) for 12 h at 4°C,
followed by an incubation with goat anti-rabbit horseradish
peroxidase (HRP)-conjugated immunoglobulin G (IgG; Alexa
Fluor® 488, 1:1,000; cat. no. ab150077; Abcam) for 2 h
at 37°C to assess the frequency of CD31 and CD69 cell subsets in
the total number of infiltrated immune cells. Stained cells were
analyzed using a FACScan flow cytometer. To assess cell apoptosis,
G422 cells (1×106) were incubated with an Annexin
V-fluorescein isothiocyanate/propidium iodide double staining kit
(Beyotime Institute of Biotechnology, Haimen, China) for 15 min at
room temperature according to the manufacturer's protocol. The
ratios of apoptotic cells were measured using a Coulter EPICS XL
Flow Cytometer and the results were analyzed using Expo32-ADC v.
1.2B software (Beckman Coulter, Inc., Brea, CA, USA).
Splenocyte collection and cytotoxic T
cell (CTL) responses
Splenocytes were obtained from the spleens of
experimental mice following treatment. The monoplast suspension was
washed three times with PBS. U251 cells were inactivated with
ethylalcohol (Sigma-Aldrich; Merck KGaA) for 30 min at 37°C.
Inactivated U251 cells were used to incubate splenocytes. IFN-γ
levels were assessed using a mouse IFN-γ Quantikine ELISA kit
(MIF00; Bio-Rad Laboratories Inc., Hercules, CA, USA) in the
supernatants obtained from cell culture fluid following a 72 h
culture at 37°C and centrifugation at 3,000 × g for 10 min at room
temperature. T cells (1×106) obtained from splenocytes
were purified (33) and co-cultured
with fresh U251 cells at 37°C for 4 h at effector:target ratios of
5:1, 15:1 and 45:1. CTL activity on target cells was determined
using MTT cytotoxicity assays as previously described (34).
Tumor cell migration and invasion
assays
G422 and U251 cells were cultured in DMEM and
treated with rNDV-EGFP or rNDV-p53. Cells were then incubated in
DMEM medium with 5% FBS for 48 h at 37°C using a Transwell insert
(BD Biosciences, Franklin Lakes, NJ, USA) instead of a Matrigel
invasion chamber to assess migration. In the invasion assay, rNDV
or rNDV-p53-treated cells were suspended at a density of
5×104 in 200 µl serum-free DMEM. DMEM medium with 5% PBS
were plated in the lower chamber of the BD BioCoat Matrigel
invasion chamber (BD Biosciences). G422 and U251 cells and then
plated in the upper chamber for 48 h at 37°C following the
manufacturer's protocol. After 48 h, the cells that invaded through
the membrane were fixed with 3% formaldehyde for 15 min at 37°C and
stained with 0.5% crystal violet for 10 min at 37°C. The invasion
and migration of tumor cells were assessed in a minimum of three
randomly selected fields using an inverted microscope (Olympus
BX51; Olympus Corporation, Tokyo, Japan) at ×40 magnification.
Immunohistochemical staining
Tumor tissues were prepared and fixed in 4%
paraformaldehyde for 2 h at 37°C. Tissues were deparaffinized in
xylene and rehydrated in a graded alcohol series. Following washing
with PBS for 15 min at room temperature, tissue sections (4 µm)
were prepared and epitope retrieval was performed using Lab Vision™
Tris-HCl buffer for heat-induced epitope retrieval (cat. no.
AP-9005-050; Thermo Fisher Scientific, Inc.). Paraffin-embedded
sections were quenched with 3% hydrogen peroxide for 15 min and
subsequently blocked with 5% bovine serum albumin (Sigma-Aldrich;
Merck KGaA) for 15 min at 37°C. Sections were then incubated with
antibodies against p53 (1:1,000; cat. no. ab1431), p21 (1:1,000;
cat. no. ab1091919), caspase-3 (1:1,000; cat. no. ab13847), B cell
lymphoma-2 (Bcl2; 1:1,000; cat. no. ab59348), Bcl-2 associated X
(Bax; 1:1,000; cat. no. ab32503), Bcl-2 like 11 (Bim; 1:1,000; cat.
no. ab32158), CD31 (1:1,000; cat. no. ab28364) and CD69 (1:1,000;
cat. no. ab202909) at 4°C for 12 h following blocking. All primary
antibodies were sourced from Abcam. All sections were washed three
times with PBS and incubated with secondary antibodies
HRP-conjugated IgG (1:5,000; PV-6001; OriGene Technologies Inc.,
Rockville, MD, USA) for 1 h at 37°C. Visualization was achieved
using peroxidase-labeled streptavidin-biotin and diaminobenzidine
(Advansta, Inc., Menlo Park, CA, USA) for at least 5 min at 37°C.
The slides were examined with a Keyence Biozero BZ8100E
fluorescence microscope (Keyence, Osaka, Japan) at a magnification
of ×40.
Western blotting
G422 and U251 cells were treated with rNDV-EGFP or
rNDV-p53 and homogenized in 10% RIPA buffer (Sigma-Aldrich; Merck
KGaA) for 1 h at 37°C lysate buffer containing a
protease-inhibitor. Cells were then centrifuged at 6,000 × g at 4°C
for 10 min and supernatants were analyzed. Protein concentration
was measured using a BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Protein samples (10 µg) were separated on 12.5%
SDS-PAGE and transferred onto polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA) SDS assays were performed as
previously described (35).
Membranes were then blocked with 5% skimmed milk for 1 h at 37°C
and then incubated with the following primary antibodies: p53
(1:1,000; cat. no. ab1431), p21 (1:1,000; cat. no. ab1091919),
caspase-3 (1:1,000; cat. no. ab13847), Bcl2 (1:1,000; cat. no.
ab59348), Bax (1:1,000; cat. no. ab32503), Bim (1:1,000; cat. no.
ab32158) and β-actin (1:1,000; cat. no. ab8226) for 12 h at 4°C.
All primary antibodies were supplied by Abcam. The membranes were
then incubated with goat anti-rabbit HRP-conjugated IgG secondary
antibodies (1:5,000; cat. no. PV-6001; OriGene Technologies, Inc.)
at 4°C for 24 h. Blots were imaged using WesternBright ECL
Chemiluminescent HRP Substrate (Advansta).
TUNEL analysis
Tumor tissue sections were fixed with 4%
paraformaldehyde solution for 2 h at 4°C. Sections were washed
three times with PBS and then permeabilized by immersing cells
slides in a 0.2% Triton X-100 solution with PBS for 14 min at 4°C.
Subsequently, sections were incubated with an equilibration buffer
for 14 min at 4°C and were then incubated with 50 µl reaction
mixture at 37°C for 60 min and washed 3 times with PBS. The tissues
or cells were incubated with 0.5 µg/ml DAPI (Sigma-Aldrich; Merck
KGaA) in a humidified chamber in the dark at 37°C for 14 min.
Following 3 washes with PBS, the terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) Apo-Green Detection kit (Biotool, Stratech Scientific,
Ltd., Suffolk, UK) was used according to the manufacturers protocol
to detect TUNEL-positive cells. Finally, tissue section images were
captured at 6 fields of view using a ZEISS LSM 510 confocal
microscope (Zeiss AG, Oberkochen, Germany).
Statistical methods
All data are presented as the mean ± standard error
of the mean. Unpaired data were analyzed using a Student's t-test.
Comparisons between multiple groups were analyzed using one-way
analysis of variance followed by Tukey's honest significance
difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characteristics of rNDV and p53
expression in rAd-p53-infected cells in vitro
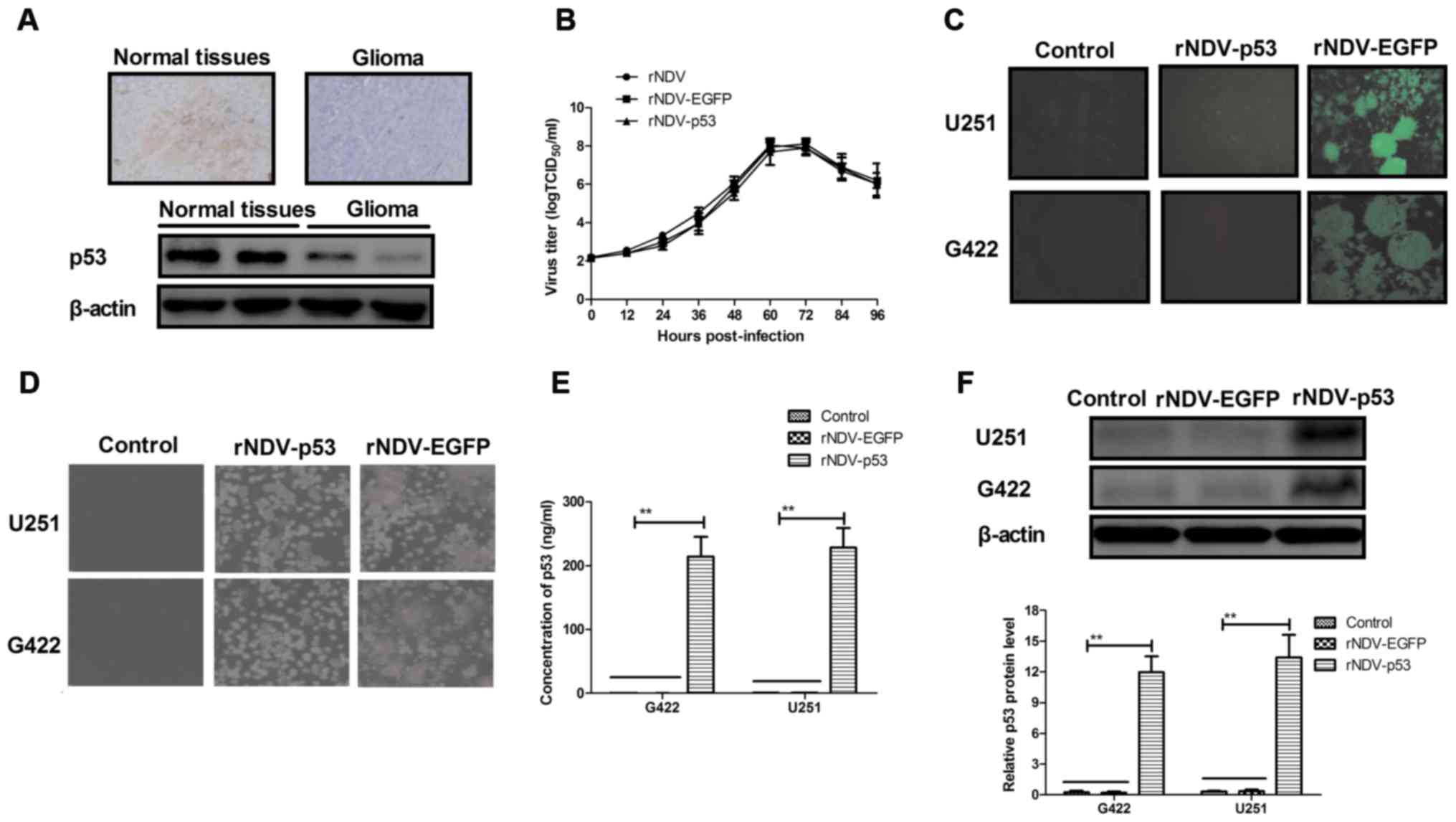
The expression of p53 was assessed in human glioma
tissues using immunohistochemistry. It was demonstrated that p53
expression was decreased in human glioma tissues compared with
normal adjacent tissue (non-tumor tissue situated around the site
of glioma; Fig. 1A). In order to
assess the efficiency of rNDV replication, a growth dynamics curve
was constructed using CEF1 cells. The results indicated that growth
was marginally affected by p53 and EGFP and viruses recovered
parental titers following 60 h (Fig.
1B). It was also demonstrated that there was an increased
expression of rNDV-EGFP in G422 and U251 cells (Fig. 1C). Increased formation of syncytium
in rNDV-p53 treated cells compared with rNDV and controls (Fig. 1D) was also observed. In addition, the
endogenous expression of p53 in G422 and U251 cells was assessed
using ELISA and western blotting. The results demonstrated that p53
was highly expressed in G422 and U251 cells (Fig. 1E and F). Collectively, these results
demonstrate that glioma cells exhibit decreased p53 expression and
that rNDV-p53 selectively replicates and expresses p53 in glioma
cell lines.
Characterization of p53 expressed by
rNDV-p53 treated glioma cells
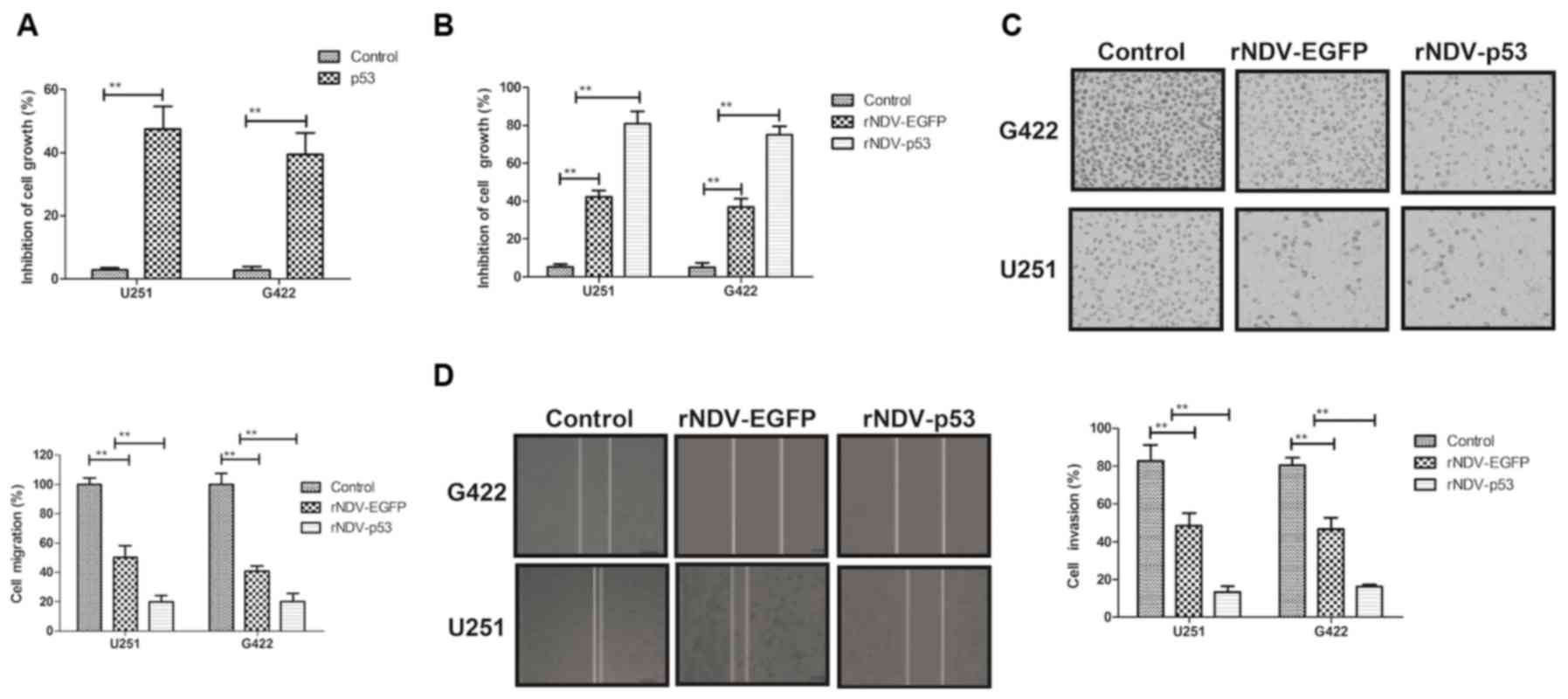
The biological activity of p53 in
rNDV-p53-transfected glioma cells was assessed. It was demonstrated
that p53 (10 mg/ml) significantly inhibited the growth of G422 and
U251 cells (P<0.01; Fig. 2A). The
inhibitory effects of rNDV-p53 on glioma cells were observed in
vitro. The rate of inhibition was significantly increased in
cells treated with rNDV-p53 compared with those that received PBS
and rNDV-EGFP (P<0.01; Fig. 2B).
The expression of p53, delivered by rNDV-p53, significantly
suppressed the migration and invasion of G422 and U251 cells
compared with those treated with rNDV-EGFP (P<0.01; Fig. 2C and D). These results indicate that
rNDV-p53 inhibits the growth and aggressiveness of glioma
cells.
rNDV-p53 downregulates anti-apoptosis
and activated pro-apoptosis proteins
It has been demonstrated that U251 is more
aggressive than other glioma cell lines (36). U251 cells were therefore utilized to
assess the mechanism of rNDV-p53-mediated apoptosis. To investigate
whether rNDV-p53 upregulates p53 downstream proteins and enhances
glioma cell apoptosis, the expression of p53, p21 and caspase-3 was
examined following treatment for 48 h (Fig. 3A). The rNDV-p53 virus markedly
enhanced the transcriptional activity of p53, p21 and caspase-3.
rNDV-p53 treatment also markedly inhibited the expression of
anti-apoptotic proteins, including Bcl-2, Bim and Bax in U251 cells
(Fig. 3B). In addition, it was
demonstrated that rNDV-p53 promotes U251 cell apoptosis at a
multiplicity of infection (MOI) of 5 for 24 h when compared with
rNDV-EGFP infected and control cells (Fig. 3C). In addition, TUNEL staining
analysis demonstrated a significantly increased number of
TUNEL-positive cells in rNDV-p53 treated cells (MOI=5) following 48
h incubation (P<0.01; Fig. 3D).
These results indicate that rNDV-p53 promotes glioma cell
apoptosis.
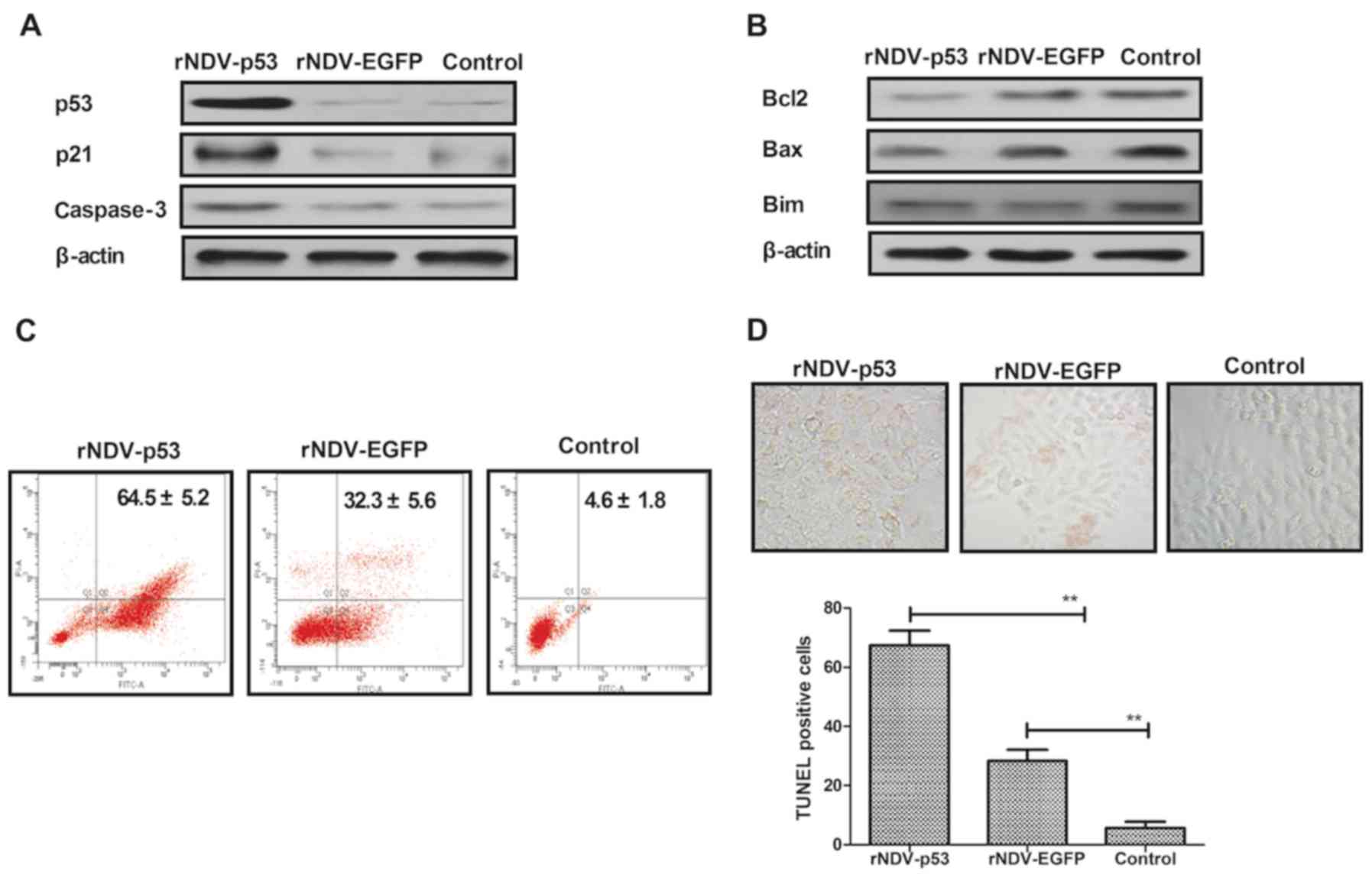 | Figure 3.rNDV-p53 promotes and activates
anti-apoptotic proteins. (A) p53, p21 and caspase-3 levels in U251
cells following treatment with rNDV-p53, rNDV-EGFP or controls. (B)
Bcl-2, Bim and Bax expression in U251 cells following treatment
with rNDV-p53, rNDV-EGFP or controls. (C) rNDV-p53 treatment for 24
h promoted U251 cell apoptosis at an MOI of 5, compared with
rNDV-EGFP and control treated cells. (D) TUNEL-positive U251 cells
incubated for 48 h with rNDV-p53, rNDV-EGFP or control (MOI=5;
magnification, ×40). **P<0.01 vs. control. rNDV, recombinant
Newcastle disease virus; EGFP, enhanced green fluorescent protein;
Bcl-2, B cell lymphoma-2; Bim, Bcl-2 like 11; Bax, Bcl-2 associated
X; MOI, multiplicity of infection. |
In vivo anti-tumor efficacy of
rNDV-p53
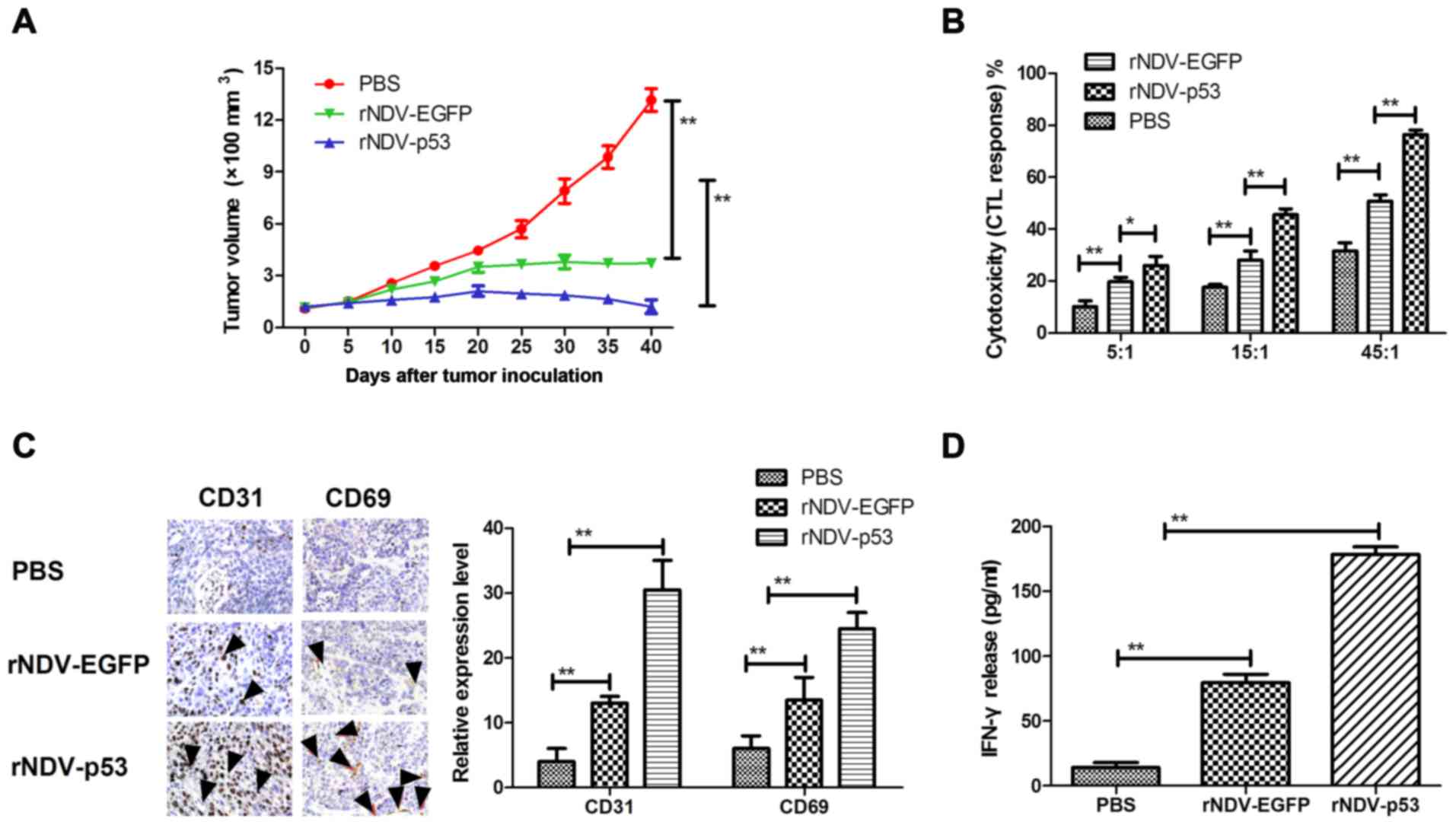
In order to assess the therapeutic effects of
rNDV-p53, mice were xenografted with glioma tissue and treated with
rNDV-p53 via intravenous injection with PBS and rAd-EGFP as a
control. The results demonstrated that rNDV-p53 significantly
inhibited glioma growth compared with rNDV-EGFP and PBS treated
mice over the 30 day observation period (P<0.01; Fig. 4A). In addition, CTL responses against
U251 cells were assessed. Mice treated with rNDV-p53 developed a
stronger CTL response against U251 cells compared with rNDV-EGFP
and PBS groups (Fig. 4B).
Furthermore, CD65 and CD31 cell infiltration was upregulated in
tumors following treatment with rNDV-EGFP (Fig. 4C). Mice treated with rNDV-p53 also
exhibited a significantly higher IFN-γ release compared with those
that received rNDV-EGFP and PBS treatment (P<0.01; Fig. 4D).
rNDV-p53 induces glioma tumor
apoptosis and prolongs survival in vivo
The obtained results indicated that rNDV-p53
inhibits the progression of U251-bearing mice. Apoptosis rates in
tumors following treatment with rNDV-p53, rNDV-EGFP or PBS was
verified using TUNEL staining (Fig.
5A) and it was demonstrated that tumor growth was inhibited.
Additionally, the downstream molecules of p53, including p21 and
caspase 3, were assessed by performing immunohistochemical
staining. Results demonstrated that rNDV-p53 treatment upregulated
these molecules in murine tumors (Fig.
5B). Further analysis revealed that rNDV-p53 treatment induced
the downregulation of anti-apoptotic proteins, including Bcl-2, Bim
and Bax compared with mice that received rNDV-EGFP and PBS
(Fig. 5C). The long-term survival
rate of mice following drug treatment was calculated to be 180
days. The results of the current study demonstrated that rNDV-p53
significantly prolonged the survival of mice compared with control
groups (P<0.01; Fig. 5D). These
data suggest that rNDV-p53 efficiently inhibits glioma and
eliminates tumor cells by initiating apoptosis, which contributes
to long-term tumor-free survival.
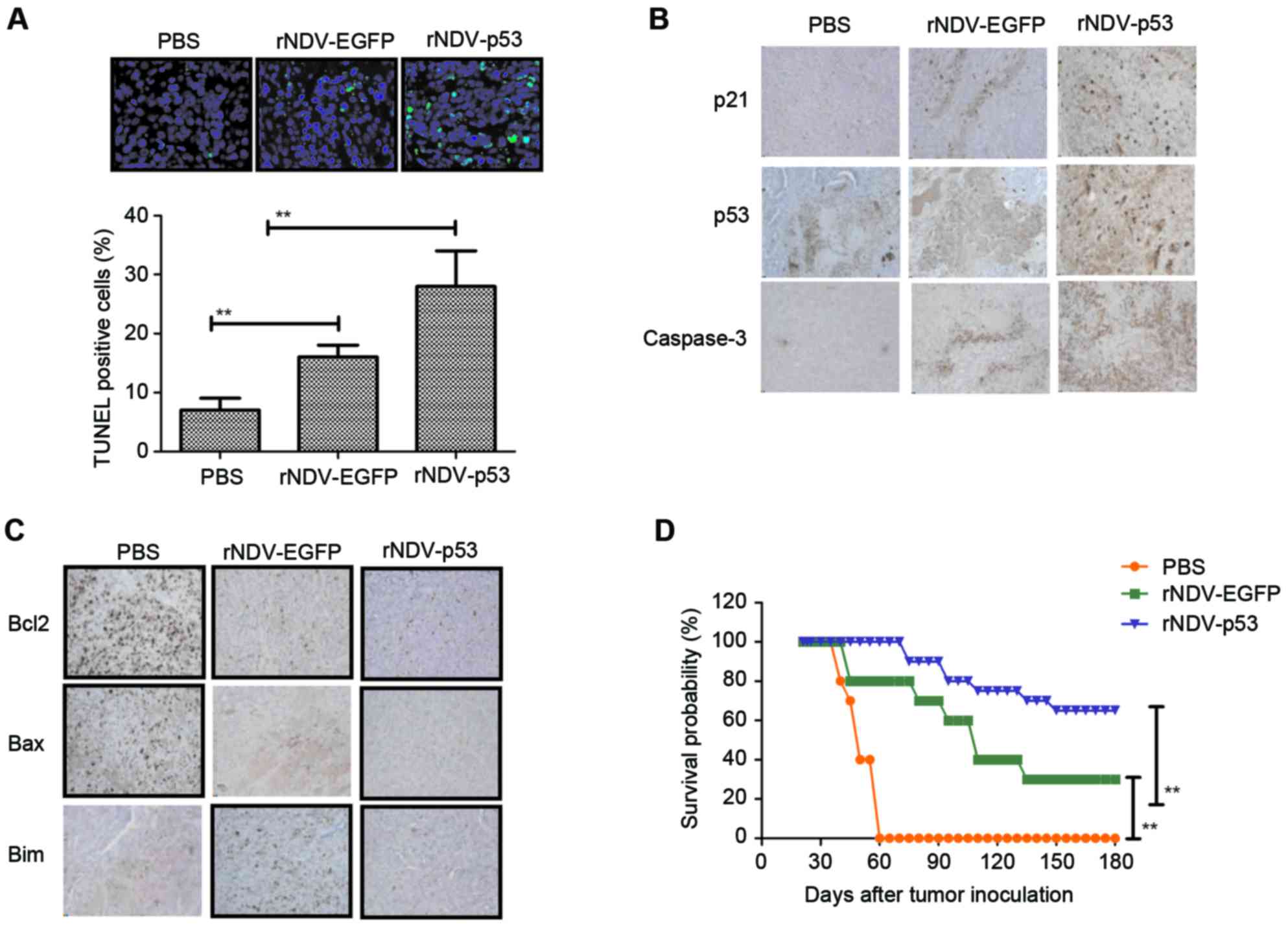 | Figure 5.Cell apoptosis and mice survival rate
in rNDV-p53 treated glioma. (A) TUNEL-positive glioma cells
following treatment with rNDV-p53, rNDV-EGFP or PBS (magnification,
×20). (B) p53, p21 and caspase3 and (C) Bcl-2, Bax and Bim
expression in tumors from experimental mice following treatment
with rNDV-p53, rNDV-EGFP or PBS (magnification, ×20). (D) Survival
rates of rNDV-p53, rNDV-EGFP or PBS treated U251-bearing mice over
a 180-day observation. **P<0.01 vs. control. rNDV, recombinant
Newcastle disease virus; EGFP, enhanced green fluorescent protein;
Bcl-2, B cell lymphoma-2; Bax, Bcl-2 associated X; Bim, Bcl-2 like
11. |
Discussion
The results of the present current study indicate
that rNDV-p53 enhances the anti-cancer potential of rNDV and p53
and exhibits strong inhibitory effects on glioma cells in
vitro and in vivo. It was also demonstrated that
rNDV-p53 induces an immune response against U251 cells.
NDV-mediated p53 gene expression exhibited increased anti-tumor
effects through NDV mediated cell death and p53-mediated apoptosis
induction. Anti-apoptotic protein expression levels were
significantly increased in glioma cells following treatment with
rNDV-p53, indicating that the apoptosis-resistance observed in
various types of cancer had been attenuated. In addition, it was
demonstrated that rNDV-p53 treatment initiates an anti-tumor immune
response, protecting mouse tissues. These results indicate that
disruption to rNDV-p53-glioma interactions may be an effective
future anti-cancer target.
NDV was utilized as an anti-tumor agent based on a
previous study that examined a patient with cervical carcinoma
(37). The oncolytic mechanism of
NDV has been examined in various carcinomas using reverse genetic
technology (38), which involves the
insertion of certain functional genes or proteins into the genome
of NDV to enhance the therapeutic effects of oncolytic virotherapy
(39). Keshelava et al
(40) assessed the efficacy of NDV
and neoadjuvant therapy in 84 patients with breast cancer. The
results of the study indicated that the LaSotha strain of NDV was
an efficient and safe from of immunotherapy and neoadjuvant
treatment. In addition, Schulze et al (41) examined NDV immunization combined with
modified colorectal cancer cells in patients with colorectal
cancer. It was demonstrated that patient survival rate improved
following treatment, indicating that this therapy may have
anti-cancer effects. Additionally, Bai et al (42) revealed that genetically engineered
NDV expressing interleukin (IL)-2 may be a potential candidate for
cancer immunotherapy in patients with hepatic carcinoma and
melanoma. Chai et al (43)
assessed the use of rNDV in patients with lung cancer, generated
using reverse genetics based on the oncolytic D90 strain and
carrying a gene encoding EGFP. Furthermore, NDV may trigger U251
glioma cell autophagy to enhance viral replication and promote the
apoptosis of tumor cells (44). It
has also been demonstrated that NDV inhibits the decrease in Rac1
gene expression exhibited in glioma tumors (45). In the present study, an rNDV encoding
p53 was constructed to assess efficacy in glioma cells and animals.
The results indicate that rNDV-p53 inhibits glioma cell growth and
aggressiveness in vitro as well as suppressing tumor growth
through the accumulation and infiltration of T lymphocytes and
apoptosis in vivo.
The p53 tumor suppressor gene is mutated in a
variety of cancers and is therefore an important area of cancer
research (46). Elucidating the
scope of p53 mutations allows for the better understanding of
cancer etiology and the molecular pathogenesis of neoplasia
(47,48). The detection of p53 abnormalities may
have diagnostic, prognostic and therapeutic implications in
patients with cancer (49,50). A previous study demonstrated that p53
mutations are important for tumor classification and gliomagenesis
(19). The most common p53 mutations
identified in glioma occur in the DNA-binding domain, specifically
within six hotspot mutation sites (51). p53 protects against neoplastic
transformation and exhibits effects on certain processes, including
cell cycle modulation, DNA repair, apoptosis, senescence,
angiogenesis and metabolism, resulting in the formation of a
complex signaling network (50,52).
However, the distinct effect of p53 in glioma therapy is yet to be
elucidated.
Immunotherapy involving anti-tumor surface antigens
that is used alongside other therapeutic methods, including
chemoradiotherapy and surgery, has been observed to have
therapeutic activities in animal models of different types of
cancer (53–55). The use of anti-neoplastic agents and
immunotherapy is an effective therapy for tumor cells with specific
recognition molecules, antigen domains or receptors (56–59). The
results of the present study indicate that rNDV-p53 stimulates the
immune system induce glioma cell death in U251-bearing mice.
Additionally, p53 gene therapy delivered by NDV is an effective
gene delivering system.
The present study demonstrates that rNDV-p53
treatment inhibits murine glioma growth by stimulating T cell
proliferation, memory T cell responses, CTL responses and IFN-γ
release targeted against tumor cells. The mechanism of
rNDV-p53-mediated anti-glioma therapy was also assessed and the
results demonstrated a significant increase in tumor cell apoptotic
rate. In conclusion, the present study indicates that rNDV-p53
possesses potential beneficial effects for the oncolytic efficacy
of NDV by expressing p53 in the treatment of glioma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science
Foundation of Shandong Province (grant no. 2013ZRB14142) and the
Development Foundation of Shandong Medical and Health Science and
Technology (grant no. 2016WS0480).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XF, HL, YC, XH analyzed and interpreted the data.
XF, XH, CH, GL performed the experiments, and GL was a major
contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in strict accordance
with the Guide for the Care and Use of Laboratory Animals of
Qianfoshan Hospital (Shandong, China). All experimental protocols
involving animals were performed in accordance with the National
Institutes of Health and approved by the Ethical Committee of
Qianfoshan Hospital of Shandong Province (Shandong, China). All
surgery and euthanasia were performed to minimize suffering. The
use of human tissue samples was also approved by the Qianfoshan
Hospital of Shandong Province and all patients signed a
study-specific written informed consent prior to inclusion.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Lu M, Zhang X, Zhang M, Chen H, Dou W, Li
S and Dai J: Non-model segmentation of brain glioma tissues with
the combination of DWI and fMRI signals. Biomed Mater Eng.
26:S1315–S1324. 2015.PubMed/NCBI
|
|
2
|
Chow KK, Naik S, Kakarla S, Brawley VS,
Shaffer DR, Yi Z, Rainusso N, Wu MF, Liu H, Kew Y, et al: T cells
redirected to EphA2 for the immunotherapy of glioblastoma. Mol
Ther. 21:629–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Englot DJ, Berger MS, Chang EF and Garcia
PA: Characteristics and treatment of seizures in patients with
high-grade glioma: A review. Neurosurg Clin N Am. 23:227–235. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siu A, Wind JJ, Iorgulescu JB, Chan TA,
Yamada Y and Sherman JH: Radiation necrosis following treatment of
high grade glioma-a review of the literature and current
understanding. Acta neurochir(Wien). 154:191–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taphoorn MJ and Bottomley A:
Health-related quality of life and symptom research in glioblastoma
multiforme patients. Expert Rev Pharmacoecon Outcomes Res.
5:763–774. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Delgado-López PD and Corrales-Garcia EM:
Survival in glioblastoma: A review on the impact of treatment
modalities. Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benjamin R, Capparella J and Brown A:
Classification of glioblastoma multiforme in adults by molecular
genetics. Cancer J. 9:82–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ortega A, Sarmiento JM, Ly D, Nuño M,
Mukherjee D, Black KL and Patil CG: Multiple resections and
survival of recurrent glioblastoma patients in the temozolomide
era. J Clin Neurosci. 24:105–111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koekkoek JA, Postma TJ, Heimans JJ,
Reijneveld JC and Taphoorn MJ: Antiepileptic drug treatment in the
end-of-life phase of glioma patients: A feasibility study. Support
Care Cancer. 24:1633–1638. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
LaRocca CJ and Davydova J: Oncolytic
virotherapy increases the detection of microscopic metastatic
disease at time of staging laparoscopy for pancreatic
adenocarcinoma. EBioMedicine. 7:15–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cassel WA and Murray DR: A ten-year
follow-up on stage II malignant melanoma patients treated
postsurgically with Newcastle disease virus oncolysate. Med Oncol
Tumor Pharmacother. 9:169–171. 1992.PubMed/NCBI
|
|
12
|
Yaacov B, Eliahoo E, Lazar I, Ben-Shlomo
M, Greenbaum I, Panet A and Zakay-Rones Z: Selective oncolytic
effect of an attenuated Newcastle disease virus (NDV-HUJ) in lung
tumors. Cancer Gene Ther. 15:795–807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang W, Wang H, Sun TM, Yao WQ, Chen LL,
Jin Y, Li CL and Meng FJ: Application of autologous tumor cell
vaccine and NDV vaccine in treatment of tumors of digestive tract.
World J Gastroenterol. 9:495–498. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song KY, Wong J, Gonzalez L, Sheng G,
Zamarin D and Fong Y: Antitumor efficacy of viral therapy using
genetically engineered Newcastle disease virus [NDV(F3aa)-GFP] for
peritoneally disseminated gastric cancer. J Mol Med (Berl).
88:589–596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Puhlmann J, Puehler F, Mumberg D, Boukamp
P and Beier R: Rac1 is required for oncolytic NDV replication in
human cancer cells and establishes a link between tumorigenesis and
sensitivity to oncolytic virus. Oncogene. 29:2205–2216. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Janke M, Peeters B, de Leeuw O, Moorman R,
Arnold A, Fournier P and Schirrmacher V: Recombinant Newcastle
disease virus (NDV) with inserted gene coding for GM-CSF as a new
vector for cancer immunogene therapy. Gene Ther. 14:1639–1649.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dastjerdi MN, Valiani A, Mardani M and Ra
MZ: Adenosine A1 receptor modifies P53 expression and apoptosis in
breast cancer cell line Mcf-7. Bratisl Lek Listy. 117:242–246.
2016.PubMed/NCBI
|
|
18
|
Ohara M, Matsuura K, Akimoto E, Noma M,
Doi M, Nishizaka T, Kagawa N and Itamoto T: Prognostic value of
Ki67 and p53 in patients with estrogen receptor-positive and human
epidermal growth factor receptor 2-negative breast cancer:
Validation of the cut-off value of the Ki67 labeling index as a
predictive factor. Mol Clin Oncol. 4:648–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karsy M, Neil JA, Guan J, Mahan MA, Colman
H and Jensen RL: A practical review of prognostic correlations of
molecular biomarkers in glioblastoma. Neurosurg Focus. 38:E42015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamanishi Y, Boyle DL, Green DR, Keystone
EC, Connor A, Zollman S and Firestein GS: p53 tumor suppressor gene
mutations in fibroblast-like synoviocytes from erosion synovium and
non-erosion synovium in rheumatoid arthritis. Arthritis Res Ther.
7:R12–R18. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ohgaki H, Eibl RH, Schwab M, Reichel MB,
Mariani L, Gehring M, Petersen I, Höll T, Wiestler OD and Kleihues
P: Mutations of the p53 tumor suppressor gene in neoplasms of the
human nervous system. Mol Carcinog. 8:74–80. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakai E, Rikimaru K, Ueda M, Matsumoto Y,
Ishii N, Enomoto S, Yamamoto H and Tsuchida N: The p53
tumor-suppressor gene and ras oncogene mutations in oral
squamous-cell carcinoma. Int J Cancer. 52:867–872. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: Clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
24
|
Rivlin N, Brosh R, Oren M and Rotter V:
Mutations in the p53 Tumor suppressor gene: Important milestones at
the various steps of tumorigenesis. Genes Cancer. 2:466–474. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yue Q, Yulong G, Liting Q, Shuai Y, Delong
L, Yubao L, Lili J, Sidang L and Xiaomei W: Mutations in and
expression of the tumor suppressor gene p53 in egg-type chickens
infected with subgroup j avian leukosis virus. Vet Pathol.
52:1052–1056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sitarek P, Skala E, Toma M, Wielanek M,
Szemraj J, Nieborowska-Skorska M, Kolasa M, Skorski T, Wysokińska H
and Śliwiński T: A preliminary study of apoptosis induction in
glioma cells via alteration of the Bax/Bcl-2-p53 axis by
transformed and non-transformed root extracts of Leonurus sibiricus
L. Tumour Biol. 37:8753–8764. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen GX, Zheng LH, Liu SY and He XH:
rAd-p53 enhances the sensitivity of human gastric cancer cells to
chemotherapy. World J Gastroenterol. 17:4289–4297. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie Q, Liang BL, Wu YH, Zhang J, Chen MW,
Liu HY, Gu XF and Xu J: Synergistic anticancer effect of rAd/P53
combined with 5-fluorouracil or iodized oil in the early
therapeutic response of human colon cancer in vivo. Gene.
499:303–308. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walker A, Taylor J, Rowe D and Summers D:
A method for generating sticky-end PCR products which facilitates
unidirectional cloning and the one-step assembly of complex DNA
constructs. Plasmid. 59:155–162. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan F, Zheng Y and Huang L:
Adenovirus-mediated combined anti-angiogenic and pro-apoptotic gene
therapy enhances antitumor efficacy in hepatocellular carcinoma.
Oncol Lett. 5:348–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bai FL, Yu YH, Tian H, Ren GP, Wang H,
Zhou B, Han XH, Yu QZ and Li DS: Genetically engineered Newcastle
disease virus expressing interleukin-2 and TNF-related
apoptosis-inducing ligand for cancer therapy. Cancer Biol Ther.
15:1226–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abd El Hafeez S, Torino C, D'Arrigo G,
Bolignano D, Provenzano F, Mattace-Raso F, Zoccali C and Tripepi G:
An overview on standard statistical methods for assessing
exposure-outcome link in survival analysis (Part II): The
kaplan-meier analysis and the cox regression method. Aging Clin Exp
Res. 24:203–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greaves MF and Brown G: Purification of
human T and B lymphocytes. J Immunol. 112:420–423. 1974.PubMed/NCBI
|
|
34
|
Zamarin D, Vigil A, Kelly K, Garcia-Sastre
A and Fong Y: Genetically engineered Newcastle disease virus for
malignant melanoma therapy. Gene Ther. 16:796–804. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wai-Hoe L, Wing-Seng L, Ismail Z and
Lay-Harn G: SDS-PAGE-Based quantitative assay for screening of
kidney stone disease. Biol Proced Online. 11:145–160. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang R, Wang R, Chen Q and Chang H:
Inhibition of autophagy using 3-methyladenine increases
cisplatin-induced apoptosis by increasing endoplasmic reticulum
stress in U251 human glioma cells. Mol Med Rep. 12:1727–1732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stoeck M, Marland-Noske C, Manasterski M,
Zawatzky R, Horn S, Möbus V, Schlag P and Schirrmacher V: In vitro
expansion and analysis of T lymphocyte microcultures obtained from
the vaccination sites of cancer patients undergoing active specific
immunization with autologous Newcastle-disease-virus-modified
tumour cells. Cancer Immunol Immunother. 37:240–244. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vigil A, Martinez O, Chua MA and
Garcia-Sastre A: Recombinant Newcastle disease virus as a vaccine
vector for cancer therapy. Mol Ther. 16:1883–1890. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schirrmacher V and Fournier P: Newcastle
disease virus: A promising vector for viral therapy, immune therapy
and gene therapy of cancer. Methods Mol Biol. 542:565–605. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Keshelava VV, Dobrovol'skaia Nlu, Chazova
NL, Bershchanskaia AM, Podol'skaia MV, Garmarnik TV and Mel'nikova
NV: Neoadjuvant therapy of breast cancer using Newcastle disease
virus. Vopr Onkol. 55:433–435. 2009.PubMed/NCBI
|
|
41
|
Schulze T, Kemmner W, Weitz J, Wernecke
KD, Schirrmacher V and Schlag PM: Efficiency of adjuvant active
specific immunization with Newcastle disease virus modified tumor
cells in colorectal cancer patients following resection of liver
metastases: Results of a prospective randomized trial. Cancer
Immunol Immunother. 58:61–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bai F, Niu Z, Tian H, Li S, Lv Z, Zhang T,
Ren G and Li D: Genetically engineered Newcastle disease virus
expressing interleukin 2 is a potential drug candidate for cancer
immunotherapy. Immunol Lett. 159:36–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chai Z, Zhang P, Fu F, Zhang X, Liu Y, Hu
L and Li X: Oncolytic therapy of a recombinant Newcastle disease
virus D90 strain for lung cancer. Virol J. 11:842014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Meng C, Zhou Z, Jiang K, Yu S, Jia L, Wu
Y, Liu Y, Meng S and Ding C: Newcastle disease virus triggers
autophagy in U251 glioma cells to enhance virus replication. Arch
Virol. 157:1011–1018. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mustafa Z, Shamsuddin HS, Ideris A,
Ibrahim R, Jaafar H, Ali AM and Abdullah JM: Viability reduction
and Rac1 gene downregulation of heterogeneous ex-vivo glioma acute
slice infected by the oncolytic Newcastle disease virus strain
V4UPM. Biomed Res Int. 2013:2485072013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bressy C, Hastie E and Grdzelishvili VZ:
Combining oncolytic virotherapy with p53 tumor suppressor gene
therapy. Mol Ther Oncolytics. 5:20–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hamzehloie T, Mojarrad M, Hasanzadeh
Nazarabadi M and Shekouhi S: The role of tumor protein 53 mutations
in common human cancers and targeting the murine double minute
2-p53 interaction for cancer therapy. Iran J Med Sci. 37:3–8.
2012.PubMed/NCBI
|
|
49
|
Fagin JA: Tumor suppressor genes in human
thyroid neoplasms: p53 mutations are associated undifferentiated
thyroid cancers. J Endocrinol Invest. 18:140–142. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Harris CC and Hollstein M: Clinical
implications of the p53 tumor-suppressor gene. N Engl J Med.
329:1318–1327. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Casson AG, Evans SC, Gillis A, Porter GA,
Veugelers P, Darnton SJ, Guernsey DL and Hainaut P: Clinical
implications of p53 tumor suppressor gene mutation and protein
expression in esophageal adenocarcinomas: Results of a ten-year
prospective study. J Thorac Cardiovasc Surg. 125:1121–1131. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kuczyk MA, Serth J, Hervatin C, Arndt H,
Derendorf L, Thon WF and Jonas U: Detection of P53
tumor-suppressor-gene protein in bladder tumors and prostate
cancer: Possible clinical implications. World J Urol. 12:345–351.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thomas AA, Ernstoff MS and Fadul CE:
Immunotherapy for the treatment of glioblastoma. Cancer J.
18:59–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Larsen CJ: Cellular immunotherapy and
glioblastoma: A hopeful treatment? Bull Cancer.
98:4572011.PubMed/NCBI
|
|
55
|
Varghese S, Rabkin SD, Nielsen GP,
MacGarvey U, Liu R and Martuza RL: Systemic therapy of spontaneous
prostate cancer in transgenic mice with oncolytic herpes simplex
viruses. Cancer Res. 67:9371–9379. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Husain SR, Behari N, Kreitman RJ, Pastan I
and Puri RK: Complete regression of established human glioblastoma
tumor xenograft by interleukin-4 toxin therapy. Cancer Res.
58:3649–3653. 1998.PubMed/NCBI
|
|
57
|
Debinski W, Gibo DM, Obiri NI, Kealiher A
and Puri RK: Novel anti-brain tumor cytotoxins specific for cancer
cells. Nat Biotechnol. 16:449–453. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bera TK, Viner J, Brinkmann E and Pastan
I: Pharmacokinetics and antitumor activity of a bivalent
disulfide-stabilized Fv immunotoxin with improved antigen binding
to erbB2. Cancer Res. 59:4018–4022. 1999.PubMed/NCBI
|
|
59
|
Ghetie MA, Richardson J, Tucker T, Jones
D, Uhr JW and Vitetta ES: Antitumor activity of Fab' and
IgG-anti-CD22 immunotoxins in disseminated human B lymphoma grown
in mice with severe combined immunodeficiency disease: Effect on
tumor cells in extranodal sites. Cancer Res. 51:5876–5880.
1991.PubMed/NCBI
|