Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed malignancy and the fourth most frequent cause of
cancer-associated mortality worldwide (1). It has recently been indicated that late
distant metastases are common in CRC, particularly liver and lung
metastases, which accounted for ~40% of all advanced patients
(2). Although notable advances have
been made in comprehensive therapy, the prognosis of metastatic CRC
remains unfavorable (3). As the
understanding of molecular mechanisms underlying tumorigenesis and
progression of CRC develops, targeted therapy has already become a
popular alternative to other, currently used treatments,
representing a significant landmark in devising individualized
treatment regimens.
It is known that epidermal growth factor receptor
(EGFR) is an important molecular target in metastatic CRC (mCRC)
(4). Furthermore, the success of
cetuximab or panitumumab, agents that target EGFR, created a new
milestone in precision medicine for mCRC (5). However, mutations of RAS genes
(including KRAS, NRAS and HRAS) or BRAF may
induce constitutive activation of downstream signaling pathways,
independent of EGFR inhibition, which is associated with
tumor proliferation and diffusion. Recent data (4) has demonstrated that KRAS exons
2, 3 and 4; NRAS exons 2 and 3; HRAS exon 2; and
BRAF exon 15 occurs in ~50% of CRC patients, and exhibits
facilitated neoplastic transformation in vitro of colorectal
cells as well as resistance to anti-EGFR therapy (6). Therefore, screening of gene mutation
profiling is important for appropriate therapeutic options and
regular surveillance. Notably, the predictive and prognostic
significance of RAS/BRAF mutations in CRC remains
controversial. A recent retrospective study (7) indicated that distant metastasis was
more likely to occur in patients with KRAS or BRAF
mutation. In addition, Morris et al (8), previously demonstrated a trend toward
lung metastasis and low survival for RAS/BRAF-mutant CRC.
Conversely, certain studies have not demonstrated that mutations in
RAS/BRAF were independent prognostic factors for CRC
(9,10). Therefore, the association of
RAS/BRAF status with late distant metastases and prognosis
of CRC requires further investigation.
The c-mesenchymal epithelial transition factor
(C-MET), a tyrosine kinase receptor for hepatocyte growth factor,
is associated with diverse biological functions ranging from
embryogenesis to wound healing (11). However, aberrant C-MET expression is
closely correlated with tumor progression and metastasis via
regulating cell proliferation, scattering and apoptosis (12). It is well known that C-MET
gene is upregulated in a variety of human malignancies, including
CRC (11). Recently, Lorenzon et
al (13), reported that in
KRAS wild-type patients with CRC, high C-MET expression
appeared as a negative predictor for disease-specific survival and
may interfere with anti-EGFR strategies, although the
patient cohort analyzed in the research was small.
Currently, use of a combination of biomarkers as a
better predictor of metastasis and prognosis in patients with CRC
has attracted more attention due to the potential of identifying
distinct tumor subtypes bearing different prognoses. However, the
clinicopathological relevance of RAS/BRAF mutations combined
with high C-MET expression in CRC is yet to be fully elucidated.
The majority of studies focused on western populations (8,11–13) and,
with few deriving data from Chinese patients (10). To improve the current knowledge, the
present study comprehensively characterized RAS/BRAF
mutations and C-MET overexpression in stage III CRC, alone and in
combination, to provide an insight into the association between
gene abnormalities and patient survival in Chinese populations.
Materials and methods
Patients and follow-up
The observational model was developed in 374 stage
III CRC samples (204 males and 170 females; age range, 23–92 years
old) and corresponding non-cancerous tissues from patients who had
undergone surgical resection at the department of gastrointestinal
surgery of Guangdong General Hospital (Guangzhou, China) between
January 2010 and October 2015. The inclusion criteria were as
follows: All patients had to have undergone complete lesion
removal, without having received any prior anticancer therapy.
Patients were also required to have normal renal and hepatic
function test results. Patients were excluded from the present
study if they exhibited inflammatory bowel disease. All patients
were classified into 4 groups: Group 1, RAS/BRAF-wild
without C-MET overexpression; group 2, RAS/BRAF-wild with
C-MET overexpression; group 3, RAS/BRAF-mutant without C-MET
overexpression; and group 4, RAS/BRAF-mutant with C-MET
overexpression. Genetic testing was performed as a part of
integrated care and information on clinicopathological data were
obtained from medical archives. Tumor grading was based on the
American Joint Committee on Cancer TNM classification and
pathological classification was in line with the World Health
Organization criteria (14,15). Overall survival (OS) or disease-free
survival (DFS) was calculated from the surgery of the primary CRC
until death/censoring or local recurrence/late distant
metastasis/censoring, respectively. Late distant metastasis was
defined as metastasis that occurred during follow-up. Of the 374
participants, 272 (72.7%) received 5-fluorouracil (5-FU)-based
postoperative adjuvant chemotherapy. An outpatient follow-up was
conducted every 3 months in accordance with Response Evaluation
Criteria in Solid Tumors 1.1 (16)
during the initial 2 years following clinical treatments and
subsequently every 6 months, until the end of a 3 year follow-up or
mortality. Written, informed consent was obtained from all
individual participants and the protocol was approved by the Ethics
Committee of Guangdong General Hospital.
Tissue sampling and mutation
assessment
Comprehensive genomic profiling was analyzed in 374
resected CRC tissue samples, which were fixed with 10% formalin
overnight at room temperature and embedded in paraffin wax. Tissues
were then sliced longitudinally to a thickness of 4 µm. Genomic DNA
was isolated from each FFPE specimen using a QIAamp DNA FFPE Tissue
Kit 56404 (Qiagen GmbH, Hilden, Germany) according to the
manufacturer's protocol. In addition, cancer cell-rich regions were
identified prior to sample DNA isolation via application of
hematoxylin and eosin (HE) staining to ascertain that all cases
exhibited enrichment of ≥70% malignant cells. HE staining was
performed according to manufacturers' instructions. Following
washing with xylene and dehydration with ethanol, the sections were
rehydrated in distilled water and then stained with the alum
haematoxylin (Shanghai XIBAO Biology Co., Ltd., Shanghai, China)
for 13 min at room temperature. After rinsing under running tap
water, slides were differentiated with 0.3% acid alcohol for 5 min
and washed in running tap water for 10 sec. Next, the tissue
sections were stained with eosin (Shanghai XIBAO Biology Co., Ltd.)
for 1 min at room temperature, dehydrated and mounted in crystal
mount. Staining was analyzed by two independent observers under an
optical microscope (magnification, ×400; CX31; Olympus Corporation,
Tokyo, Japan). Ultimately, extracted DNA concentration was
determined using an ND-1000 spectrophotometer (NanoDrop; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA).
Each tumor specimen was examined for KRAS
exon 2, 3 and 4; NRAS exon 2 and 3; HRAS exon 2; and
BRAF exon 15 (codon 600). AmpliSeq Designer v.1.2.6 software
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to
design primer pairs for PCR amplification of each gene region of
interest (17). DNA was amplified
using GoTaq Hot Start Polymerase (Promega Corporation, Madison, WI,
USA) and 0.2 µM each primer on the GeneAmp PCR System 9700 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Cycling conditions
were as previously described (18).
Amplicons were finally Sanger sequenced bidirectionally on an ABI
3730XL genetic analyzer (Invitrogen; Thermo Fisher Scientific,
Inc.). Primers and procedures were the same as previously reported
(19).
Immunohistochemical (IHC) analysis of
C-MET protein expression
Immunohistochemistry was performed as described
previously (11). Briefly, slides
were dewaxed, rehydrated and antigens were retrieved with EDTA (pH
8) by microwave heating at 95°C. Following the inhibition of
endogenous peroxidase activity and blocking non-specific antibody
binding, sections were incubated with lyophilized primary antibody
against C-MET (1:100; EP1454Y; BD Biosciences, Franklin Lakes, NJ)
overnight at 4°C. Following a 30-min incubation at room temperature
with secondary antibodies (cat. no. sc-3699; 1:200; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), immunoreaction was
visualized using the streptavidin-biotin peroxidase complex method.
Subsequently, slides were examined under an optical microscope
(magnification, ×400, CX31; Olympus Corporation). C-MET staining
was assessed according to Hercep Test guidelines (20) as follows: 0, no membrane staining or
membrane staining in <10% of tumor cells; 1+, faint membrane
staining; 2+, moderate and smooth membrane staining; 3+, strong and
granular membrane staining in ≥10% of tumor cells. C-MET
overexpression was defined as IHC 2+/3+. The results were judged by
two independent pathologists.
Statistical analysis
Data analysis was performed using SPSS version 19.0
(SPSS, Inc., Chicago, IL, USA). Pearson's Chi-square
(χ2) test was used to compare the correlation between
RAS/BRAF mutations and clinicopathological variables.
Kruskal-Wallis test or Mann Whitney U test were performed to
compare treatment response. Survival curves of OS and DFS were
plotted via Kaplan-Meier analysis with significance assessed using
log-rank test. Univariate and multivariate proportional Cox models
were performed to assess independent prognostic factors. Logistic
regression using a backward stepwise method and receiver operating
characteristic (ROC) analysis were performed to evaluate
synchronous liver metastasis of patients with CRC. P<0.05 was
considered to indicate a statistically significant difference.
Results
Frequencies of gene mutations and
C-MET status in stage III CRC patients
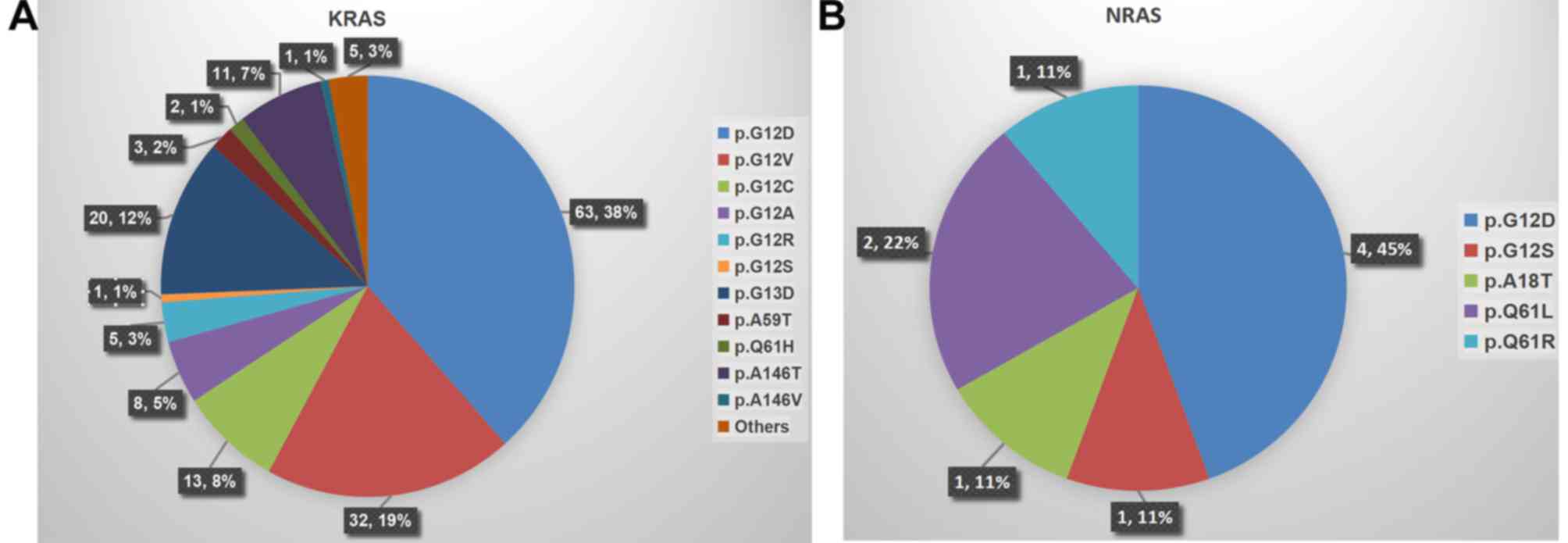
Mutations in KRAS, NRAS and HRAS were
observed in 43.9% (164/374), 2.4% (9/374) and 0.3% (1/374) of
patients, respectively. In addition, as another vital component of
the EGFR pathway, BRAF mutations were observed in
5.9% (22/374) cases. Mapping correlations between molecular
biomarkers demonstrated that 4 patients carried concurrent
KRAS and NRAS mutations (combinations were
p.G12D/p.G12D, p.G12D/p.A18T and p.A146T/p.Q61L), and in another 4
patients, KRAS and BRAF mutations (combinations were
all p.G12D/p.V600E) were concomitantly observed. However, no
co-mutations of NRAS with BRAF were observed in the
present study. Notably, the most prevalent mutation occurred in
exon 2 (codons 12 and 13) of KRAS (38.0%, 142/374). The
detailed distribution of KRAS and NRAS mutation
subtypes is presented in Fig. 1A and
B.
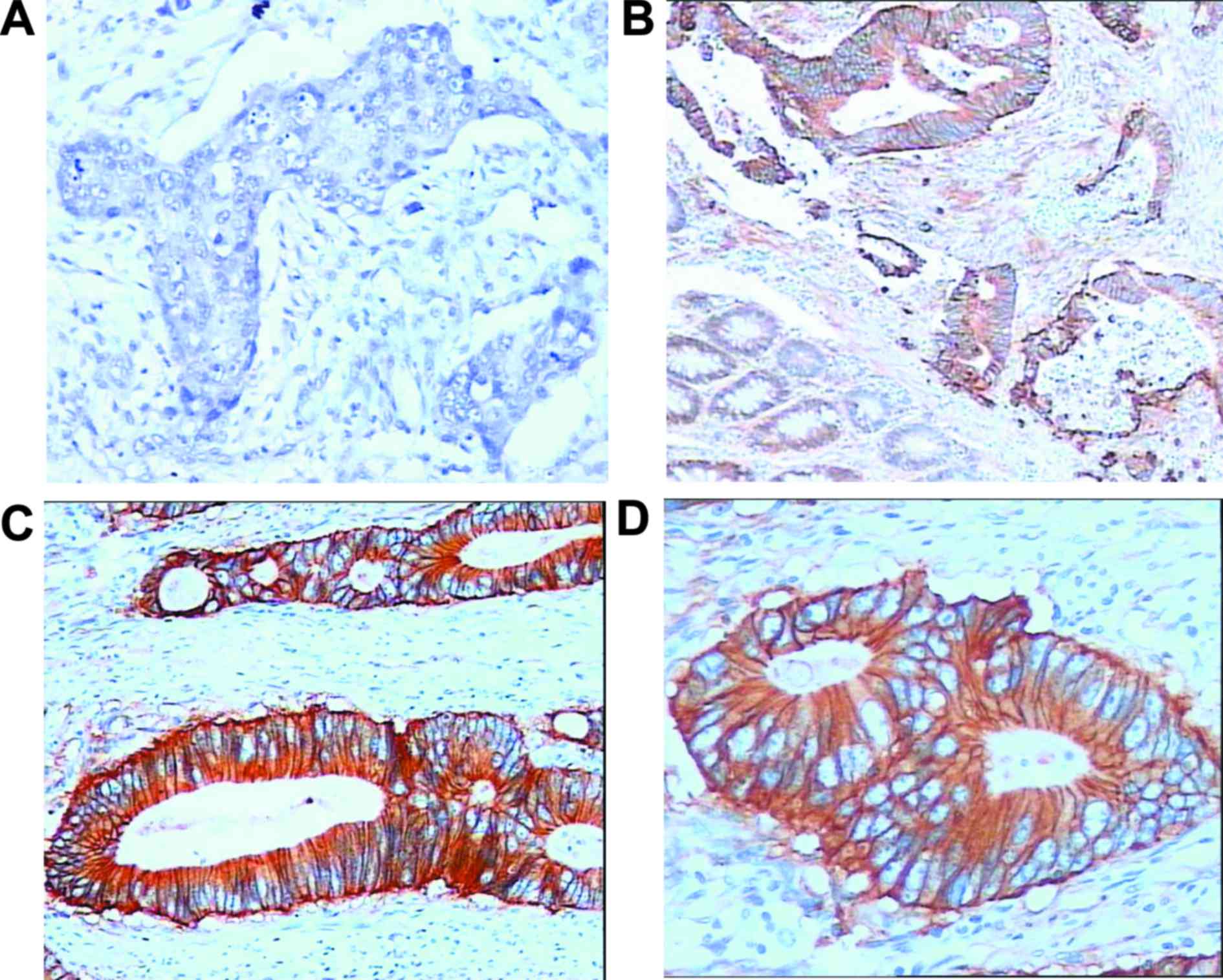
In addition, the status of C-MET protein in all
stage III CRC biopsies were investigated via IHC assay (Fig. 2). It was observed that 269 (71.9%)
cases exhibited C-MET overexpression (Fig. 2B-D). In paired non-tumorous
specimens, C-MET staining was either absent or present in the
membrane of only a few cells (Fig.
2A).
Associations between RAS/BRAF
mutations and C-MET overexpression with clinicopathological
features
The present study evaluated the correlations of
RAS/BRAF and C-MET status, alone or in combination, with the
clinicopathological characteristics in patients with stage III CRC.
Briefly, KRAS mutations were significantly correlated with
vascular invasion (P<0.001) and late distant metastasis,
particularly lung metastases (P=0.001). NRAS mutations were
more likely to exhibit low COX-2 expression (P=0.001). Furthermore,
BRAF exhibited a higher mutation rate in female patients
than males (P<0.001) and right colon than other tumor locations
(P=0.002; Table I). The present
study demonstrated that, compared with low C-MET expression, C-MET
overexpression was more likely to occur in cases with late nodal
stage (P=0.019), vascular invasion (P=0.023) and late distant
metastases, particularly lung and liver metastases (P<0.001;
Table II). Considering both
RAS/BRAF mutations and C-MET status, there were
significant differences in the clinicopathological features
distribution among different groups. For patients in group 4,
vascular invasion (P=0.001), high carcino-embryonic antigen level
(P=0.031) and late distant metastases (P<0.001) were observed at
significantly higher levels than in the other groups (Table III).
 | Table I.Correlation between mutation profile
and clinicopathological features in 374 patients with stage III
colorectal cancer. |
Table I.
Correlation between mutation profile
and clinicopathological features in 374 patients with stage III
colorectal cancer.
|
|
| KRAS status | BRAF status | NRAS status |
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Clinicopathological
features | Patients, n | Wild-type
(n=210) | Mutation
(n=164) | P-value | Wild-type
(n=352) | Mutation
(n=22) | P-value | Wild-type
(n=365) | Mutation (n=9) | P-value | All wild-type
(n=186) | Any mutation
(n=188) | P-value |
|---|
| Sex |
|
|
| 0.470 |
|
| <0.001 |
|
| 0.951 |
|
| 0.076 |
|
Male | 204 | 118 (57.8) | 86 (42.2) |
| 200 (98.0) | 4 (2.0) |
| 199 (97.5) | 5 (2.5) |
| 110 (53.9) | 94 (46.1) |
|
|
Female | 170 | 92 (54.1) | 78 (45.9) |
| 152 (89.4) | 18 (10.6) |
| 166 (97.6) | 4 (2.4) |
| 76 (44.7) | 94 (55.3) |
|
| Age, years |
|
|
| 0.355 |
|
| 0.074 |
|
| 0.089 |
|
| 0.079 |
|
<65 | 188 | 110 (58.5) | 78 (41.5) |
| 181 (96.3) | 7 (3.7) |
| 186 (98.9) | 2 (1.1) |
| 102 (54.3) | 86 (45.7) |
|
|
≥65 | 186 | 100 (53.8) | 86 (46.2) |
| 171 (91.9) | 15 (8.1) |
| 179 (96.2) | 7 (3.8) |
| 84 (45.2) | 102 (54.8) |
|
| Tumor location |
|
|
| 0.360 |
|
| 0.002 |
|
| 0.622 |
|
| 0.300 |
| Left
colon | 166 | 100 (60.2) | 66 (39.8) |
| 152 (91.6) | 14 (8.4) |
| 162 (97.6) | 4 (2.4) |
| 84 (50.6) | 82 (49.4) |
|
| Right
colon | 46 | 24 (52.2) | 22 (47.8) |
| 40 (87.0) | 6 (13.0) |
| 44 (95.7) | 2 (4.3) |
| 18 (39.1) | 28 (60.9) |
|
|
Rectum | 162 | 86 (53.1) | 76 (46.9) |
| 160 (98.8) | 2 (1.2) |
| 159 (98.1) | 3 (1.9) |
| 84 (51.9) | 78 (48.1) |
|
|
Differentiation |
|
|
| 0.609 |
|
| 0.068 |
|
| 0.372 |
|
| 0.611 |
|
Well/Moderate | 238 | 136 (57.1) | 102 (42.9) |
| 220 (92.4) | 18 (7.6) |
| 231 (97.1) | 7 (2.9) |
| 116 (48.7) | 122 (51.3) |
|
|
Poor | 136 | 74 (54.4) | 62 (45.6) |
| 132 (97.1) | 4 (2.9) |
| 134 (98.5) | 2 (1.5) |
| 70 (51.5) | 66 (48.5) |
|
| Depth of
invasion |
|
|
| 0.406 |
|
| 0.712 |
|
| 0.404 |
|
| 0.360 |
| T1 | 2 | 0 (0.0) | 2 (100.0) |
| 2 (100.0) | 0 (0.0) |
| 2 (100.0) | 0 (0.0) |
| 0 (0.0) | 2 (100.0) |
|
| T2 | 24 | 14 (58.3) | 10 (41.7) |
| 22 (91.7) | 2 (8.3) |
| 24 (100.0) | 0 (0.0) |
| 12 (50.0) | 12 (50.0) |
|
| T3 | 284 | 158 (55.6) | 126 (44.4) |
| 266 (93.7) | 18 (6.3) |
| 275 (96.8) | 9 (3.2) |
| 138 (48.6) | 146 (51.4) |
|
| T4 | 64 | 38 (59.4) | 26 (40.6) |
| 62 (96.9) | 2 (3.1) |
| 64 (100.0) | 0 (0.0) |
| 36 (56.3) | 28 (43.8) |
|
| Nodal stage |
|
|
| 0.299 |
|
| 0.941 |
|
| 0.489 |
|
| 0.126 |
| N1 | 260 | 148(56.9) | 112 (43.1) |
| 244(93.8) | 16 (6.2) |
| 255 (98.1) | 5 (1.9) |
| 132 (50.8) | 128 (49.2) |
|
|
N2a | 74 | 44 (59.5) | 30 (40.5) |
| 70 (94.6) | 4 (5.4) |
| 72 (97.3) | 2 (2.7) |
| 40 (54.1) | 34 (45.9) |
|
|
N2b | 40 | 18 (45.0) | 22 (55.0) |
| 38 (95.0) | 2 (5.0) |
| 38 (95.0) | 2 (5.0) |
| 14 (35.0) | 26 (65.0) |
|
| Vascular
invasion |
|
|
| <0.001 |
|
| 0.222 |
|
| 0.160 |
|
| 0.001 |
| No | 308 | 186 (60.4) | 122 (39.6) |
| 292 (94.8) | 16 (5.2) |
| 299 (97.1) | 9 (2.9) |
| 166 (53.9) | 142 (46.1) |
|
|
Yes | 66 | 24 (36.4) | 42 (63.6) |
| 60 (90.9) | 6 (9.1) |
| 66 (100.0) | 0 (0.0) |
| 20 (30.3) | 46 (69.7) |
|
| Initial CEA,
ng/ml |
|
|
| 0.613 |
|
| 0.350 |
|
| 0.757 |
|
| 0.950 |
|
<20 | 100 | 54 (54.0) | 46 (46.0) |
| 96 (96.0) | 4 (4.0) |
| 98 (98.0) | 2 (2.0) |
| 50 (50.0) | 50 (50.0) |
|
|
≥20 | 274 | 156 (56.9) | 118 (43.1) |
| 256 (93.4) | 18 (6.6) |
| 267 (97.4) | 7 (2.6) |
| 136 (49.6) | 138 (50.4) |
|
| Late distant
metastases |
|
|
| 0.001 |
|
| 0.628 |
|
| 0.574 |
|
| 0.001 |
| No | 46 | 36 (78.3) | 10 (21.7) |
| 44 (95.7) | 2 (4.3) |
| 46 (100.0) | 0 (0.0) |
| 34 (73.9) | 12 (26.1) |
|
|
Liver | 126 | 76 (60.3) | 50 (39.7) |
| 116 (92.1) | 10 (7.9) |
| 121 (96.0) | 5 (4.0) |
| 61 (48.4) | 65 (51.6) |
|
|
Lung | 68 | 29 (42.6) | 39 (57.4) |
| 63 (92.6) | 5 (7.4) |
| 67 (98.5) | 1 (1.5) |
| 24 (35.3) | 44 (64.7) |
|
|
Abdomen | 72 | 42 (58.3) | 30 (41.7) |
| 69 (95.8) | 3 (4.2) |
| 70 (97.2) | 2 (2.8) |
| 41 (56.9) | 31 (43.1) |
|
|
Others | 62 | 27 (43.5) | 35 (56.5) |
| 60 (96.8) | 2 (3.2) |
| 61 (98.4) | 1 (1.6) |
| 26 (41.9) | 36 (58.1) |
|
| COX-2
expression |
|
|
| 0.080 |
|
| 0.180 |
|
| <0.001 |
|
| 0.126 |
|
Negative/Weak | 32 | 24 (75.0) | 8 (25.0) |
| 31 (96.9) | 1 (3.1) |
| 28 (87.5) | 4 (12.5) |
| 21 (65.6) | 11 (34.4) |
|
|
Moderate | 66 | 36 (54.5) | 30 (45.5) |
| 59 (89.4) | 7 (10.6) |
| 66 (100.0) | 0 (0.0) |
| 29 (43.9) | 37 (56.1) |
|
|
Strong | 276 | 150 (54.3) | 126 (45.7) |
| 262 (94.9) | 14 (5.1) |
| 271 (98.2) | 5 (1.8) |
| 136 (49.3) | 140 (50.7) |
|
| MSI |
|
|
| 0.466 |
|
| 0.111 |
|
| 0.448 |
|
| 0.979 |
|
MSI-H | 22 | 14 (63.6) | 8 (36.4) |
| 19 (86.4) | 3 (13.6) |
| 22 (100.0) | 0 (0.0) |
| 11 (50.0) | 11 (50.0) |
|
|
MSI-L/MSS | 352 | 196 (55.7) | 156 (44.3) |
| 333 (94.6) | 19 (5.4) |
| 343 (97.4) | 9 (2.6) |
| 175 (49.7) | 177 (50.3) |
|
 | Table II.Correlation between C-MET
overexpression and clinicopathological features in 374 patients
with stage III colorectal cancer. |
Table II.
Correlation between C-MET
overexpression and clinicopathological features in 374 patients
with stage III colorectal cancer.
|
|
| C-MET
overexpression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Patients, n | No (n=105) | Yes (n=269) | P-value |
|---|
| Gender |
|
|
| 0.690 |
|
Male | 204 | 59 (28.9) | 145 (71.1) |
|
|
Female | 170 | 46 (27.1) | 124 (72.9) |
|
| Age, years |
|
|
| 0.610 |
|
<65 | 188 | 55 (29.3) | 133 (70.7) |
|
|
≥65 | 186 | 50 (26.9) | 136 (73.1) |
|
| Tumor location |
|
|
| 0.699 |
| Left
colon | 166 | 50 (30.1) | 116 (69.9) |
|
| Right
colon | 46 | 13 (28.3) | 33 (71.7) |
|
|
Rectum | 162 | 42 (25.9) | 120 (74.1) |
|
|
Differentiation |
|
|
| 0.103 |
|
Well/Moderate | 238 | 60 (25.2) | 178 (74.8) |
|
|
Poor | 136 | 45 (33.1) | 91 (66.9) |
|
| Depth of
invasion |
|
|
| 0.251 |
| T1 | 2 | 0 (0.0) | 2 (100.0) |
|
| T2 | 24 | 6 (25.0) | 18 (75.0) |
|
| T3 | 284 | 75 (26.4) | 209 (73.6) |
|
| T4 | 64 | 24 (37.5) | 40 (62.5) |
|
| Nodal stage |
|
|
| 0.019 |
| N1 | 260 | 84 (32.3) | 176 (67.7) |
|
|
N2a | 74 | 15 (20.3) | 59 (79.7) |
|
|
N2b | 40 | 6 (15.0) | 34 (85.0) |
|
| Vascular
invasion |
|
|
| 0.023 |
| No | 308 | 94 (30.5) | 214 (69.5) |
|
|
Yes | 66 | 11 (16.7) | 55 (83.3) |
|
| Initial CEA,
ng/ml |
|
|
| 0.072 |
|
<20 | 100 | 35 (35.0) | 65 (65.0) |
|
|
≥20 | 274 | 70 (25.5) | 204 (74.5) |
|
| Late distant
metastases |
|
|
| <0.001 |
| No | 46 | 23 (50.0) | 23 (50.0) |
|
|
Liver | 126 | 27 (21.4) | 99 (78.6) |
|
|
Lung | 68 | 7 (10.3) | 61 (89.7) |
|
|
Abdomen | 72 | 26 (36.1) | 46 (63.9) |
|
|
Others | 62 | 22 (35.5) | 40 (64.5) |
|
| COX-2
expression |
|
|
| 0.490 |
|
Negative/Weak | 32 | 10 (31.2) | 22 (68.8) |
|
|
Moderate | 66 | 22 (33.3) | 44 (66.7) |
|
|
Strong | 276 | 73 (26.4) | 203 (73.6) |
|
| MSI |
|
|
| 0.167 |
|
MSI-H | 22 | 9 (40.9) | 13 (59.1) |
|
|
MSI-L/MSS | 352 | 96 (27.3) | 256 (72.7) |
 | Table III.Association of combinational status
of RAS/BRAF genes and C-MET protein with clinicopathological
features. |
Table III.
Association of combinational status
of RAS/BRAF genes and C-MET protein with clinicopathological
features.
| Clinicopathological
features | Patients, n | Group 1 (n=62) | Group 2
(n=124) | Group 3 (n=43) | Group 4
(n=145) | P-value |
|---|
| Gender |
|
|
|
|
| 0.053 |
|
Male | 204 | 32 (51.6) | 77 (62.1) | 27 (62.8) | 68 (46.9) |
|
|
Female | 170 | 30 (48.4) | 47 (37.9) | 16 (37.2) | 77 (53.1) |
|
| Age, years |
|
|
|
|
| 0.068 |
|
<65 | 188 | 39 (62.9) | 63 (50.8) | 16 (32.6) | 70 (50.3) |
|
|
≥65 | 186 | 23 (37.1) | 61 (49.2) | 27 (67.4) | 75 (49.7) |
|
| Tumor location |
|
|
|
|
| 0.190 |
| Right
colon | 46 | 8 (12.9) | 10 (8.1) | 5 (11.6) | 23 (15.9) |
|
| Left
colon/Rectum | 328 | 54 (87.1) | 114 (91.9) | 38 (88.4) | 112 (84.1) |
|
|
Differentiation |
|
|
|
|
| 0.293 |
|
Well/Moderate | 238 | 33 (53.2) | 83 (66.9) | 27 (62.8) | 95 (65.5) |
|
|
Poor | 136 | 29 (46.8) | 41 (33.1) | 16 (37.2) | 50 (34.5) |
|
| Depth of
invasion |
|
|
|
|
| 0.310 |
|
T1+T2 | 26 | 2 (3.2) | 12 (9.7) | 4 (9.3) | 8 (5.5) |
|
|
T3+T4 | 348 | 60 (96.8) | 112 (90.3) | 39 (90.7) | 137 (94.5) |
|
| Nodal stage |
|
|
|
|
| 0.054 |
| N1 | 260 | 50 (80.6) | 82 (66.1) | 34 (79.1) | 94 (64.8) |
|
| N2 | 114 | 12 (19.4) | 42 (33.9) | 9 (20.9) | 51 (35.2) |
|
| Vascular
invasion |
|
|
|
|
| 0.001 |
| No | 308 | 57 (91.9) | 109 (87.9) | 37 (86.0) | 105 (72.4) |
|
|
Yes | 66 | 5 (8.1) | 15 (12.1) | 6 (14.0) | 40 (27.6) |
|
| Initial CEA
(ng/ml) |
|
|
|
|
| 0.031 |
|
<20 | 100 | 16 (25.8) | 34 (27.4) | 19 (44.2) | 31 (21.4) |
|
|
≥20 | 274 | 46 (74.2) | 90 (72.6) | 24 (55.8) | 114 (78.6) |
|
| Late distant
metastases |
|
|
|
|
| <0.001 |
| No | 46 | 18 (29.0) | 16 (12.9) | 5 (11.6) | 7 (4.8) |
|
|
Yes | 328 | 44 (71.0) | 108 (87.1) | 38 (88.4) | 138 (95.2) |
|
| COX-2
expression |
|
|
|
|
| 0.657 |
|
Negative/Weak | 32 | 4 (6.5) | 12 (9.7) | 2 (4.7) | 14 (9.7) |
|
|
Moderate/Strong | 342 | 58 (93.5) | 112 (90.3) | 41 (95.3) | 131 (90.3) |
|
| MSI |
|
|
|
|
| 0.523 |
|
MSI-H | 22 | 5 (8.1) | 5 (4.0) | 4 (9.3) | 8 (5.5) |
|
|
MSI-L/MSS | 352 | 57 (91.9) | 119 (96.0) | 39 (90.7) | 137 (94.5) |
|
Survival analysis
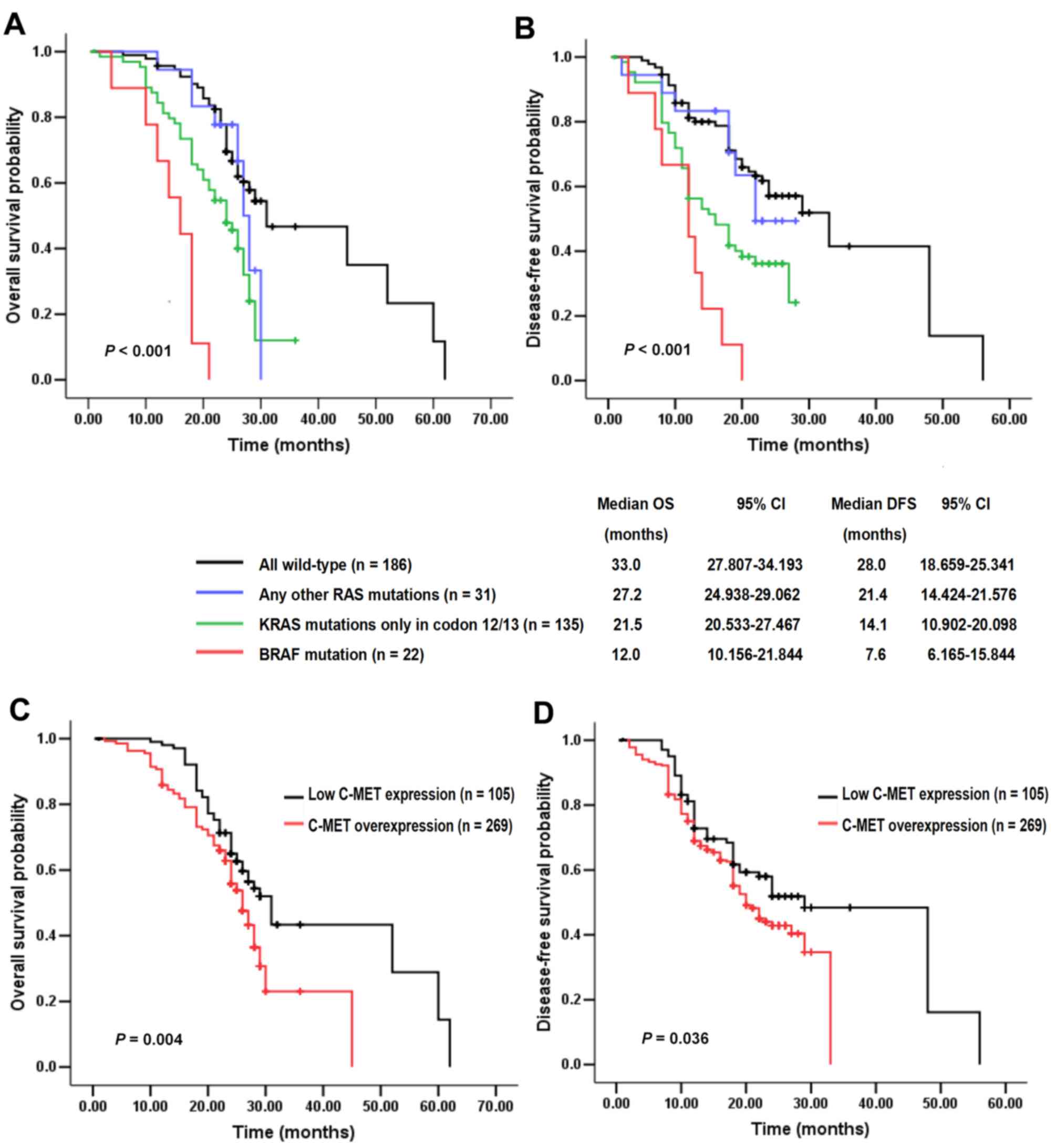
By May 1, 2017, the end of follow-up period, 68.4%
(256/374) of patients had succumbed. The median follow-up duration
was 32.0 months (range, 0.6–76.3 months) and 19 (5.1%) patients
were lost to follow-up. The potential influence of RAS/BRAF
mutations and C-MET status on survival was analyzed. In the entire
study cohort, it was concluded that OS and DFS for RAS/BRAF
mutant patients, particularly those exhibiting BRAF
mutation, were significantly reduced compared with those of cases
with all wild-type. The any-other-KRAS/NRAS-mutated group
exhibited longer median OS and DFS (27.2 and 21.4 months,
respectively) than the other two mutational groups (Fig. 3A and B). As compared with C-MET low
expression cancers (median OS and DFS, 38.7 and 32.3 months,
respectively), C-MET overexpression cases (median OS and DFS, 26.4
and 21.2 months, respectively) were correlated with worse OS
(P=0.004) and DFS (P=0.036; Fig. 3C and
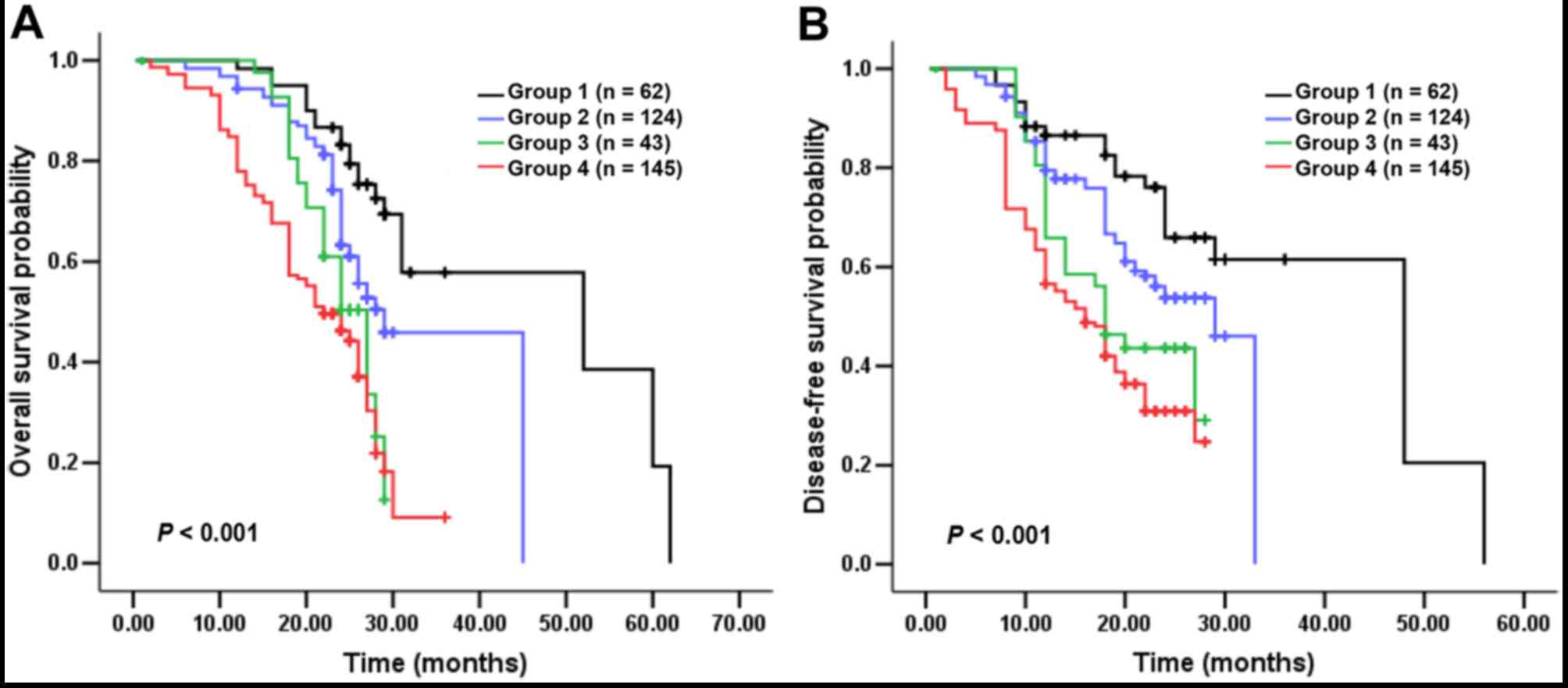
D). Notably, patients in Group 2 exhibited a more favorable
survival than those in Group 3, indicating that tumors which harbor
single RAS/BRAF mutations demonstrate higher malignant
potential in comparison with cases carrying a single C-MET
overexpression. Therefore RAS/BRAF mutations may have a more
powerful impact on OS and DFS than elevated C-MET (Fig. 4A and B).
Furthermore, the Cox proportional hazards model was
applied to estimate prognostic factors. As confirmed by
multivariate analyses, RAS/BRAF mutations emerged as
independent risk factors for OS [hazard ratio (HR), 2.045; 95%
confidence interval (CI), 1.276–3.279; P=0.003)] and DFS (HR,
1.976; 95% CI, 1.230–3.175; P=0.005), whereas C-MET overexpression
only exerted a significant prognostic effect on OS (HR, 2.837; 95%
CI, 1.103–6.053; P=0.031; Table
IV).
 | Table IV.Univariate and multivariate analyses
of OS and DFS for 374 patients. |
Table IV.
Univariate and multivariate analyses
of OS and DFS for 374 patients.
|
|
| OS univariate
analysis | OS multivariate
analysis | DFS univariate
analysis | DFS multivariate
analysis |
|---|
|
|
|
|
|
|
|
|---|
| Parameter | Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender | Male vs.
female | 1.041
(0.701–1.545) | 0.843 |
|
| 1.061
(0.714–1.576) | 0.771 |
|
|
| Age, years | <65 vs. ≥65 | 1.258
(0.845–1.874) | 0.258 |
|
| 1.048
(0.706–1.554) | 0.817 |
|
|
| Tumor location | Left/right colon
vs. rectum | 0.911
(0.623–1.377) | 0.658 |
|
| 1.076
(0.871–1.330) | 0.496 |
|
|
|
Differentiation | Well/moderate vs.
poor | 1.062
(0.702–1.605) | 0.776 |
|
| 1.061
(0.085–1.000) | 0.771 |
|
|
| Depth of
invasion | T1+T2 vs.
T3+T4 | 1.011
(0.818–1.250) | 0.916 |
|
| 1.140
(0.765–1.700) | 0.520 |
|
|
| Nodal stage | N0+N1 vs.
N2a+N2b | 1.042
(0.806–1.347) | 0.752 |
|
| 1.123
(0.868–1.453) | 0.377 |
|
|
| Vascular
invasion | No vs. yes | 0.982
(0.782–1.234) | 0.879 |
|
| 0.968
(0.772–1.214) | 0.779 |
|
|
| Initial CEA,
ng/ml | <20 vs. ≥20 | 1.154
(0.890–1.497) | 0.281 |
|
| 1.186
(0.916–1.536) | 0.195 |
|
|
| Late distant
metastases | No vs. yes | 3.334
(2.139–5.197) | <0.001 | 2.678
(1.655–4.334) | <0.001 | 3.291
(2.092–5.178) | <0.001 | 2.782
(1.678–4.435) | <0.001 |
| COX-2
expression | Negative/weak vs.
moderate/strong | 0.991
(0.758–1.294) | 0.946 |
|
| 0.991
(0.759–1.293) | 0.946 |
|
|
| MSI | MSI-H vs.
MSI-L/MSS | 0.713
(0.345–1.471) | 0.360 |
|
| 0.619
(0.300–1.277) | 0.194 |
|
|
| C-MET
overexpression | No vs. yes | 3.032
(1.323–6.948) | 0.009 | 2.837
(1.103–6.053) | 0.031 | 2.642
(1.154–6.045) | 0.021 | 2.382
(0.892–4.753) | 0.083 |
| RAS/BRAF
mutations | No vs. yes | 2.459
(1.617–3.739) | <0.001 | 2.045
(1.276–3.279) | 0.003 | 2.222
(1.460–3.382) | <0.001 | 1.976
(1.230–3.175) | 0.005 |
| Anti-EGFR
therapy | No vs. yes | 0.497
(0.229–1.080) | 0.077 |
|
| 0.396
(0.182–0.864) | 0.020 | 1.055
(0.411–2.710) | 0.911 |
Predictive value of RAS/BRAF mutations
and C-MET overexpression to late metastasis in patients with
CRC
As distant metastasis was significantly associated
with malignant progression and poor survival in patients with CRC,
the potential predictors for late metastasis were investigated
using unconditional logistic regression and ROC curves. Items that
were verified to be statistically significant were regarded as
independent variables. It was observed that RAS/BRAF
mutations [yes=1, no=0; odds ratio (OR), 2.544; P=0.002], C-MET
overexpression (yes=1, no=0; OR, 3.408; P=0.003) and depth of
invasion (T3+T4=1, T1+T2=0; OR, 3.363; P<0.001) were all
significantly correlated with the occurrence of late distant
metastases (Table V).
 | Table V.Logistic regression analysis of
factors associated with late distant metastases in patients with
colorectal cancer. |
Table V.
Logistic regression analysis of
factors associated with late distant metastases in patients with
colorectal cancer.
|
Characteristics | OR | 95% CI | P-value |
|---|
| Depth of invasion:
T3+T4 vs. T1+T2 | 3.363 | 1.911–5.916 | <0.001 |
| RAS/BRAF mutations:
Yes vs. no | 2.544 | 1.402–4.613 | 0.002 |
| C-MET
overexpression: Yes vs. no | 3.408 | 1.527–7.604 | 0.003 |
| Constant | 0.001 |
|
|
The number of cases included the whole study
population. With ROC curve analysis, the sensitivity and
specificity of RAS/BRAF mutations alone, C-MET
overexpression alone, depth of invasion alone, or their combination
for predicting late distant metastasis among patients with CRC were
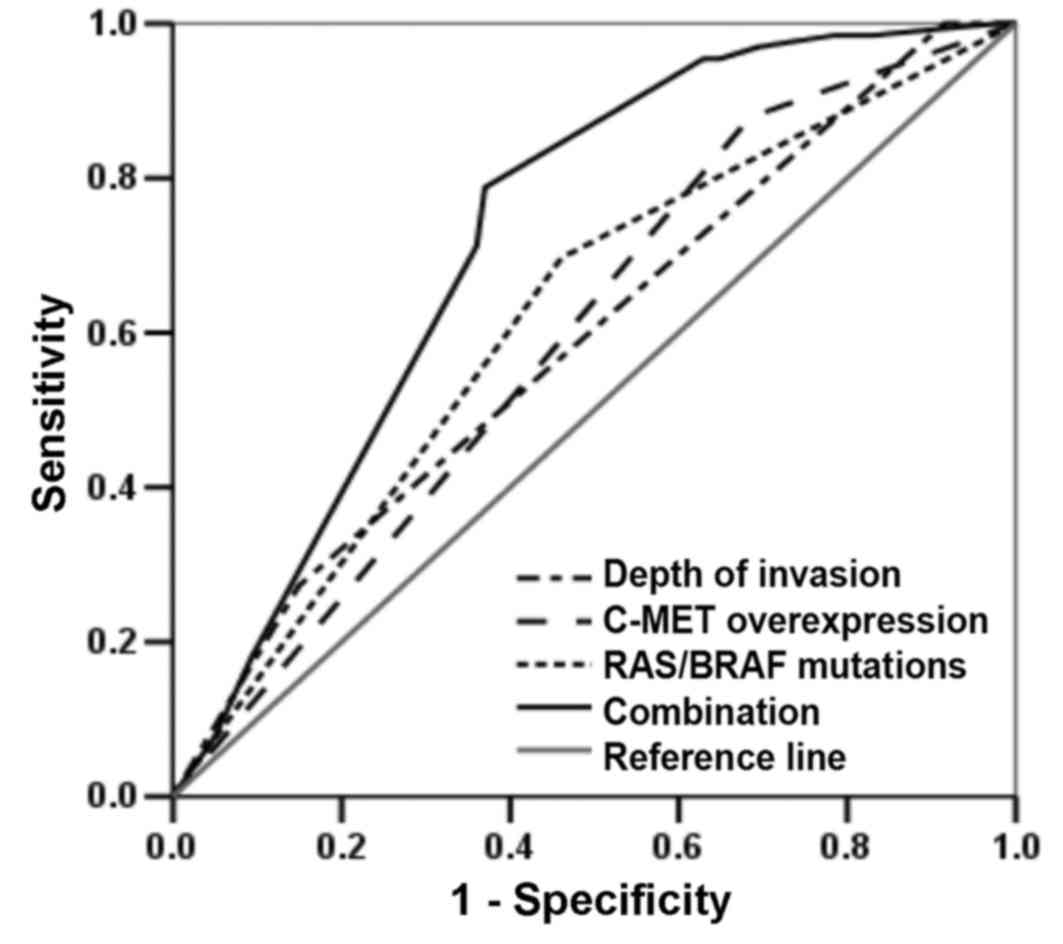
evaluated. The predictive findings presented in Fig. 5, demonstrated that the combination of
RAS/BRAF mutations, C-MET overexpression and depth of
invasion [area under curve (AUC), 0.734; 95% CI, 0.672–0.797;
P<0.001] exhibited a better predictive value compared with
single RAS/BRAF mutations (AUC, 0.618; 95% CI, 0.545–0.691;
P=0.003), C-MET overexpression (AUC, 0.600; 95% CI, 0.531–0.670;
P=0.011) or depth of invasion (AUC, 0.628; 95% CI, 0.553–0.702;
P=0.001).
Efficacy of anti-EGFR therapies
In the present study, 342 patients suffered from
late distant metastasis and/or recurrence during the follow-up
period, 46 of whom received cetuximab combined with first-line
FOLFIRI (irinotecan/5-Fu/leucovorin) or FOLFOX6
(oxaliplatin/5-Fu/leucovorin) chemotherapy, including 1 patient in
group 1, 41 in group 2 and 4 in group 4. No instances of patient
complete response (CR) were observed; 1 case in group 1 and 7 cases
in group 2 exhibited partial response (PR); 24 cases in group 2
exhibited stable disease (SD), whereas 4 cases in group 4 exhibited
all progressive disease (PD) for the first response evaluation at 3
months. The disease control rate (including CR, PR and SD) was
69.6% (32/46). Therefore, the efficacy of anti-EGFR therapy
in RAS/BRAF wild-type patients were better than that in
mutant counterparts, although no statistical significance was
observed. However, the influence of C-MET status on
anti-EGFR therapies were not assessed due to the low number
of suitable cases.
Discussion
CRC is a clinically and pathologically heterogeneous
malignancy, presenting high incidence of metastasis and a
consequent poor clinical outcome on account of its invasive nature
(1). Despite the complexity of
carcinogenesis, a number of molecular studies have been performed
in search of more specific and feasible markers with predictive and
prognostic significance. As a result, multiple genes, such as
vascular endothelial growth factor, cyclooxygenase-2,
PIK3CA, protein kinase B and ERBB2 (7,21), have
been considered as biomarkers of the aggressiveness of CRC. In
recent years, increasing attention has been given to extended
RAS and C-MET status, whose abnormalities have been
demonstrated to contribute to uncontrolled cell growth and
malignant transformation in CRC (18,22). To
the best of our knowledge, this is the first study where a combined
analysis of RAS/BRAF mutations plus C-MET overexpression was
performed, which clarified their clinical value in a large cohort
of Chinese patients with stage III CRC.
According to the present data, mutations in KRAS,
NRAS, HRAS, BRAF and C-MET overexpression were observed in
43.9% (164/374), 2.4% (9/374), 0.3% (1/374), 5.9% (22/374) and
71.9% (269/374) of cases, respectively. The prevalence of genetic
abnormalities was in accordance with previous publications
(7,23–26).
Different from intra-tumoral heterogeneity of KRAS mutations
and rare NRAS or HRAS mutations, BRAF
aberrance exhibited relative intra-tumoral homogeneity. In
addition, the present study also demonstrated that mutations in
RAS/BRAF oncogenes were not mutually exclusive, although the
findings conflicted with several reports from other populations
(27–29). One likely explanation for this may be
the disparity of sample sources (Chinese vs. European population).
Notably, emerging studies (30,31) have
observed a high concordance of RAS/BRAF mutations between
primary CRCs and corresponding metastases, indicating that these
genetic changes existed early in tumorigenesis, and maintained
their status during development (21). However, the level of concordance for
C-MET expression was controversial (22,32).
Shoji et al (31), previously
indicated that c-MET protein was more highly expressed in liver
metastases than in paired primary tumors. In contrast, another
study (33) revealed that C-MET
expression in late metastases tended to be decreased, which
supported the outcome of the present study. Therefore, more studies
in ethnically-diverse populations are required.
In the present study, the association between
combinational status of RAS/BRAF plus C-MET and
clinicopathological features were investigated. Briefly, it was
indicated that KRAS mutations and C-MET overexpression, or
their combination, may be important indicators to identify subsets
of CRC with vascular invasion and late distant metastases.
Particularly, 35% of patients in the present study developed liver
metastases during their disease course and >50% of cases
exhibited metastases in other sites, including lung metastases. Of
the cases with liver metastases, 39.7% had KRAS mutations
and 78.6% exhibited high C-MET expression. By contrast, genetic
abnormalities were more closely associated with lung metastases. In
addition, NRAS mutations were correlated with low
COX-2 expression, suggesting the reduced aggression of
tumors carrying NRAS mutations compared with those with
other RAS/BRAF mutations. This is in accordance with
previous studies (10,23). Recently, a retrospective study
(34) reported that BRAF
mutations were observed more frequently in right colon and female
patients, which supported the conclusions of the present study.
Numerous experimental model systems have confirmed RAS/BRAF
mutations and upregulated C-MET collaboration, or their
interactions, contributed to cell proliferation and the
invasion-metastasis cascade, which may yield tumor aggressiveness
and distant organ involvement (6,35).
Furthermore, Bradley et al (22), recently illustrated that small
interfering RNA-mediated knockdown of c-MET inhibited the migration
and invasion potential of CRC cells, thereby suppressing tumor
progression and metastasis in vivo. These outcomes indicated
that genetic abnormalities are important in promoting CRC
malignancy.
The initiation and development of CRC is a complex,
multi-step process that is accompanied by the accumulation of
diverse gene alterations (3,6). RAS/BRAF mutations are typically
the most frequent driver mutations in CRC (36), C-MET overexpression is regarded as
adjuvant pro-metastatic marker, both of which represent the
principle aspect of somatic genetic changes (37,38).
Another focus of the present study was further exploring the
predictive value of RAS/BRAF mutations and C-MET status. In
one prior study (39), KRAS
exon 2-mutated CRC patients exhibited a marked propensity for lung
metastases. Similar results have also been described by Morris
et al (8), in which all
RAS/BRAF mutant cases harbored the trend towards distant
metastases. The present data highlighted that RAS/BRAF
mutations combined with C-MET overexpression were significant
predictors for higher risk of late distant metastasis, suggesting
their importance in distinguishing CRCs with highly aggressive
behavior from low metastatic lesions. The results also demonstrated
that these mutations provide powerful insights into the complexity
of tumor foci genotype and provide a rationale for the combination
therapeutic strategies. Previous studies have proposed that the
block of C-MET, the HDAC inhibitor and CDK1 inhibition may markedly
attenuate CRC development (40–42).
Previously, KRAS mutation was regarded as an
adverse prognostic indicator in 1990 (43). Only in the last several years has the
prognostic value of extended RAS mutations in CRC received
more attention. Conversely, high C-MET expression has been
documented to be associated with lower survival in diverse human
tumors (12,32). A previous study (31) has demonstrated that C-MET
overexpression indicated a poor outcome in terms of the risk of
recurrence and mortality in patients with mCRC following
metastasectomy. Similarly, the present data also revealed that
C-MET overexpression and RAS/BRAF mutations, particularly
BRAF mutation, were significantly associated with shorter OS
and DFS in the entire study population. Notably, compared with
C-MET overexpression, RAS/BRAF mutations appeared to be more
powerful prognostic markers of a short interval to low survival and
late metastasis following surgery. Furthermore, as the National
Comprehensive Cancer Network recommends patients with mCRC and
RAS/BRAF wild-type for anti-EGFR treatment (44), the present results also illustrated
wild-type cases may gain survival benefits from cetuximab.
Regarding C-MET status, Inno et al (32) previously proposed that C-MET
overexpression was significantly associated with a worse outcome
and anti-EGFR resistance; whereas in the present study, too small
sample size in low C-MET expression patients treated with cetuximab
prevented the elucidation of potential therapeutic importance of
C-MET. A focus on this issue is required in future studies.
In view of the retrospective nature of the current
methodology, there has been an inevitable selection bias in the
present outcomes. Firstly, certain participants and their medical
record documentation may have been lost to follow-up, particularly
for those who were not hospitalized following first-line
chemotherapy. Secondly, the patients were heterogeneous and
selected according to the availability of genetic detection, which
limited data analyses. Therefore, further prospective studies are
required to confirm the present conclusions.
In conclusion, the status of RAS/BRAF and
C-MET may serve as significant predictors for metastatic behavior
and refining prognosis in CRC. Accordingly, radiological diagnosis
in combination with RAS/BRAF and C-MET detection may help in
the prognostic evaluation for postoperative stage III CRC cases, as
well as devised appropriate individualized medicine in the
future.
Acknowledgements
Not applicable.
Funding
The present work was supported by a grant from the
Program of Health and Family Planning Commission Foundation of
Guangzhou City, Guangdong Province, China (grant no. A2017418); and
a grant from the Program of Science and Technology Commission
Foundation of Guangzhou City, Guangdong Province, China (grant no.
20140705).
Availability of data and materials
The data in the present study are available from
Guangdong General Hospital (Guangzhou, China).
Authors' contributions
JL designed the study, analyzed the data, wrote the
present manuscript and gave final approval of the manuscript to be
published. CH analyzed data, WZ conducted the follow-up, JW
performed IHC and Sanger sequencing, LX performed survival analysis
and DM designed the experiments.
Ethics approval and consent to
participate
Written, informed consent was obtained from all
individual participants and the protocol was approved by the Ethics
Committee of Guangdong General Hospital.
Consent for publication
Written informed consent was obtained from each
participant.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kawai M, Komiyama H, Hosoya M, Okubo H,
Fujii T, Yokoyama N, Sato C, Ueyama T, Okuzawa A, Goto M, et al:
Impact of chromosome 17q deletion in the primary lesion of
colorectal cancer on liver metastasis. Oncol Lett. 12:4773–4778.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zong Z, Zhou T, Rao L, Jiang Z, Li Y, Hou
Z, Yang B, Han F and Chen S: Musashi2 as a novel predictive
biomarker for liver metastasis and poor prognosis in colorectal
cancer. Cancer Med. 5:623–630. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scaltriti M and Baselga J: The epidermal
growth factor receptor pathway: A model for targeted therapy. Clin
Cancer Res. 12:5268–5272. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maughan TS, Adams RA, Smith CG, Meade AM,
Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL,
et al: Addition of cetuximab to oxaliplatin-based first-line
combination chemotherapy for treatment of advanced colorectal
cancer: Results of the randomised phase 3 MRC COIN trial. Lancet.
377:2103–2114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McCubrey JA, Steelman LS, Abrams SL, Lee
JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA,
D'Assoro AB, et al: Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT
pathways in malignant transformation and drug resistance. Adv
Enzyme Regul. 46:249–279. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li ZZ, Wang F, Zhang ZC, Wang F, Zhao Q,
Zhang DS, Wang FH, Wang ZQ, Luo HY, He MM, et al: Mutation
profiling in chinese patients with metastatic colorectal cancer and
its correlation with clinicopathological features and anti-EGFR
treatment response. Oncotarget. 7:28356–28368. 2016.PubMed/NCBI
|
|
8
|
Morris VK, Lucas FA, Overman MJ, Eng C,
Morelli MP, Jiang ZQ, Luthra R, Meric-Bernstam F, Maru D, Scheet P,
et al: Clinicopathologic characteristics and gene expression
analyses of non-KRAS 12/13, RAS-mutated metastatic colorectal
cancer. Ann Oncol. 25:2008–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang CW, Tsai HL, Chen YT, Huang CM, Ma
CJ, Lu CY, Kuo CH, Wu DC, Chai CY and Wang JY: The prognostic
values of EGFR expression and KRAS mutation in patients with
synchronous or metachronous metastatic colorectal cancer. BMC
Cancer. 13:5992013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen Y, Han X, Wang J, Wang S, Yang H, Lu
SH and Shi Y: Prognostic impact of mutation profiling in patients
with stage II and III colon cancer. Sci Rep. 6:243102016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bottaro DP, Rubin JS, Faletto DL, Chan AM,
Kmiecik TE, Vande Woude GF and Aaronson SA: Identification of the
hepatocyte growth factor receptor as the c-met proto-oncogene
product. Science. 251:802–804. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Elliott VA, Rychahou P, Zaytseva YY and
Evers BM: Activation of c-Met and upregulation of CD44 expression
are associated with the metastatic phenotype in the colorectal
cancer liver metastasis model. Plos One. 9:e974322014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lorenzon L, Ricca L, Pilozzi E, Lemoine A,
Riggio V, Giudice MT, Mallel G, Fochetti F and Balducci G: Tumor
regression grades, K-RAS mutational profile and c-MET in colorectal
liver metastases. Pathol Res Pract. 213:1002–1009. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao P, Song YX, Wang ZN, Xu YY, Tong LL,
Sun JX, Yu M and Xu HM: Is the prediction of prognosis not improved
by the seventh edition of the TNM classification for colorectal
cancer? Analysis of the surveillance, epidemiology, and end results
(SEER) database. BMC Cancer. 13:1232013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yin H, Xu L and Yao HW: New opinions of
colorectal cancer in 2010. Chin J Practical Surg. 30:764–768.
2010.
|
|
16
|
Aras M, Erdil TY, Dane F, Gungor S, Ones
T, Dede F, Inanir S and Turoglu HT: Comparison of WHO, RECIST 1.1,
EORTC, and PERCIST criteria in the evaluation of treatment response
in malignant solid tumors. Nucl Med Commun. 37:9–15.
2016.PubMed/NCBI
|
|
17
|
Lupini L, Bassi C, Mlcochova J, Musa G,
Russo M, Vychytilova-Faltejskova P, Svoboda M, Sabbioni S, Nemecek
R, Slaby O and Negrini M: Prediction of response to anti-EGFR
antibody-based therapies by multigene sequencing in colorectal
cancer patients. BMC Cancer. 15:8082015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie G, Xie F, Wu P, Yuan X, Ma Y, Xu Y, Li
L, Xu L, Yang M and Shen L: The mutation rates of EGFR in non-small
cell lung cancer and KRAS in colorectal cancer of Chinese patients
as detected by pyrosequencing using a novel dispensation order. J
Exp Clin Cancer Res. 34:632015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao J, Wu H, Wang L, Zhang H, Duan H, Lu J
and Liang Z: Validation of targeted next-generation sequencing for
RAS mutation detection in FFPE colorectal cancer tissues:
Comparison with Sanger sequencing and ARMS-Scorpion real-time PCR.
BMJ Open. 6:article e009532. 2016. View Article : Google Scholar
|
|
20
|
Ma PC, Tretiakova MS, MacKinnon AC,
Ramnath N, Johnson C, Dietrich S, Seiwert T, Christensen JG,
Jagadeeswaran R, Krausz T, et al: Expression and mutational
analysis of MET in human solid cancers. Genes Chromosomes Cancer.
47:1025–1037. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fearon ER and Vogelstein B: A genetic
model for colorectal. tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bradley CA, Dunne PD, Bingham V, McQuaid
S, Khawaja H, Craig S, James J, Moore WL, McArt DG, Lawler M, et
al: Transcriptional upregulation of c-MET is associated with
invasion and tumor budding in colorectal cancer. Oncotarget.
7:78932–78945. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ogura T, Kakuta M, Yatsuoka T, Nishimura
Y, Sakamoto H, Yamaguchi K, Tanabe M, Tanaka Y and Akagi K:
Clinicopathological characteristics and prognostic impact of
colorectal cancers with NRAS mutations. Oncol Rep. 32:50–56. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yokota T, Ura T, Shibata N, Takahari D,
Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, et
al: BRAF mutation is a powerful prognostic factor in advanced and
recurrent colorectal cancer. Br J Cancer. 104:856–862. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Russo AL, Borger DR, Szymonifka J, Ryan
DP, Wo JY, Blaszkowsky LS, Kwak EL, Allen JN, Wadlow RC, Zhu AX, et
al: Mutational analysis and clinical correlation of metastatic
colorectal cancer. Cancer. 120:1482–1490. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen Y, Wang J, Han X, Yang H, Wang S, Lin
D and Shi Y: Effectors of epidermal growth factor receptor pathway:
The genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in
colorectal cancer characteristics and personalized medicine. PLoS
One. 8:e816282013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hawkes E and Cunningham D: Relationship
between colorectal cancer biomarkers and response to epidermal
growth factor receptor monoclonal antibodies. J Clin Oncol.
28:e529–e531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kawazoe A, Shitara K, Fukuoka S, Kuboki Y,
Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, et al: A
retrospective observational study of clinicopathological features
of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with
metastatic colorectal cancer. BMC Cancer. 15:2582015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kafatos G, Niepel D, Lowe K,
Jenkins-Anderson S, Westhead H, Garawin T, Traugottová Z, Bilalis
A, Molnar E, Timar J, et al: RAS mutation prevalence among patients
with metastatic colorectal cancer: A meta-analysis of real-world
data. Biomark Med. 10.2217/bmm-2016-0358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fujiyoshi K, Yamamoto G, Takahashi A, Arai
Y, Yamada M, Kakuta M, Yamaguchi K, Akagi Y, Nishimura Y, Sakamoto
H, et al: High concordance rate of KRAS/BRAF mutations and MSI-H
between primary colorectal cancer and corresponding metastases.
Oncol Rep. 37:785–792. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shoji H, Yamada Y, Taniguchi H, Nagashima
K, Okita N, Takashima A, Honma Y, Iwasa S, Kato K, Hamaguchi T, et
al: Clinical impact of c-MET expression and genetic mutational
status in colorectal cancer patients after liver resection. Cancer
Sci. 105:1002–1007. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Inno A, Di Salvatore M, Cenci T, Martini
M, Orlandi A, Strippoli A, Ferrara AM, Bagalà C, Cassano A, Larocca
LM, et al: Is there a role for IGF1R and c-MET pathways in
resistance to cetuximab in metastatic colorectal cancer? Clin
Colorectal Cancer. 10:325–332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsui S, Osada S, Tomita H, Komori S,
Mori R, Sanada Y, Takahashi T, Yamaguchi K and Yoshida K: Clinical
significance of aggressive hepatectomy for colorectal liver
metastasis, evaluated from the HGF/c-Met pathway. Int J Oncol.
37:289–297. 2010.PubMed/NCBI
|
|
34
|
Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu
T, Sun J, Jiang H, Zhu Y, Zheng Y, et al: Molecular spectrum of
KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer
patients: analysis of 1,110 cases. Sci Rep. 5:186782015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mendelsohn J and Baselga J: Epidermal
growth factor receptor targeting in cancer. Semin Oncol.
33:369–385. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Organ SL and Tsao MS: An overview of the
c-MET signaling pathway. Ther Adv Med Oncol. 3 1 Suppl:S7–S19.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim MJ, Lee HS, Kim JH, Kim YJ, Kwon JH,
Lee JO, Bang SM, Park KU, Kim DW, Kang SB, et al: Different
metastatic pattern according to the KRAS mutational status and
site-specific discordance of KRAS status in patients with
colorectal cancer. BMC Cancer. 12:3472012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun Y, Sun L, An Y and Shen X:
Cabozantinib, a Novel c-Met Inhibitor, Inhibits Colorectal Cancer
Development in a Xenograft Model. Med Sci Monit. 21:2316–2321.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Carson R, Celtikci B, Fenning C, Javadi A,
Crawford N, Carbonell LP, Lawler M, Longley DB, Johnston PG and Van
Schaeybroeck S: HDAC inhibition overcomes acute resistance to MEK
inhibition in BRAF-mutant colorectal cancer by downregulation of
c-FLIPL. Clin Cancer Res. 21:3230–3240. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Costa-Cabral S, Brough R, Konde A, Aarts
M, Campbell J, Marinari E, Riffell J, Bardelli A, Torrance C, Lord
CJ, et al: CDK1 Is a synthetic lethal target for KRAS mutant
tumours. PloS One. 11:e01490992016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Slebos RJ, Kibbelaar RE, Dalesio O,
Kooistra A, Stam J, Meijer CJ, Wagenaar SS, Vanderschueren RG, van
Zandwijk N, Mooi WJ, et al: K-ras oncogene activation as a
prognostic marker in adenocarcinoma of the lung. N Engl J Med.
323:561–565. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
NCCN Clinical Practice Guidelines in
Oncology_Colon Cancer, . Rectal Cancer Version 1. 2015.http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf29–08.
2014
|