Introduction
Acute lymphoblastic leukemia (ALL) is a
heterogeneous disease, which is characterized by massive
proliferation, extensive infiltration and inhibition of normal
hematopoiesis (1). ALL is the most
common cancer in children, accounting for 25% of cancer diagnosed
among children <15 years of age (2). Lymphocyte phenotyping reveals that ALL
has two subtypes: B cell (B-) and T cell (T-) ALL, with 85% of
cases being B-ALL and 15% of T-ALL (3). Various therapeutic protocols have been
applied in treatment and management of ALL, including chemotherapy,
targeted therapy and bone marrow transplantation (4). Among children with ALL, ~95% of
patients achieved complete remission following targeted therapy and
15-20% achieved an initial remission followed by a relapse
(5). The etiology and pathogenesis
of ALL is yet to be fully elucidated (1,3).
Previous studies have demonstrated that the malignant proliferation
of B-ALL cells was closely associated with a low level of
anti-tumor immunity (1–3). However, the molecular mechanism of
antitumor immune dysfunction remains unclear (1). It may be associated with the emergence
and accumulation of immune regulatory cells, including regulatory T
regulatory cells (Tregs) suppressing anti-cancer immunity (6–8).
CD4+CD25+ Tregs have been
discovered recently as a subpopulation of T cells, characterized by
low reactive, immune suppression and expression of forkhead box P3
(FoxP3) (9,10). Tregs are produced in the thymus
during T-cell maturation and are generated in the peripheral blood
from naive CD4+ T cells (11). Previously studies revealed that
numerous cancers induced the generation of Tregs from naïve T cells
and promoted their proliferation, resulting in the accumulation of
these cells in the tumor microenvironment and peripheral blood,
leading to the suppression of tumor-specific T cells and regulation
of antitumor responses (6–8).
To gain insight into this potential mechanism of
B-ALL pathogenesis, the present study investigated the changes of
Treg cells in pediatric patients and the possible mechanism of
differentiation and regulation, with the objective to further
elucidate the tumorigenesis of B-ALL.
Materials and methods
Patients
A total of 18 newly diagnosed pediatric patients
with B-ALL and admitted to Shenzhen Children's Hospital (Shenzhen,
China) between July 2012 and February 2013 were enrolled in the
current study. The cohort consisted of 13 males and 5 females, aged
2.3-11.5 years, with a mean age of 5.1 years. All patients with
B-ALL were examined and diagnosis was confirmed using clinical
examination, bone marrow cell morphology and immunophenotyping by
flow cytometry (12). The diagnosis
was confirmed in accordance with the Recommendation of Diagnosis of
Pediatric Acute Lymphoblastic Leukemia (3rd Amendment Draft)
(13). Inclusion criteria of
patients with B-ALL: Age >12 months and <13 years; confirmed
new diagnosis of B-ALL with ≥25% blasts in the bone marrow; no
prior therapy. Exclusion criteria: Age ≥13 years at the time of
consent; relapsed or refractory B-ALL; prior therapy; known HIV
positive. Blood samples were collected prior to chemotherapy. A
total of 15 outpatients (10 males, 5 females; age range, 3.2-9.6
years; mean age, 4.8 years) recruited between June 2012 and March
2013, received a physical examination in the Department of
Pediatrics in Shenzhen Children's Hospital and served as the
healthy control group. No significant differences in age or gender
were identified between the two groups. Blood samples of patients
with B-ALL and control group were analyzed immediately and without
mitogen stimulation to avoid interference of the activation of
immunocompetent cells. Informed consent was obtained from family
members of all subjects and the study was approved by the
Biomedical Ethics Committee of Shenzhen Children's Hospital.
Isolation of peripheral blood cluster
of differentiation (CD)4+ T cells
Anti-coagulant EDTA was used to collect 3 ml sterile
venous blood from each patient in the current study. Peripheral
blood mononuclear cells were isolated using density gradient
centrifugation at 500 × g for 20 min at 4°C with
fructose-diatrizoate (P=1.077; GE Healthcare Life Sciences, Little
Chalfont, UK). Peripheral CD4+ T cells were isolated
using immunomagnetic beads according to manufacturer's instructions
(DynalBeads CD4 kit; cat. no. 111.45D; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cell activity was determined
by light microscopic examination (BX41; Olympus Corporation, Tokyo,
Japan), following staining with 4% trypan blue at room temperature
for 5 sec. Cell purity was determined using flow cytometry and Diva
V6.1.3 software (BD Biosciences, Franklin Lakes, NJ, USA) following
staining with CD4-fluorescein isothiocyanate (FITC) antibody (10
µg/ml; cat. no. 11-0049-80; Invitrogen; Thermo Fisher Scientific,
Inc.) for 1 h at 4°C. Cells were then prepared completely and
immediately used in further experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from isolated peripheral
CD4+ T cells using the RNAqueous kit (cat. no. AM1912;
Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RNA was quantified using a UV
spectrophotometer. cDNA was synthesized by reverse transcription
using a RevertAid H Minus First Strand cDNA Synthesis kit (cat. no.
K1632; Thermo Fisher Scientific, Inc., Waltham, MA, USA) following
the manufacturer's protocol. cDNA (1 µl) was used as a template and
PCR amplification was conducted as follows: First cycle at 95°C for
15 min, followed by 35-50 cycles at 95°C for 15 sec, 54 to 62°C for
15 sec and 72°C for 25 sec. Primers were designed using the mRNA
sequence of target genes from Genebank (https://www.ncbi.nlm.nih.gov/genbank/), as presented
in Table I. All primers were
synthesized by Shanghai Yingjun Biotechnology Co., Ltd. (Shanghai,
China). The amplification products (each 10 µl) of forkhead box 3
(Foxp3), cytotoxic T lymphocyte-associated antigen 4 (CTLA4),
glucocorticoid-induced tumor necrosis factor receptor (GITR),
lymphocyte activation gene 3 (LAG3), interleukin (IL)-2 receptor
(R)β/γ, IL-6Rα/β, mothers against decapentaplegic homolog
(Smad)3/4, runt-related transcription factor (RUNX)1/3 and β-actin
were loaded into 2% agarose gel. Electrophoresis was conducted at
90 v for 30 min. The gel was then recovered and purified for
sequencing at Shanghai Yingjun Biotechnology Co., Ltd. The
sequencing results were compared with the mRNA sequence of the
target genes obtained from Genebank. All products of amplification,
including Foxp3, CTLA4, GITR, LAG3, IL-2Rβ/γ, IL-6Rα/β, Smad3/4,
RUNX1/3 and β-actin were identical to the mRNA sequences presented
in Genebank. The cDNA synthesized in RT-qPCR was detected using a
SYBR Green kit (cat. no. DRR820S; Takara Biotechnology Co., Ltd.,
Dalian, China) and an RT-PCR cycler (LightCycle 480II; Roche
Applied Science, Penzberg, Germany). Results were analyzed using
the 2−ΔΔCq method with LightCycler Software V1.5 (Roche
Applied Science) (14). The results
were expressed as the ratio of tested gene to β-actin. This
procedure was performed following the manufacturer's protocol.
 | Table I.Primers utilized in reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers utilized in reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer
sequence | Renaturation
temperature (°C) | Number of
cycles | Amplification
position | Length of products
(bp) |
|---|
| Foxp3 | Sense:
5′-GTGGCATCATCCGACAAGG-3′ | 58 | 50 | 1,002-1,167 | 166 |
|
| Antisense:
5′-TGTGGAGGAACTCTGGGAAT-3′ |
|
|
|
|
| CTLA4 | Sense:
5′-GTCCGGGTGACAGTGCTTCG-3′ | 58 | 59 | 360-579 | 220 |
|
| Antisense:
5′-CCAGGTAGTATGGCGGTGGG-3′ |
|
|
|
|
| GITR | Sense:
5′-ACACGCACTTCACCTGGGTCG-3′ | 58 | 50 |
18-146 | 129 |
|
| Antisense:
5′-TGTGCCATGCTCGGGTTTCA-3′ |
|
|
|
|
| LAG3 | Sense:
5′-CCTCACTGTTCTGGGTCTGG-3′ | 56 | 48 | 1,117-1,355 | 239 |
|
| Antisense:
5′-GGATATGGCAGGTGTAGGTC −3′ |
|
|
|
|
| IL-2Rβ | Sense:
5′-AAGCCCTTTGAGAACCTTCG-3′ | 56 | 42 | 504-593 | 90 |
|
| Antisense:
5′-GATTTCCCAGCTTATGTTGC-3′ |
|
|
|
|
| IL-2Rγ | Sense:
5′-ATTGGAAGCCGTGGTTATCT-3′ | 56 | 42 |
869-1013 | 145 |
|
| Antisense:
5′-AAAGTTCCCGTGGTATTCAG-3′ |
|
|
|
|
| IL-6Rα | Sense:
5′-TAGTGTCGGGAGCAAGTTCAG-3′ | 58 | 40 | 1,025-1,116 | 92 |
|
| Antisense:
5′-CGGCAGTGACTGTGATGTTGG-3′ |
|
|
|
|
| IL-6Rβ | Sense:
5′-AGTCGTGCCTGTTTGCTTAG-3′ | 56 | 40 | 2,176-2,338 | 163 |
|
| Antisense:
5′-ATTGTGCCTTGGAGGAGTGT-3′ |
|
|
|
|
| TGF-βRI | Sense:
5′-AATGGGCTTAGTATTCTGGG-3′ | 55 | 35 | 1,285-1,437 | 153 |
|
| Antisense:
5′-ATATTTGGCCTTAACTTCTG-3′ |
|
|
|
|
| Smad3 | Sense:
5′-GGGCTTTGAGGCTGTCTACC-3′ | 56 | 45 | 1,372-1,458 | 87 |
|
| Antisense:
5′-TGTCTCCTGTACTCCGCTCC-3′ |
|
|
|
|
| Smad4 | Sense:
5′-GGATACGTGGACCCTTCTGG-3′ | 55 | 45 | 1,592-1,667 | 76 |
|
| Antisense:
5′-CAATGGCTTCTGTCCTGTGG-3′ |
|
|
|
|
| RUNX1 | Sense:
5′-CATCGCTTTCAAGGTGGTGG-3′ | 58 | 50 | 1,836-1,948 | 113 |
|
| Antisense:
5′-TGGCTGCGGTAGCATTTCTC-3′ |
|
|
|
|
| RUNX3 | Sense:
5′-CCAGGAAAGCACCTACAGAC-3′ | 56 | 50 | 2,447-2,644 | 198 |
|
| Antisense:
5′-AATGATCCCTCACCTCAATG −3′ |
|
|
|
|
| β-actin | Sense:
5′-GAGCTACGAGCTGCCTGACG-3′ | 56-61 | 50 | 787-906 | 120 |
|
| Antisense:
5′-GTAGTTTCGTGGATGCCACAG-3′ |
|
|
|
|
Cytometric bead array
A total of 2 ml peripheral blood was collected from
patients with B-ALL and healthy control subjects, following 6 h of
fasting. Heparin was added for anti-coagulation. Samples were
centrifuged at 500 × g for 10 min at room temperature and plasma
from the upper layer was separated. The plasma concentrations of
IL-6 and transforming growth factor (TGF)-β were measured using a
cytometric bead array (eBioscience; Thermo Fisher Scientific,
Inc.). The procedure was performed according to the manufacturer's
protocol, using FlowCytomix Pro v3.0 (eBioscience; Thermo Fisher
Scientific, Inc.).
Flow cytometry
The percentage of
CD4+CD25highFoxP3+ T cells was
detected using a whole blood counting method. According the
instruction from the Foxp3 Staining Buffer set (cat. no.
00-5523-00; Invitrogen; Thermo Fisher Scientific, Inc.), cells were
gated with CD4-FITC (10 µg/ml; cat. no. 11-0049-80; Invitrogen;
Thermo Fisher Scientific, Inc.) for 30 min at 4°C, fixed and
permeabilized with Foxp3/Transcription Factor Staining Buffer set
(cat. no. 00-5523-00; Invitrogen; Thermo Fisher Scientific, Inc.)
for 60 min at 4°C. Samples were blocked with normal mouse serum
(cat. no. 24-5524-94; Invitrogen; Thermo Fisher Scientific, Inc.)
for 15 min at 4°C and incubated with anti-CD25-phycoerythrin (PE)
(1.25 µg/ml; cat. no. 12-0259-41; Invitrogen; Thermo Fisher
Scientific, Inc.) and anti-Foxp3-allophycocyanin (APC) (1.25 µg/ml;
cat. no. 17-4777-42; Invitrogen; Thermo Fisher Scientific, Inc.)
antibodies for 30 min at 4°C. Cells were also incubated with
anti-phosphorylated (p)-signal transducer and activator of
transcription factor (STAT)3-PerCP-Cy5.5 (as supplied; cat. no.
560114; BD Bioscience, Inc., San Jose, CA, USA) and
anti-pSTAT5-Alexa Fluor647 (as supplied; cat. no. 612599; BD
Bioscience, Inc.) for 30 min at 4°C, to detect the protein mean
fluorescence intensity (MFI) of pSTAT3 and pSTAT5 in
CD4+T cells. To detect the MFI of TGF-βRII and IL-2Rα on
CD4+T, peripheral blood samples were stained and gated
with CD4-eFlour450 (2.5 µg/ml; cat. no. 48-0049-42; Invitrogen;
Thermo Fisher Scientific, Inc.) for 30 min at 4°C, fixed and
permeabilized with intracellular fixation and permeabilization
buffer set (cat. no. 88-8824-00; Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min at room temperature and stained with
anti-TGF-βRII-FITC (as supplied; cat. no. FAB241F-100; R&D
System, Inc., Minneapolis, MN, USA) and anti-IL-2Rα-PE (1.25 µg/ml;
cat. no. 12-0259-41; Invitrogen; Thermo Fisher Scientific, Inc.)
for 30 min at 4°C. Cell counting was conducted using a CantoII flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were
obtained and analyzed using Diva V6.1.3 software.
Statistical analysis
SPSS v19.0 statistical software (IBM Corp., Armonk,
NY, USA) was used for all statistical analyses. Continuous
variables were represented as the mean ± standard deviation. A
two-tailed t-test was used for the comparison of continuous
variables between two groups. P<0.05 was considered to indicate
a statistically significant result. Associations between the
expression of pSTAT3 and CD4+CD25highFoxp3+
Treg in children with B-ALL were analyzed using a Pearson's
correlation test.
Results
Detection of
CD4+CD25highFoxp3+ cells
The percentage of Tregs and the expression of Treg
associated molecules were detected using flow cytometry (Fig. 1A-D) and RT-qPCR (Fig. 1E-H). The percentage of
CD4+CD25highFoxp3+ cells
(P<0.0001; Table II and Fig. 1A-D) and the expression of Foxp3 were
significantly increased (P<0.05; Fig.
1E) in peripheral blood samples of pediatric patients with
B-ALL compared with the healthy controls. The expression of
inhibitory signaling molecules CTLA4, GITR and LAG3 was also
significantly higher in pediatric patients with B-ALL compared with
the control group (P<0.05; Fig.
1F-H).
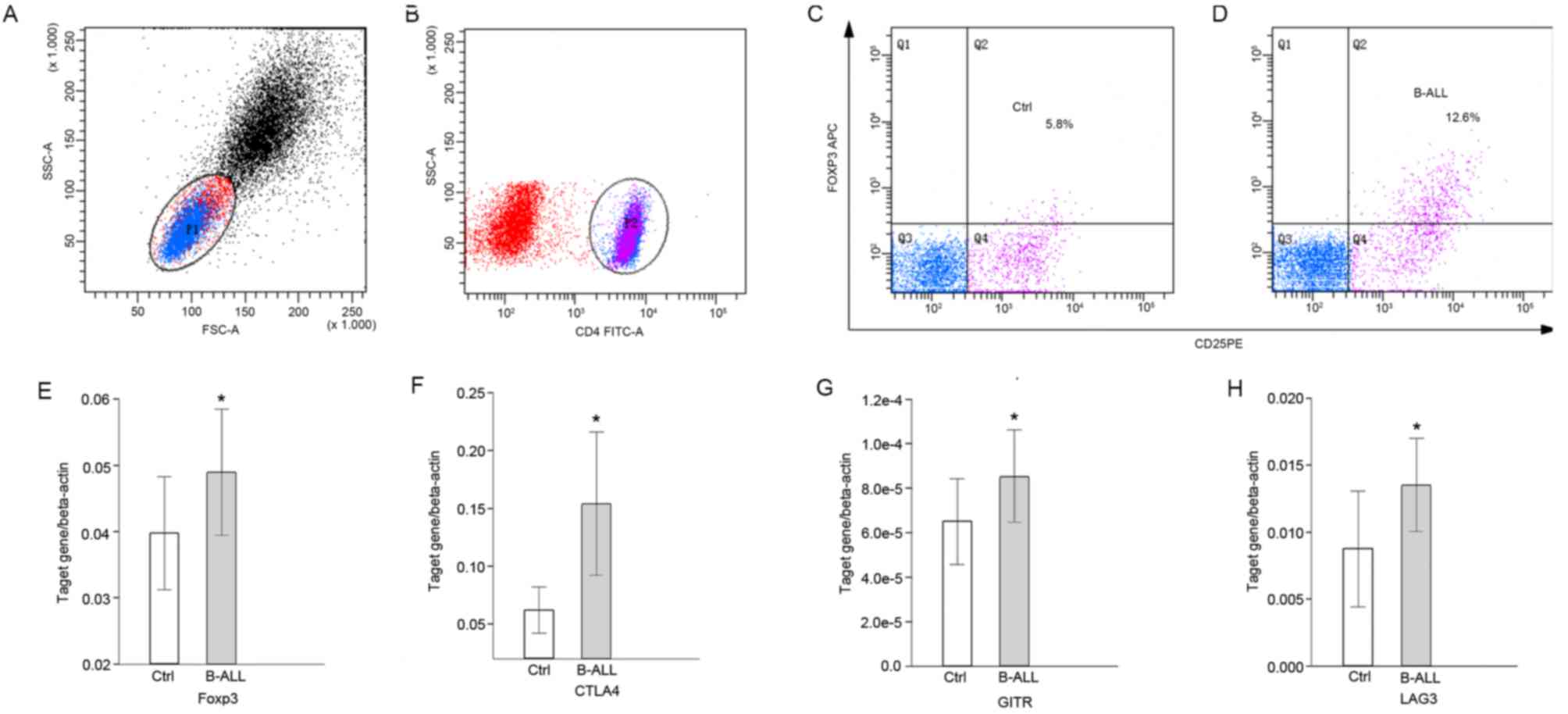 | Figure 1.Proportions of Treg cells and
expression of molecules associated with the suppressor function of
Tregs in pediatric patients with B-ALL, using flow cytometry and
reverse-transcription quantitative polymerase chain reaction. (A)
Dot-plot representing the lymphocytes gated by FSC and SSC; (B)
dot-plot representing CD4+ T cells gated by CD4-FITC
antibody. Dot-plots representing the
CD4+CD25+FOXP3high Treg gated by
(C) CD25-PE and (D) FOXP3-APC antibodies. Transcription levels of
(E) Foxp3, (F) CTLA4, (G) GITR and (H) LAG3 relative to β-actin.
*P<0.05 vs. control. P1, lymphocytes gated by FSC and SSC; P2,
CD4+ T cells gated by CD4-FITC antibody; Q1-4, quadrants
representing cells of single positive for the antibody representing
the x-axis, double positive, double negative and single positive
for the antibody representing the y-axis; Ctrl, control; B-ALL,
B-cell acute lymphocytic leukemia; SSC-A, side scatter area; FSC-A,
forward scatter area; CD, cluster of differentiation; FITC,
fluorescein isothiocyanate; Foxp3, forkhead box protein 3; APC,
allophycocyanin; Ctrl, control; PE, phycoerythrin; CTLA4, cytotoxic
T-lymphocyte associated protein 4; GITR, glucocorticoid-induced
tumor necrosis factor receptor; LAG3, lymphocyte activation gene
3. |
 | Table II.Comparison of associated factors
between patients with B-ALL and controls. |
Table II.
Comparison of associated factors
between patients with B-ALL and controls.
|
| Group |
|
|
|---|
|
|
|
|
|
|---|
| Variables | B-ALL (n=18) | Controls
(n=15) | T-value | P-value |
|---|
|
CD25highFOXP3+/CD4+ | 9.62±4.35% |
4.87±2.61%a | 3.71 | P<0.001 |
|
IL-2Rα/CD4+ | 120.89±37.93 |
79.62±20.22a | 9.79 | P<0.05 |
|
pSTAT3/CD4+ | 29.61±6.85 |
41.92±17.12a | 2.79 | P<0.05 |
|
pSTAT5/CD4+ | 45.83±14.17 | 34.01±9.04 | 2.90 | P<0.05 |
|
TGF-βRII/CD4+ | 50.78±18.87 |
31.39±9.02a | 3.65 | P<0.05 |
| RUNX1 | 2.38±1.44 |
3.07±1.17a | 2.87 | P<0.05 |
| Smad3 | 6.58±4.41 |
4.77±2.38a | 6.81 | P<0.05 |
| IL-6 (pg/ml) | 27.32±8.12 |
16.39±5.78a | 4.51 | P<0.05 |
| TGF-β (ng/ml) | 23.53±13.28 |
8.61±6.10a | 4.01 | P<0.05 |
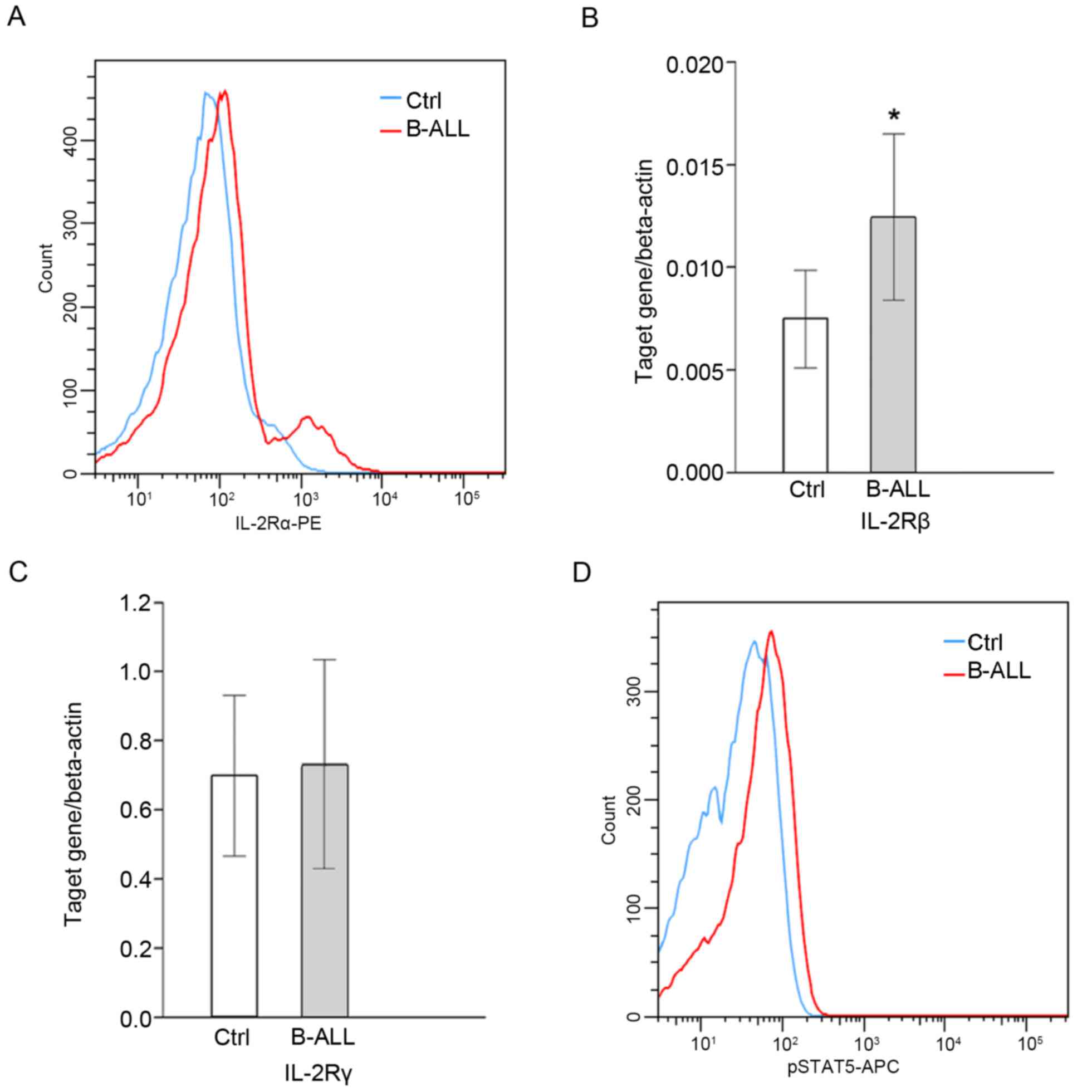
Detection of IL-2 signaling
molecules
The expression of IL-2Rα/β in CD4+ T
cells in patients with B-ALL was significantly upregulated when
compared with healthy controls (P<0.05; Fig. 2A and B); however, no significant
difference in IL-2Rγ was identified (Fig. 2C). Further investigation into the
activation of downstream molecules associated with the IL-2 signal
transduction pathway and the effect of IL-2 signaling on Treg cell
differentiation in B-ALL patients reveled that the expression of
pSTAT5 was significantly higher in patients with B-ALL compared
with healthy controls (P<0.05; Table III; Fig.
2D). Furthermore, pSTAT5 expression was positively correlated
with the percentage of
CD4+CD25highFoxp3+ cells (r=0.17;
P<0.05; Table III).
 | Table III.Correlation between the expression of
differentiation associated factors and
CD4+CD25highFoxp3+ T regulatory
cells in pediatric patients with B-cell acute lymphocytic
leukemia. |
Table III.
Correlation between the expression of
differentiation associated factors and
CD4+CD25highFoxp3+ T regulatory
cells in pediatric patients with B-cell acute lymphocytic
leukemia.
|
|
CD4+CD25highFoxp3+ |
|---|
|
|
|
|---|
| Variables | R-value | P-value |
|---|
|
pSTAT3/CD4+ | −0.39 |
6.73×10−3 |
|
pSTAT5/CD4+ | 0.17 |
9.62×10−4 |
| RUNX1 | 0.60 |
7.24×10−3 |
| Smad3 | 0.87 |
9.59×10−5 |
Detection of IL-6/TGF-β signaling
molecules
The peripheral concentration of TGF-β and the
expression of TGF-βRI/II in CD4+ T cells were
significantly upregulated in patients with B-ALL compared with
healthy controls (P<0.05; Fig. 3A and
B). The expression of the downstream signaling molecules
Smad3/4 was also significantly increased (P<0.05; Fig. 3C and D, respectively). However, the
expression of RUNX1/3 was significantly lower than the control
group (P<0.05; Fig. 3E and F,
respectively). Expression levels of Smad3 and RUNX1 were positively
correlated with CD4+CD25highFoxp3+
cell percentage (r=0.87 and 0.60, respectively; P<0.05; Table III). The changes of IL-6 signaling
were further analyzed using flow cytometry and RT-qPCR. The
concentration of IL-6 in the peripheral blood of patients with
B-ALL was significantly higher than the control group, but no
significant difference was observed in the expression of IL-6Rα/β
in CD4+T cells (Fig. 3G and
H, respectively). In addition, the expression of downstream
pSTAT3 was significantly decreased (P<0.05; Fig. 3I) and the expression of pSTAT3 was
negatively correlated with
CD4+CD25highFoxp3+ cell percentage
(r=−0.39, P<0.05; Table
III).
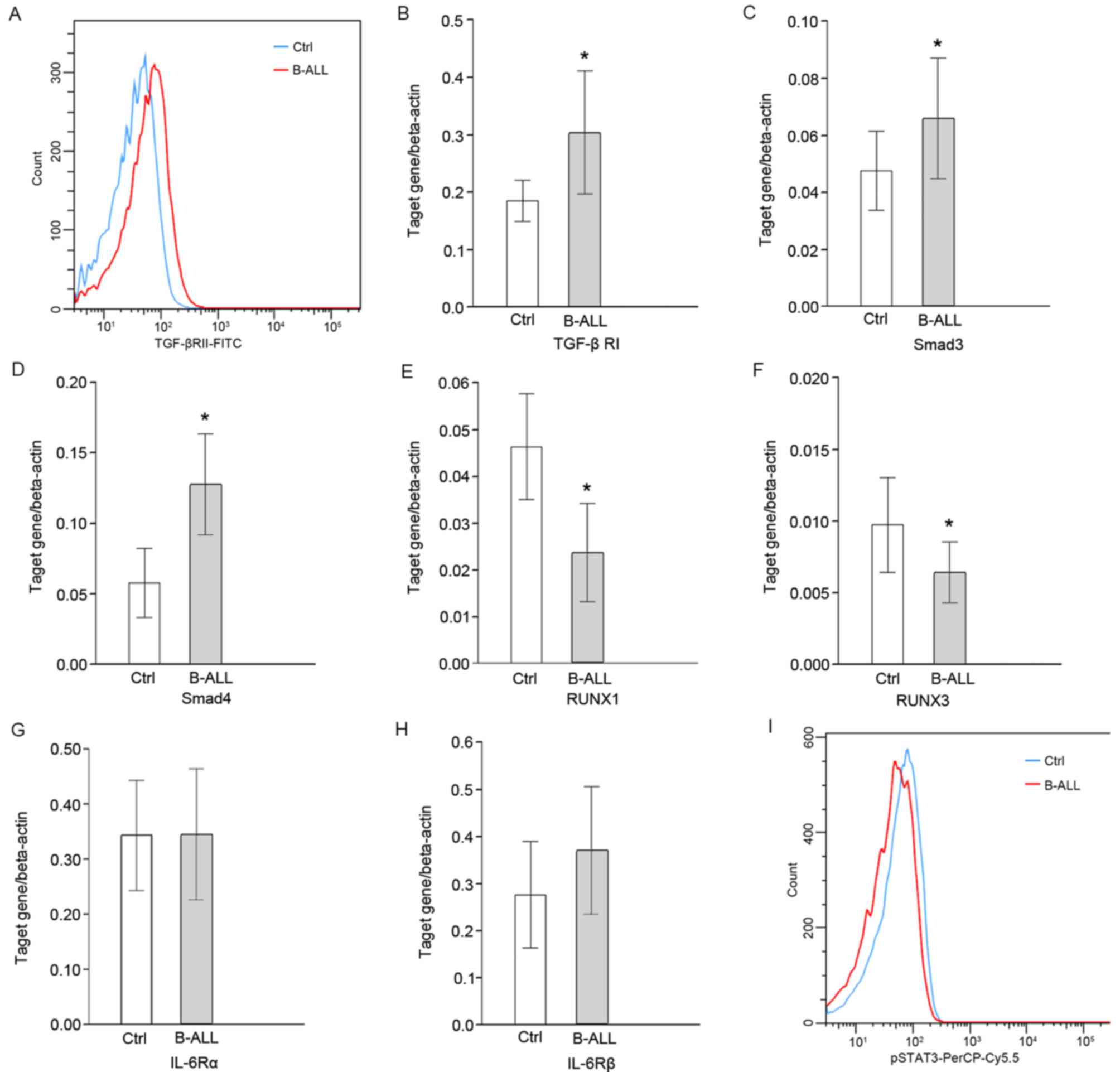 | Figure 3.Protein and mRNA levels of molecules
associated with IL-6/TGF-β signaling in patients with B-ALL
determined using flow cytometry and reverse-transcription
quantitative polymerase chain reaction. (A) Histogram representing
the protein levels of TGF-βRII on the surface of CD4+ T
cells. Transcription levels of (B) TGF-βRI, (C) Smad3, (D) Smad4,
(E) RUNX1, (F) RUNX3, (G) IL-6Rα and (H) IL-6Rβ relative to
β-actin. (I) Histogram representing the protein level of p-STAT3 on
CD4+ T cells. *P<0.05 vs. control. Ctrl, control;
B-ALL, B-cell acute lymphocytic leukemia; TGF-βR, transforming
growth factor-β receptor; RUNX, runt-related transcription factor;
IL, interleukin; R, receptor; p, phosphorylated; STAT, signal
transducer and activator of transcription factor; FITC, fluorescein
isothiocyanate; PerCP-Cy5.5, peridinin chlorophyll protein complex
with cyanine-5.5. |
Discussion
ALL is a heterogeneous disease, primarily caused by
primitive and immature lymphocytic malignant clones, which exhibit
increased cell proliferation, extensive infiltration and the
inhibition of normal hematopoiesis (1). However, the etiology and immune
pathogenesis of ALL remains unclear (1–3). A
healthy immune system effectively identifies and removes abnormal
cells to maintain tumor immune tolerance. Through cell contact or
cytokine secretion, Tregs inhibit the development and activation of
anti-tumor effects, which may lead to tumor immune escape (10,11). The
transcription factor Foxp3 is primarily expressed in Tregs, serving
key roles in differentiation, maturation and cell function
maintenance; thus, Foxp3 is considered to be a specific marker of
Tregs (7). Previous studies have
demonstrated that abnormal Treg cell number and percentage occurs
in certain tumors, including non-small cell lung cancer and ovarian
cancer (15–18), which indicates that the
immunosuppressive effect of Tregs is closely associated with
tumorigenesis. However, the function and status of Tregs in
patients with ALL is yet to be fully elucidated (19,20).
This may be due to the disease exhibiting various subtypes and
durations, as well as the different methods used for detection. In
the present study, under conditions that represented the in
vivo active status of immunocompetent cells without mitogen
stimulation, it was determined that
CD4+CD25highFoxp3+ cell percentage
and Foxp3 expression were higher in patients with B-ALL compared
with healthy controls. In addition, the expression of inhibitory
molecules, including CTLA4, GITR and LAG3 were elevated, suggesting
that overactivation of Tregs may be a factor contributing to tumor
immune escape in B-ALL.
The induction of Treg cell differentiation still
remains unclear. Previous studies have demonstrated that IL-2, IL-6
and TGF-β signaling serve important roles in Treg differentiation,
proliferation and function (21–26).
However, interaction of IL-2 with IL-2R, may trigger various signal
conduction pathways of IL-2; the fast-conducting janus tyrosine
kinase (JAK)/STAT pathway remaining the most predominant (27). IL-2 and STAT5 signals may ensure the
consistent expression of Foxp3 in induced Tregs and may further its
suppressive function (28). The
IL-2R signaling conduction pathway modulates Treg function by
activating STAT5 to upregulate the expression of Foxp3 (29). IL-2 facilitates Treg cell development
and maintenance in peripheral blood, and its proliferation
(29). The knockout of IL-2
signaling may lead to significant Treg cell deficiency, which may
result in autoimmune disease (22,23). The
present study determined that the expression of IL-2Rα/β on the
surface of CD4+T cells and the downstream signaling
molecule pSTAT5 were upregulated when compared with controls.
Furthermore, pSTAT5 expression was positively correlated with
CD4+CD25highFoxp3+ percentage,
indicating that the overactivation of Treg cells in patients with
B-ALL may be associated with an abnormal IL-2 signal.
Naïve CD4+ T cells can be induced by
cytokines into different subpopulations of T helper cells, which
mutually transform to each other with various cytokine
concentrations (6). The TGF-β
cytokine inhibits cellular mitosis, proliferation and migration
(30,31). In early stage tumors, TGF-β exerts an
inhibitory function; however various changes occur within certain
components of the TGF-β signaling pathway, leading to the loss of
TGF-β inhibitory function, resulting in uncontrollable cell
proliferation and tumor progression (32,33). The
transcription factor RUNX is one of the primary targets of TGF-β,
including RUNX 1, 2 and 3. RUNX1 and RUNX3 have important
implications to T lymphocyte differentiation; any functional
changes that occur within RUNX impacts the transduction of the
TGF-β signaling pathway. A previous study has demonstrated that
RUNX and Foxp3 form a feedback loop, such that RUNX proteins
facilitate Foxp3 expression and are associated with the
co-modulation of downstream target gene expression with Foxp3
proteins (34). TGF-β binds to the
cell surface receptor TGF-βRI/II, and triggers Smad and RUNX
signaling (24–26). The former induces the demethylation
of the Foxp3 promoter and initiates its expression (24,25); the
latter interacts with the Smad signal, upregulates the Foxp3
expression and facilitates the differentiation of primary
CD4+ T cells into Tregs (25,26).
IL-6 suppresses the TGF-β induced expression of Foxp3 by
methylating the Foxp3 upstream enhancer through STAT3 signaling
(21). In the present study,
peripheral concentrations of TGF-β, TGF-βI/II and Smad3/4 in
CD4+ T cells were upregulated in patients with B-ALL
when compared with healthy controls, whereas the expression of
RUNX1/3 was decreased. Correlation analysis also revealed that
Smad3 and RUNX1 expression were positively correlated with
CD4+CD25highFoxp3+ cells. In
addition, it was determined that although the concentration of IL-6
in the peripheral blood of patients with B-ALL was higher than that
of the control group, the expression of downstream pSTAT3 was
decreased and negatively correlated with
CD4+CD25highFoxp3+ cell
percentage. These results suggested that TGF-β/Smad signal
overactivation and the lack of pSTAT3 expression may lead to an
abnormal increase of Tregs in pediatric patients with B-ALL.
However, other factors may be associated with the regulation of
TGF-β/RUNX and IL-6/pSTAT3 signaling. The interaction of IL-6 with
IL-6R activates various signal transduction pathways, including
JAK/STAT (35). The activation of
IL-6 enables the phosphorylation of JAK to activate STAT3
transcription factors (36).
Phosphorylated STAT3 then forms dimers that are transduced into
nucleus, further activating or modulating the transcription
capacity of genome (36). Thus, IL-6
leads to the methylation of Foxp3 gene enhancers through STAT3
signaling (21). The present study
revealed that although pediatric patients with acute B-ALL exhibit
higher IL-6 concentrations than the healthy control group, the
expression of their downstream signaling factor, pSTAT3, is
significantly lower. Considering that variations in IL-6, its
receptors and the signaling factors are inconsistent, it is
speculated that the reason for insufficiency of pSTAT3 and abnormal
proliferation of Treg cell in patients with acute B-ALL may involve
other factors, which may further participate in modulating
IL-6/pSTAT3 signal. Therefore, further study into the mechanism of
modulation is required.
In conclusion, the overactivation of Tregs in
patients with B-ALL may be associated with the overactivation or
insufficient expression of a variety of regulatory cytokine
signaling molecules. The overactivation of IL-2/pSTAT5 and
TGF-β/Smad, and the insufficient expression of pSTAT3 served an
important role in the regulation of Tregs in pediatric patients
with B-ALL. However, further investigation into the molecular
mechanism of aforementioned abnormal signaling, as well as the
factors participating in the regulation of IL-6 and TGF-β signaling
is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Projects from the Science Technology and Innovation
Committee of Shenzhen Municipality (grant no.
JCYJ20140416141331552), Nature Science Foundation of Guangdong
Province (grant no. 2015A030313759), Sanming Project of Medicine in
Shenzhen (SZSM201512033) and Shenzhen Public Service Platform of
Molecular Medicine in Pediatric Hematology and Oncology.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FW and CL contributed to the conception of the
study. HX, GW and XC contributed significantly to the analysis and
manuscript preparation; HM, XY and GL contributed to the clinical
diagnosis and data management, and obtained the informed consents.
SL performed the data analyses and wrote the manuscript. All
authors reviewed and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Informed consent was collected from family members
of all subjects and the study was approved by the Biomedical Ethics
Committee of Shenzhen Children's Hospital (Shenzhen, China).
Patient consent for publication
Informed consent was collected from family members
of all subjects.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Onciu M: Acute lymphoblastic leukemia.
Hematol Oncol Clin North Am. 23:655–674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woo JS, Alberti MO and Tirado CA:
Childhood B-acute lymphoblastic leukemia: A genetic update. Exp
Hematol Oncol. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiarini F, Lonetti A, Evangelisti C,
Buontempo F, Orsini E, Evangelisti C, Cappellini A, Neri LM,
McCubrey JA and Martelli AM: Advances in understanding the acute
lymphoblastic leukemia bone marrow microenvironment: From biology
to therapeutic targeting. Biochim Biophys Acta. 1863:449–463. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pui CH, Carroll WL, Meshinchi S and Arceci
RJ: Biology, risk stratification, and therapy of pediatric acute
leukemias: An update. J Clin Oncol. 29:551–565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hangai M, Watanabe K, Shiozawa R, Hiwatari
M, Ida K and Takita J: Relapsed acute lymphoblastic leukemia with
unusual multiple bone invasions: A case report. Oncol Lett.
7:991–993. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Colombo MP and Piconese S:
Regulatory-T-cell inhibition versus depletion: The right choice in
cancer immunotherapy. Nat Rev Cancer. 7:880–887. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oleinika K, Nibbs RJ, Graham GJ and Fraser
AR: Suppression, subversion and escape: The role of regulatory T
cells in cancer progression. Clin Exp Immunol. 171:36–45. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Facciabene A, Motz GT and Coukos G:
T-regulatory cells: Key players in tumor immune escape and
angiogenesis. Cancer Res. 72:2162–2171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu J, Yamane H and Paul WE:
Differentiation of effector CD4 T cell populations (*). Annu Rev
Immunol. 28:445–489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rudensky AY: Regulatory T cells and Foxp3.
Immunol Rev. 241:260–268. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shevach EM: Mechanisms of foxp3+ T
regulatory cell-mediated suppression. Immunity. 30:636–645. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peters JM and Ansari MQ: Multiparameter
flow cytometry in the diagnosis and management of acute leukemia.
Arch Pathol Lab Med. 135:44–54. 2011.PubMed/NCBI
|
|
13
|
Subspecialty Group of Hematology Diseases;
The Society of Pediatrics; Chinese Medical Association; Editorial
Board; Chinese Journal of Pediatrics. Recommendations for diagnosis
and treatment of acute lymphoblastic leukemia in childhood (3rd
revised version). Zhonghua Er Ke Za Zhi. 44:392–395. 2006.(In
Chinese). PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto T, Yanagimoto H, Satoi S,
Toyokawa H, Hirooka S, Yamaki S, Yui R, Yamao J, Kim S and Kwon AH:
Circulating CD4+CD25+ regulatory T cells in patients with
pancreatic cancer. Pancreas. 41:409–415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beyer M, Classen S, Endl E, Kochanek M,
Weihrauch MR, Debey-Pascher S, Knolle PA and Schultze JL:
Comparative approach to define increased regulatory T cells in
different cancer subtypes by combined assessment of CD127 and
FOXP3. Clin Dev Immunol. 2011:7340362011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shenghui Z, Yixiang H, Jianbo W, Kang Y,
Laixi B, Yan Z and Xi X: Elevated frequencies of CD4+
CD25+ CD1271o regulatory T cells is associated to poor
prognosis in patients with acute myeloid leukemia. Int J Cancer.
129:1373–1381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weiss L, Melchardt T, Egle A, Grabmer C,
Greil R and Tinhofer I: Regulatory T cells predict the time to
initial treatment in early stage chronic lymphocytic leukemia.
Cancer. 117:2163–2169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li AH, Qiu GQ, Gu WY, Ling Y, Weng KZ, Tan
Q and Cao XS: Expression of CD4+ CD25+ regulatory T cells in the
patients with acute lymphocytic leukemia. Xi Bao Yu Fen Zi Mian Yi
Xue Za Zhi. 23:439–442. 2007.(In Chinese). PubMed/NCBI
|
|
20
|
Wu CP, Qing X, Wu CY, Zhu H and Zhou HY:
Immunophenotype and increased presence of CD4(+)CD25(+) regulatory
T cells in patients with acute lymphoblastic leukemia. Oncol Lett.
3:421–424. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lal G, Zhang N, van der Touw W, Ding Y, Ju
W, Bottinger EP, Reid SP, Levy DE and Bromberg JS: Epigenetic
regulation of Foxp3 expression in regulatory T cells by DNA
methylation. J Immunol. 182:259–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng G, Yu A, Dee MJ and Malek TR: IL-2R
signaling is essential for functional maturation of regulatory T
cells during thymic development. J Immunol. 190:1567–1575. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rouse M, Nagarkatti M and Nagarkatti PS:
The role of IL-2 in the activation and expansion of regulatory
T-cells and the development of experimental autoimmune
encephalomyelitis. Immunobilogy. 218:674–682. 2013. View Article : Google Scholar
|
|
24
|
Schlenner SM, Weigmann B, Ruan Q, Chen Y
and von Boehmer H: Smad3 binding to the foxp3 enhancer is
dispensable for the development of regulatory T cells with the
exception of the gut. J Exp Med. 209:1529–1535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rahimi RA and Leof EB: TGF-beta signaling:
A tale of two responses. J Cell Biochem. 102:593–608. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klunker S, Chong MM, Mantel PY, Palomares
O, Bassin C, Ziegler M, Rückert B, Meiler F, Akdis M, Littman DR
and Akdis CA: Transcription factors RUNX1 and RUNX3 in the
induction and suppressive function of Foxp3+ inducible regulatory T
cells. J Exp Med. 206:2701–2715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao W, Lin JX and Leonard WJ:
Interleukin-2 at the crossroads of effector responses, tolerance,
and immunotherapy. Immunity. 38:13–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Q, Kim YC, Laurence A, Punkosdy GA
and Shevach EM: IL-2 controls the stability of Foxp3 expression in
TGF-beta-induced Foxp3+ T cells in vivo. J Immunol. 186:6329–6337.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mahmud SA, Manlove LS and Farrar MA:
Interleukin-2 and STAT5 in regulatory T cell development and
function. JAKSTAT. 2:e231542013.PubMed/NCBI
|
|
30
|
Classen S, Zander T, Eggle D, Chemnitz JM,
Brors B, Büchmann I, Popov A, Beyer M, Eils R, Debey S and Schultze
JL: Human resting CD4+ T cells are constitutively inhibited by TGF
beta under steady-state conditions. J Immunol. 178:6931–6940. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Delisle JS, Giroux M, Boucher G, Landry
JR, Hardy MP, Lemieux S, Jones RG, Wilhelm BT and Perreault C: The
TGF-β-Smad3 pathway inhibits CD28-dependent cell growth and
proliferation of CD4 T cells. Genes Immun. 14:115–126. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Seoane J and Gomis RR: TGF-β family
signaling in tumor suppression and cancer progression. Cold Spring
Harb Perspect Biol. 9:a0222772017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Principe DR, Doll JA, Bauer J, Jung B,
Munshi HG, Bartholin L, Pasche B, Lee C and Grippo PJ: TGF-β:
Duality of function between tumor prevention and carcinogenesis. J
Natl Cancer Inst. 106:djt3692014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bruno L, Mazzarella L, Hoogenkamp M,
Hertweck A, Cobb BS, Sauer S, Hadjur S, Leleu M, Naoe Y, Telfer JC,
et al: Runx proteins regulate Foxp3 expression. J Exp Med.
206:2329–2337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neurath MF and Finotto S: IL-6 signaling
in autoimmunity, chronic inflammation and inflammation-associated
cancer. Cytokine Growth Factor Rev. 22:83–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun Q, Liu Q, Zheng Y and Cao X: Rapamycin
suppresses TLR4-triggered IL-6 and PGE(2) production of colon
cancer cells by inhibiting TLR4 expression and NF-kappaB
activation. Mol Immunol. 45:2929–2936. 2008. View Article : Google Scholar : PubMed/NCBI
|