Introduction
Alzheimer's disease (AD) is a devastating
neurodegenerative disease that causes progressive damage to neurons
(1). It is characterized by the
impairment of cognition, memory and learning, and causes >80% of
cases of dementia in the world's rapidly growing aging population
(2). Currently, AD treatment is
enormously expensive, and no curative treatment for AD is available
because the etiology of AD is poorly understood. Non-coding RNAs,
including microRNAs (miRs) and long non-coding RNAs (lncRNAs), are
regulatory molecules associated with a wide variety of biological
processes and disease states (3).
miR-339-5p levels are significantly reduced in brain specimens
isolated from AD patients, and miR-339-5p regulates expression of
β-secretase 1 (BACE1), a crucial enzyme in the pathophysiology of
AD, in human brain cells (4).
Emerging evidence has demonstrated that lncRNAs have an important
role in many neurological diseases, such as AD, Parkinson's disease
and Huntington's disease (5).
Certain differentially expressed lncRNAs associated with AD have
been identified (6,7). For example, gene set enrichment
analysis identified a downregulated lncRNA n341006 in association
with protein ubiquitination pathway, and significantly upregulated
lncRNA n336934 associated with cholesterol homeostasis in AD
patients (8). Massone et al
(9) previously demonstrated that
lncRNA 17A was upregulated in cerebral tissues derived from AD
patients, and that it could enhance the secretion of amyloid β (Aβ)
peptide and the Aβ1-42/Αβ1–40 peptide ratio.
The BACE1-antisense transcript (BACE1-AS) has been
identified as a conserved non-coding antisense BACE1. BACE1-AS can
positively regulate BACE1 mRNA and thus BACE1 protein expression
in vitro and in vivo (10). In addition, silencing lncRNA BACE1-AS
expression with short interfering RNA (siRNA) in senile plaque AD
SH-SY5Y cells attenuates the ability of BACE1 to cleave amyloid
precursor protein (APP) and reduce the production of Aβ1–42
oligomers (11). However, whether
BACE1-AS can regulate memory and learning behaviors remains
unknown. The aim of the present study was to elucidate the role of
lncRNA BACE1-AS in memory and learning.
Materials and methods
Blood samples
Peripheral blood samples of AD patients (n=30;
male/female, 17/13; age range, 60–82 years) and age-matched normal
subjects (n=36; male/female, 20/16; age range, 65–79 years) without
notable illness, including diabetes, heart disease, stroke or
cancer were collected at the Department of Neurology, Hefei
Affiliated Hospital of Anhui Medical University (Hefei, China)
between March 2015 and May 2016. Samples were stored at −80°C prior
to further use. The present study was approved by the Ethics
Committee of Hefei Affiliated Hospital of Anhui Medical University.
All participants provided written informed consent.
Animals
Male SAMR1 (age, 6 months; weight range, 23–30 g;
n=8) and male SAMP8 (age, 6 months; weight range, 22–32 g; n=32; 8
mice per group) mice were obtained from the Animal Center of
Beijing University Medical Department (Beijing, China). SAMP8 is an
AD animal model with age-related learning and memory deficits
(12) and SAMR1 mice served as a
healthy control. Mice were fed ad libitum and housed in a
12-h light/dark cycle at 25±1°C and 50% humidity. To knockdown
BACE1-AS in hippocampus, SAMP8 mice anesthetized with chloral
hydrate (40 mg/kg; cat. no. 47335-U; Merck KGaA, Darmstadt,
Germany) were positioned in a stereotaxic apparatus with bregma and
lambda at a horizontal level, and administered with 1.5 µl
1×109 BACE1-AS siRNA lentivirus (2×109 titer
units/ml diluted 10× with enhance infected solution) or an empty
lentivirus (Shanghai GeneChem Co., Ltd., Shanghai, China) into
bilateral hippocampi using the following coordinates:
Anteroposterior −3.50 mm relative to bregma; lateral ± 1.50 mm;
dorsoventral 3.5 mm from the skull, as previously described
(13). The administration lasted 5
min, allowing slow diffusion. SAMP8 mice injected with empty
lentivirus were used as negative control (NC). SAMP8 mice received
an equal volume of vehicle were used as Control. Mice were allowed
to survive for 3 weeks. Brains were harvested for further analysis
following Y-maze and Morris water maze test behavioral tests
(14,15). The experimental protocol was approved
by the Animal Care and Use Committee of Hefei Affiliated Hospital
of Anhui Medical University, in compliance with the National
Institutes of Health Guide for the Care and Use of Laboratory
Animals.
Primary hippocampal neuron
culture
A pregnant SAMP8 mouse (age, 3 months; weight 50 g;
n=1; gestational age, 2 weeks) was obtained from the Animal Center
of Beijing University Medical Department (Beijing, China) and was
fed ad libitum and housed in a 12-h light/dark cycle at
25±1°C and 50% humidity prior to experiments. The mouse was
anesthetized with chloral hydrate (40 mg/kg; cat. no. 47335-U;
Merck KGaA, Darmstadt, Germany) and 8 embryos were harvested.
Primary hippocampal neurons were obtained from embryonic day-15
hippocampi of SAMP8 mice. Briefly, the hippocampi were mechanically
removed and cut into 1 mm3 pieces and treated with
trypsin and 0.05 mg/ml DNase (cat. no. AMPD1-1KT; Merck KGaA,
Darmstadt, Germany) for 15 min at 37°C in serum-free Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Hippocampal cells were washed with DMEM
containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and resuspended in completed culture medium [DMEM
supplemented with 10% FBS, penicillin (50 U/ml), streptomycin (50
U/ml) and glutamine (0.5 mmol/l)]. The cells were cultured in a
humidified atmosphere containing 5% CO2 at 37°C. To
knockdown BACE1-AS in primary hippocampal neurons, the cells were
infected with BACE1-AS siRNA lentivirus (10×107 titer
units/ml) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. After 72 h, cells were
photographed using light microscopy (magnification, ×100) to
observe morphological alterations.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
An Ultrapure RNA kit (cat. no. CW0581M; CWBio,
Beijing, China) was used to extract RNA from peripheral blood
samples and hippocampi tissues from SAMR1 and SAMP8 mice peripheral
blood samples from patients with AD and normal subjects or primary
hippocampal neurons from embryos according to the manufacturer's
instructions. Maxima First Strand cDNA Synthesis kit (cat. no.
K1642; Thermo Fisher Scientific, Inc.) was used for reverse
transcription according to the manufacturer's protocol. Expression
of BACE1-AS was detected via UltraSYBR Mixture (cat. no. CW2602;
CWBio). Expression of β-actin was used as an endogenous control.
The following primers were used: BACE-AS1 forward,
5′-TCTGGGCAGTAGGGGGTTAC-3′ and reverse, 5′-GACTACCTGCCCACCCAAGA-3′;
and β-actin forward, 5′-GCCCTATAAAACCCAGCGGC-3′ and reverse,
5′-TCGATGGGGTACTTCAGGGT-3′. Amplification conditions were as
follows: 95°C for 3 min, 40 cycles of denaturation at 95°C for 15
sec and annealing at 58°C for 45 sec. Data were quantified using
the 2−ΔΔCq method (16).
ELISA determination of Aβ1–40 and
1–42
Hippocampi tissues were homogenized using a Tissue
Protein Extraction kit containing protease inhibitor cocktail (cat.
no. CW0891; CWBio) and centrifuged at 12,000 × g for 30 min at 4°C.
Supernatants were used for ELISA quantification using a Mouse
Aβ1–40 ELISA kit (cat. no. CSB-E10787m; Cusabio Biotech Co., Ltd.,
Wuhan, China) and a Mouse Aβ1–42 ELISA kit (cat. no. CSB-E08300m;
Cusabio Biotech Co., Ltd.) according to the manufacturer's
instructions.
Western blot analysis
Total protein was extracted from hippocampus tissues
using a Cold Tissue Protein Extraction kit containing protease
inhibitor cocktail (cat. no. CW0891; CWBio). A BCA Protein Assay
kit (cat. no. CW0014S; CWBio) was used to determine the protein
concentration. Equal protein samples (60 µg) were then separated by
12% SDS-PAGE and transferred to a 0.22 µm nitrocellulose membrane
(cat. no. CW2002S; CWBio). The membrane was blocked in 5% non-fat
dried milk in TBS-Tween-20 for 2 h at room temperature and
incubated with the following primary antibodies: Anti-BACE1 (cat.
no. ab183612; dilution, 1:500), anti-APP (cat. no. ab12266;
dilution, 1:500), anti-phosphorylated (p)-tau (cat. no. ab81268;
dilution, 1:500), anti-tau (cat. no. ab64193; dilution, 1:500) and
anti-GAPDH (cat. no. ab8245; dilution, 1:500; all Abcam, Cambridge,
UK) overnight at 4°C. The membrane was washed and incubated with
secondary antibodies: Goat anti-rabbit IgG (HRP; cat. no. ab6721;
dilution, 1:3,000; Abcam, Cambridge, UK) or goat anti-mouse IgG
(HRP; cat. no. ab205719; dilution, 1:3,000; Abcam) for 2 h at room
temperature. The signal on the membrane was visualized using
enhanced chemiluminescence reagent (EMD Millipore, Billerica, MA,
USA) and densitometry analysis was performed using Image-Pro plus
software 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Cell counting kit (CCK)-8 assay
At 24 h prior to the experiment, cells were plated
in 96-well plates at a density of 1,000 cells in 100 µl medium per
well at 37°C. The cell viability was assessed via CCK-8 assay
(Beyotime Institute of Biotechnology, Haimen, China) according to
the manufacturers' instructions. The assay was repeated three times
in triplicate wells.
Y-maze test
The Y-maze based on place or object exploration is
used to assess spatial recognition memory (17). The Y-maze has been used to study
learning and memory under certain conditions, such as chronic
stress (18). Shin et al
(15) previously measured spatial
learning and memory using the Y-maze and Morris water maze in rats
stimulated with Neuropep-1. The Y-maze test was performed as
previously described (19). Briefly,
mice were initially placed at the end of one arm and allowed to
move freely for 10 min. The series of arm entries was recorded by a
video camera. Spontaneous alternation was defined as successive
entries into the three arms in overlapping triplet sets. The
alternation percentage was determined as the ratio of actual
alternations to maximum alternations.
Morris water maze test
The Morris water maze test was performed as
described previously (14). Briefly,
a circular pool divided into four quadrants with fixed visual cues
was filled with opaque water at a constant temperature (22°C), and
was monitored by a video camera. Each mouse was trained via four
visible platform (10 cm in diameter) tests prior to the behavioral
experiment. Each mouse was placed into the water facing the pool
wall (back to platform) and given 60 sec to swim freely and climb
onto the visible platform, once daily for 4 days to observe and
record the time needed to find and climb onto the platform (escape
latency). On day 5, hidden platform trials were performed four
times per day for 6 days. The next day, a spatial probe trial was
performed. The number of times of crossing the original platform
location in the pool within 90 sec was recorded using a Morris
water maze image automatic monitoring system (Gene and I Co., Ltd.,
Beijing, China). Following the experiment, mice were sacrificed and
the brains were removed.
Statistical analyses
Statistical analyses were performed using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA), and data were
presented as the mean + or ± standard error of the mean. Unpaired
two-tailed Student's t-test was used to analyze differences between
two groups, and one-way analysis of variance with a post hoc
Bonferroni test was used to analyze differences between three or
more groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
BACE1-AS levels are increased in blood
of patients with AD
The expression level of BACE1-AS was detected via
RT-qPCR in AD patients (n=30) and age-matched normal controls
(n=36). The present results demonstrated that BACE1-AS was
significantly increased in peripheral blood of AD patients compared
with controls (Fig. 1A). In
addition, BACE1-AS expression was measured in the peripheral blood
and hippocampus from SAMR1 (control) and SAMP8 mice. Compared with
controls, the expression of BACE1-AS was significantly increased in
peripheral blood and hippocampus tissues of SAMP8 mice, suggesting
that BACE1-AS may be associated with age-related cognitive decline
in AD.
Knockdown of BACE1-AS by siRNA
promotes the survival of primary neurons
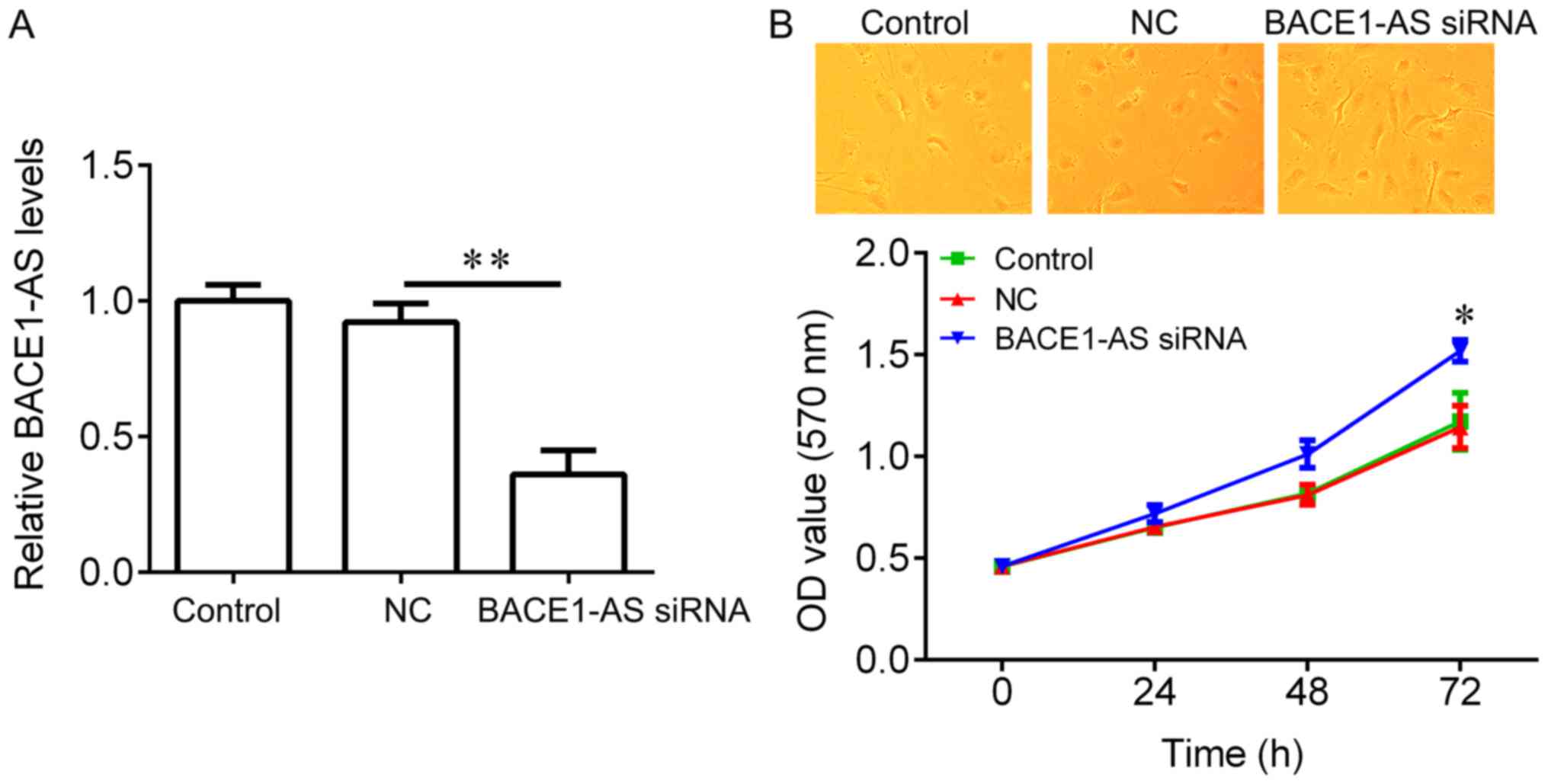
To test if BACE1-AS regulates hippocampal neurons
proliferation, BACE1-AS was knocked down in hippocampal neurons and
an CCK-8 assay was performed. BACE1-AS expression was significantly
reduced in hippocampal neurons infected with BACE1-AS siRNA
lentivirus compared with negative controls (Fig. 2A). No significant changes in the
morphology of hippocampal neurons between the three groups were
observed (Fig. 2B, upper panel).
Compared with the negative control group, BACE1-AS siRNA
transfection exhibited a significant promotion on cell
proliferation (Fig. 2B, lower
panel). These results suggested that BACE1-AS downregulation
promotes hippocampal neurons proliferation.
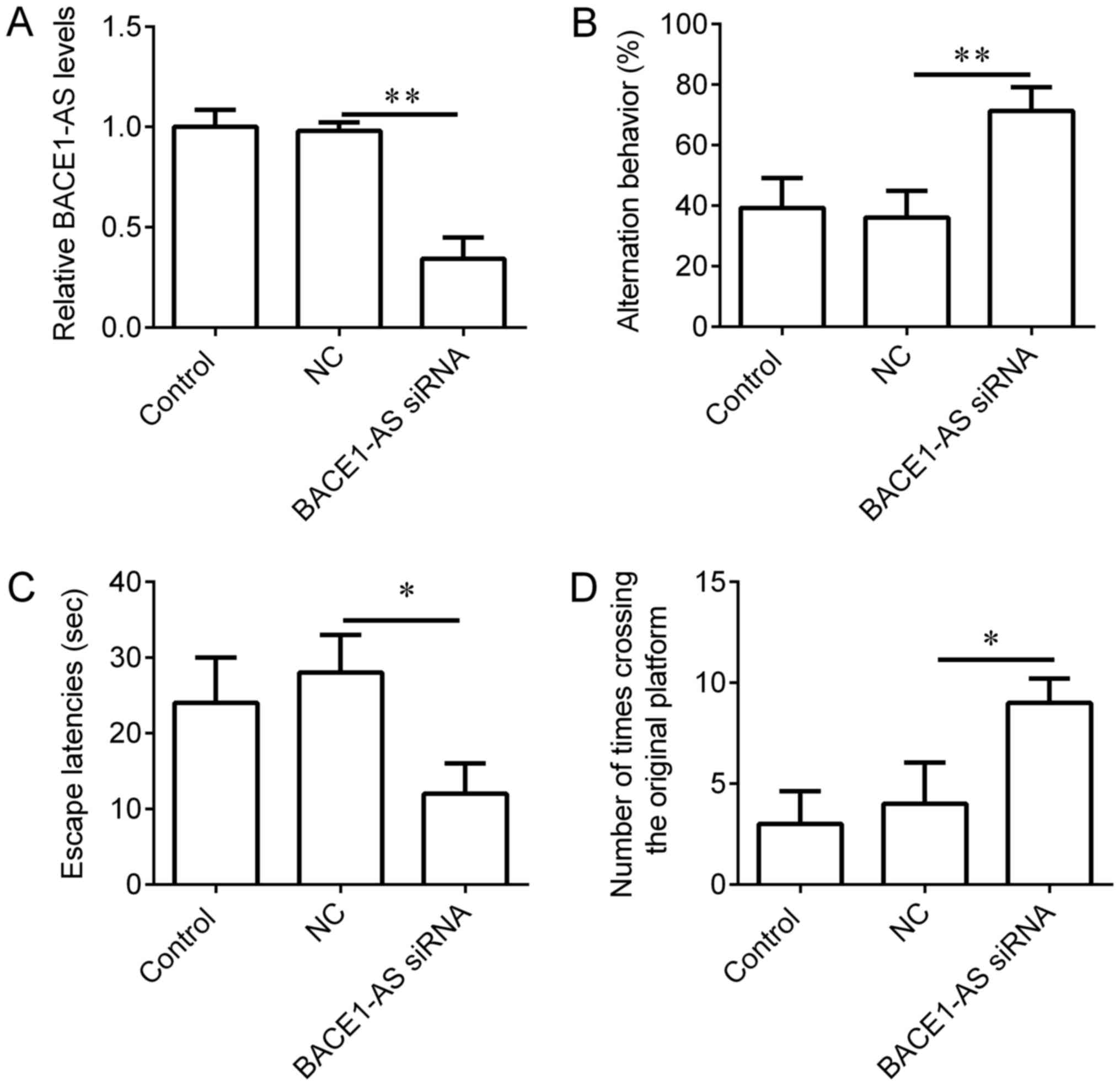
Knockdown of BACE1-AS in hippocampi
improves learning and memory behaviors of SAMP8 mice
To evaluate the functions of BACE1-AS on learning
and memory behaviors in vivo, BACE1-AS siRNA lentivirus was
administered to the hippocampi of SAMP8 mice. SAMR1 mice, which
received an equal volume of vehicle, were used as controls and mice
injected with empty lentivirus were used as negative controls. The
expression of BACE1-AS in mice injected with BACE1-AS siRNA
lentivirus was significantly decreased compared with negative
control (Fig. 3A). Following 3 weeks
of BACE1-AS siRNA lentivirus infection, downregulation of BACE1-AS
in hippocampi significantly increased successive entries in the
Y-maze test (Fig. 3B), reduced the
escape latencies in the Morris water maze (Fig. 3C) and increased instances of crossing
the original platform in the Morris water maze (Fig. 3D) in comparison with negative
controls, indicating that BACE1-AS downregulation improves the
learning and memory behaviors of SAMP8 mice.
Knockdown of BACE1-AS decreases BACE1
and Aβ levels in vivo
It was examined whether BACE1-AS could regulate
several important proteins for AD, including BACE1, APP, tau and
Aβ. Aβ 1–40 and Aβ 1–42 levels were measured by ELISA, and BACE1,
APP, p-tau and tau expression was measured via western blotting. It
was demonstrated that, compared with negative controls, BACE1-AS
knockdown significantly inhibited BACE1, APP and p-tau expression
(Fig. 4A), and also reduced the
concentration of Aβ 1–40 and Aβ 1–42 in hippocampi treated with
BACE1-AS siRNA (Fig. 4B and C).
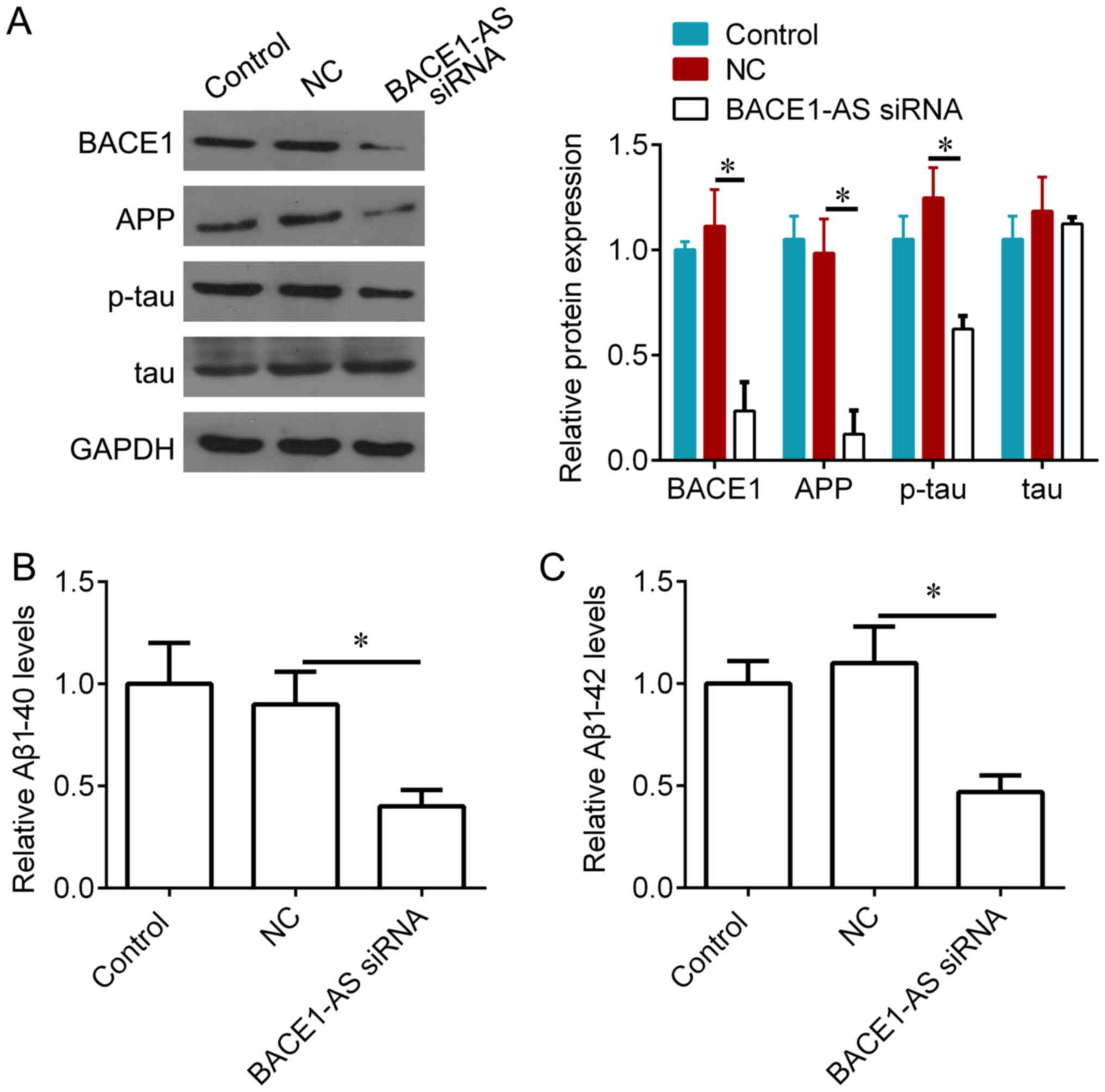 | Figure 4.Knockdown of BACE1-AS decreases BACE1
and APP accumulation, and phosphorylation of tau protein in
hippocampus of SAMP8 mice. (A) Western blot analysis for BACE1,
APP, p-tau and total tau protein in hippocampal tissues from SAMP8
mice following 3 weeks lentivirus injection, and quantification of
the bands. (B) ELISA analysis for Aβ1–40 in hippocampal tissues
from SAMP8 mice following 3 weeks lentivirus injection. (C) ELISA
analysis for Aβ1–42 in hippocampal tissues from SAMP8 mice
following 3 weeks lentivirus injection. *P<0.05 vs. NC.
BACE1-AS, β-secretase 1-antisense transcript; APP, amyloid
precursor protein; p, phosphorylated; Aβ, amyloid β; NC, negative
control; siRNA, short interfering RNA. |
Discussion
In the present study, it was demonstrated that
lncRNA BACE1-AS expression was highly expressed in blood samples
from AD patients, and also upregulated in peripheral blood samples
and hippocampi from an AD animal model. Knockdown of BACE1-AS by
siRNA increased the primary hippocampal neurons proliferation in
vitro, and improved the memory and learning behaviors in SAMP8
mice by inhibiting BACE1 and APP production, and phosphorylation of
tau protein.
Aβ peptide recurrently is accepted as the culprit in
the pathogenesis of AD (20). BACE1
is required for the production of Aβ peptide (21). This suggests that the inhibition of
BACE1 and subsequent reduction of Aβ may cure or prevent AD.
Although the precise mechanisms that trigger Aβ accumulation remain
unclear, much effort has focused on screening BACE1 inhibitors
(22). Recent studies in which BACE1
activity is specifically inhibited in animal models with knockout
technology, virus-delivered siRNAs and bioavailable small-molecule
agents support the use of therapeutic BACE1 inhibition (23,24).
Genetic BACE1 inhibition may be a promising treatment strategy for
AD (25). Non-coding RNAs were
demonstrated to control BACE1 expression and Aβ production
(26). Kim et al demonstrated
that a reduction in miR-186 levels during aging may lead to the
upregulation of BACE1 in the brain, thus increasing the risk of AD
in elderly individuals (27).
miR-195 negatively regulated by nuclear factor-κB-mediated Aβ
aggregation and tau hyperphosphorylation in chronic brain
hypoperfusion (28). In addition,
lncRNAs also have critical roles in progression of AD by regulating
BACE1 (29). Neuroblastoma
differentiation marker 29 (NDM29) is a non-coding RNA that is
dose-dependently induced by inflammatory stimulation (30). NDM29 can promote the cleavage
activities of BACE to increase Aβ formation and the Aβx-42/Aβx-40
ratio (30).
BACE1-AS is a crucial enzyme in AD pathophysiology
that was originally identified as a conserved non-coding antisense
transcript for BACE1 (10). BACE1-AS
transcript was increased in the parietal lobes and cerebellum from
postmortem brains of AD patients. BACE1-AS can regulate BACE1 mRNA
and protein expression in vitro and in vivo, and Aβ
1–42 stimulation also can elevate the expression of BACE1-AS,
increasing BACE1 mRNA stability and generating additional Aβ 1–42
through a post-transcriptional feed-forward mechanism (10). In addition, downregulation of lncRNA
BACE1-AS expression in SH-SY5Y cells by siRNA silencing attenuates
the ability of BACE1 to cleave APP and delays the induction of
senile plaque formation in a senile plaque AD cell model (11). BACE1-AS levels were associated with
HuD, a primarily neuronal RNA-binding protein that is implicated in
learning and memory. BACE1-AS level was higher in the brain of
HuD-overexpressing mice (31). HuD
can interact with the 3′untranslated regions of BACE1 mRNA to
increase the half-life of this mRNA (31). In addition, dysregulation of the
BACE1/BACE1-AS/β-amyloid axis was also relevant in heart failure
pathogenesis (32). BACE1-AS also
has a role in cancer (33). BACE1-AS
was significantly increased in anisomycin-treated ovarian cancer
stem cells. Elevation of lncRNA BACE1-AS expression can suppress
human ovarian cancer stem cells proliferation and invasion
(34).
The present study suggests that BACE1-AS levels are
significantly upregulated in peripheral blood samples of patients
with AD, suggesting that BACE1-AS might be an indicator for
progression of AD. Knockdown of BACE1-AS by siRNA improves memory
and learning behaviors, possibly via increasing the hippocampal
neurons growth, and decreasing BACE1 and Aβ accumulation, and
phosphorylation of tau protein in hippocampus of SAMP8 mice.
Therefore, BACE1-AS may be a potential target for management of
memory loss related disease, such as AD.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WX and MX designed the study. WZ and HZ collected
the patient data and samples. QW, WZ and HZ performed cell
biological experiments. QW, WX and MX performed qPCR, ELISA and
western blot. WX and MX performed the animal experiments. All
authors contributed to writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the Animal Care
and Use Committee of Hefei Affiliated Hospital of Anhui Medical
University in compliance with the National Institutes of Health
Guide for the Care and Use of Laboratory Animals. Experiments using
human tissue were approved by the Ethics Committee of Hefei
Affiliated Hospital of Anhui Medical University. All participants
provided written informed consent.
Patient consent for publication
All participants provided written informed
consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Maoz R, Garfinkel BP and Soreq H:
Alzheimer's disease and ncRNAs. Adv Exp Med Biol. 978:337–361.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ritchie C, Smailagic N, Noel-Storr AH,
Ukoumunne O, Ladds EC and Martin S: CSF tau and the CSF tau/ABeta
ratio for the diagnosis of Alzheimer's disease dementia and other
dementias in people with mild cognitive impairment (MCI). Cochrane
Database Syst Rev. 3:D108032017.
|
|
3
|
Awan HM, Shah A, Rashid F and Shan G:
Primate-specific long Non-coding RNAs and MicroRNAs. Genomics
Proteomics Bioinformatics. 15:187–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Long JM, Ray B and Lahiri DK:
MicroRNA-339-5p down-regulates protein expression of β-site amyloid
precursor protein-cleaving enzyme 1 (BACE1) in human primary brain
cultures and is reduced in brain tissue specimens of Alzheimer
disease subjects. J Biol Chem. 289:5184–5198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu P, Zuo X, Deng H, Liu X, Liu L and Ji
A: Roles of long noncoding RNAs in brain development, functional
diversification and neurodegenerative diseases. Brain Res Bull.
97:69–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang M, Zhang P, Zhao Y and Liu X:
Bioinformatics and co-expression network analysis of differentially
expressed lncRNAs and mRNAs in hippocampus of APP/PS1 transgenic
mice with Alzheimer disease. Am J Transl Res. 9:1381–1391.
2017.PubMed/NCBI
|
|
7
|
Magistri M, Velmeshev D, Makhmutova M and
Faghihi MA: Transcriptomics profiling of Alzheimer's disease reveal
neurovascular defects, altered amyloid-β homeostasis, and
deregulated expression of long noncoding RNAs. J Alzheimers Dis.
48:647–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou X and Xu J: Identification of
Alzheimer's disease-associated long noncoding RNAs. Neurobiol
Aging. 36:2925–2931. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Massone S, Vassallo I, Fiorino G,
Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E,
Russo C, et al: 17A, a novel non-coding RNA, regulates GABA B
alternative splicing and signaling in response to inflammatory
stimuli and in Alzheimer disease. Neurobiol Dis. 41:308–317. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu T, Huang Y, Chen J, Chi H, Yu Z, Wang
J and Chen C: Attenuated ability of BACE1 to cleave the amyloid
precursor protein via silencing long noncoding RNA BACE1-AS
expression. Mol Med Rep. 10:1275–1281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Butterfield DA and Poon HF: The
senescence-accelerated prone mouse (SAMP8): A model of age-related
cognitive decline with relevance to alterations of the gene
expression and protein abnormalities in Alzheimer's disease. Exp
Gerontol. 40:774–783. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo YW, Xu Y, Cao WY, Zhong XL, Duan J,
Wang XQ, Hu ZL, Li F, Zhang JY, Zhou M, et al: Insulin-like growth
factor 2 mitigates depressive behavior in a rat model of chronic
stress. Neuropharmacology. 89:318–324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ye S, Wang TT, Cai B, Wang Y, Li J, Zhan
JX and Shen GM: Genistein protects hippocampal neurons against
injury by regulating calcium/calmodulin dependent protein kinase IV
protein levels in Alzheimer's disease model rats. Neural Regen Res.
12:1479–1484. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shin MK, Kim HG and Kim KL: A novel
trimeric peptide, Neuropep-1-stimulating brain-derived neurotrophic
factor expression in rat brain improves spatial learning and memory
as measured by the Y-maze and Morris water maze. J Neurochem.
116:205–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dellu F, Mayo W, Cherkaoui J, Le Moal M
and Simon H: A two-trial memory task with automated recording:
Study in young and aged rats. Brain Res. 588:132–139. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wright RL and Conrad CD: Chronic stress
leaves novelty-seeking behavior intact while impairing spatial
recognition memory in the Y-maze. Stress. 8:151–154. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang N, Lu S, Liu XG, Zhu J, Wang YJ and
Liu RT: PLGA nanoparticles modified with a BBB-penetrating peptide
co-delivering Aβ generation inhibitor and curcumin attenuate memory
deficits and neuropathology in Alzheimer's disease mice.
Oncotarget. 8:81001–81013. 2017.PubMed/NCBI
|
|
20
|
Watts JC and Prusiner SB: β-Amyloid prions
and the pathobiology of Alzheimer's disease. Cold Spring Harb
Perspect Med. 8:pii: a023507. 2018. View Article : Google Scholar
|
|
21
|
Munro KM, Nash A, Pigoni M, Lichtenthaler
SF and Gunnersen JM: Functions of the Alzheimer's disease protease
BACE1 at the synapse in the central nervous system. J Mol Neurosci.
60:305–315. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moussa CE: Beta-secretase inhibitors in
phase I and phase II clinical trials for Alzheimer's disease.
Expert Opin Investig Drugs. 26:1131–1136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ohno M: Alzheimer's therapy targeting the
β-secretase enzyme BACE1: Benefits and potential limitations from
the perspective of animal model studies. Brain Res Bull.
126:183–198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nigam SM, Xu S, Ackermann F, Gregory JA,
Lundkvist J, Lendahl U and Brodin L: Endogenous APP accumulates in
synapses after BACE1 inhibition. Neurosci Res. 109:9–15. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kandalepas PC and Vassar R: The normal and
pathologic roles of the Alzheimer's β-secretase, BACE1. Curr
Alzheimer Res. 11:441–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren RJ, Zhang YF, Dammer EB, Zhou Y, Wang
LL, Liu XH, Feng BL, Jiang GX, Chen SD, Wang G and Cheng Q:
Peripheral blood MicroRNA expression profiles in Alzheimer's
disease: Screening, validation, association with clinical phenotype
and implications for molecular mechanism. Mol Neurobiol.
53:5772–5781. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim J, Yoon H, Chung DE, Brown JL,
Belmonte KC and Kim J: imR-186 is decreased in aged brain and
suppresses BACE1 expression. J Neurochem. 137:436–445. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun LH, Ban T, Liu CD, Chen QX, Wang X,
Yan ML, Hu XL, Su XL, Bao YN, Sun LL, et al: Activation of Cdk5/p25
and tau phosphorylation following chronic brain hypoperfusion in
rats involves microRNA-195 down-regulation. J Neurochem.
134:1139–1151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo Q and Chen Y: Long noncoding RNAs and
Alzheimer's disease. Clin Interv Aging. 11:867–872. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Massone S, Ciarlo E, Vella S, Nizzari M,
Florio T, Russo C, Cancedda R and Pagano A: NDM29, a RNA polymerase
III-dependent non coding RNA, promotes amyloidogenic processing of
APP and amyloid β secretion. Biochim Biophys Acta. 1823:1170–1177.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang MJ, Abdelmohsen K, Hutchison ER,
Mitchell SJ, Grammatikakis I, Guo R, Noh JH, Martindale JL, Yang X,
Lee EK, et al: HuD regulates coding and noncoding RNA to induce
APP→Aβ processing. Cell Rep. 7:1401–1409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Greco S, Zaccagnini G, Fuschi P,
Voellenkle C, Carrara M, Sadeghi I, Bearzi C, Maimone B,
Castelvecchio S, Stellos K, et al: Increased BACE1-AS long
noncoding RNA and β-amyloid levels in heart failure. Cardiovasc
Res. 113:453–463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh
HJ, Lee JH, Nam SW and Lee EK: A long non-coding RNA snaR
contributes to 5-fluorouracil resistance in human colon cancer
cells. Mol Cells. 37:540–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Q, Liu X, Xu L, Wang Y, Wang S, Li Q,
Huang Y and Liu T: Long non-coding RNA BACE1-AS is a novel target
for anisomycin-mediated suppression of ovarian cancer stem cell
proliferation and invasion. Oncol Rep. 35:1916–1924. 2016.
View Article : Google Scholar : PubMed/NCBI
|