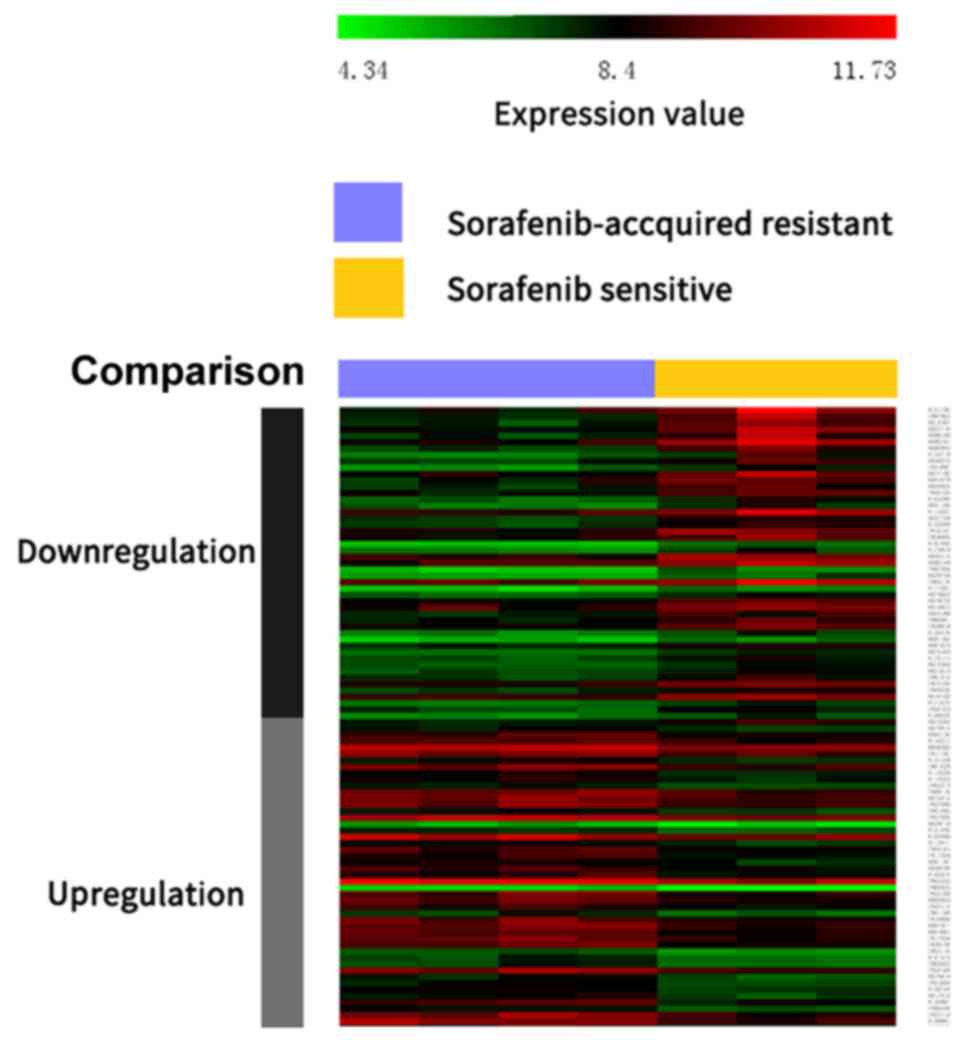
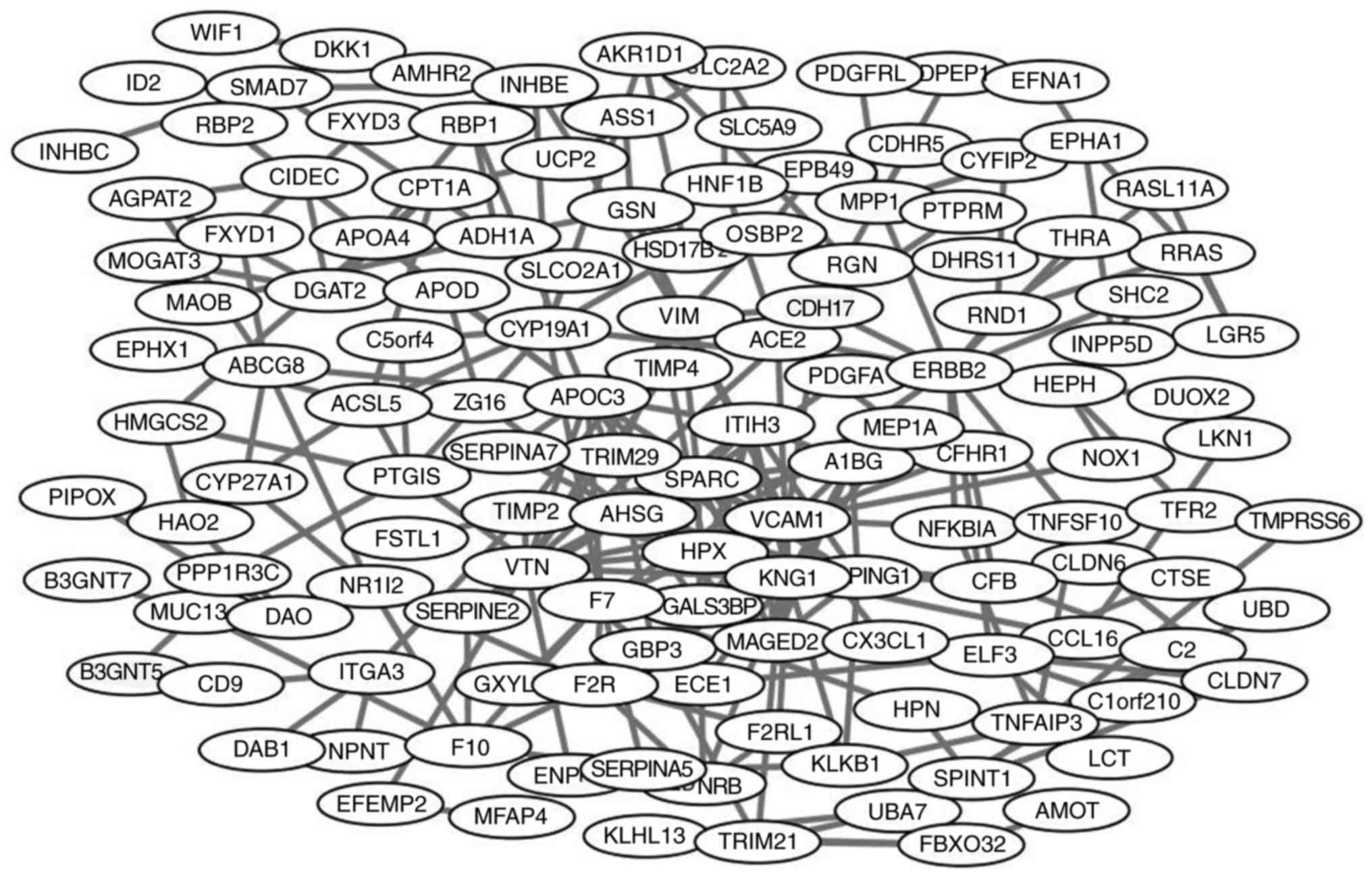
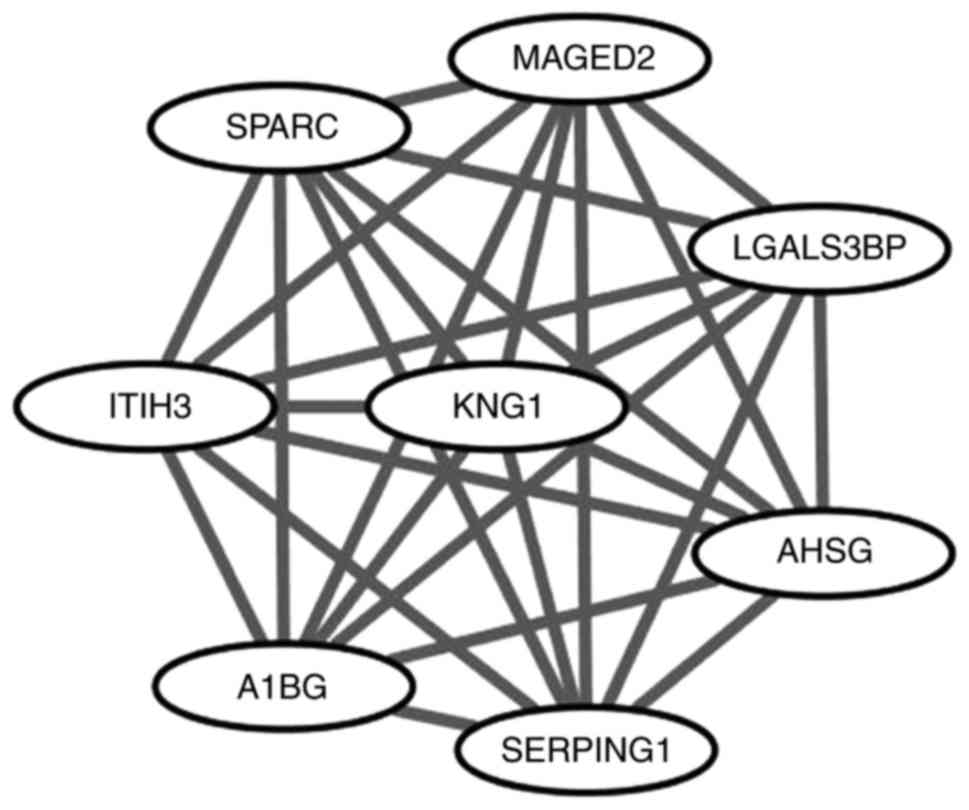
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Le Grazie M, Biagini MR, Tarocchi M,
Polvani S and Galli A: Chemotherapy for hepatocellular carcinoma:
The present and the future. World J Hepatol. 9:907–920. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tovar V, Cornella H, Moeini A, Vidal S,
Hoshida Y, Sia D, Peix J, Cabellos L, Alsinet C, Torrecilla S, et
al: Tumour initiating cells and IGF/FGF signalling contribute to
sorafenib resistance in hepatocellular carcinoma. Gut. 66:530–540.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J, Takayama T, Mazzaferro V, Chau
GY, Yang J, Kudo M, Cai J, Poon RT, Han KH, Tak WY, et al: Adjuvant
sorafenib for hepatocellular carcinoma after resection or ablation
(STORM): A phase 3, randomised, double-blind, placebo-controlled
trial. Lancet Oncol. 16:1344–1354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lencioni R, Llovet JM, Han G, Tak WY, Yang
J, Guglielmi A, Paik SW, Reig M, Kim DY, Chau GY, et al: Sorafenib
or placebo plus TACE with doxorubicin-eluting beads for
intermediate stage HCC: The SPACE trial. J Hepatol. 64:1090–1098.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen J, Jin R, Zhao J, Liu J, Ying H, Yan
H, Zhou S, Liang Y, Huang D, Liang X, et al: Potential molecular,
cellular and microenvironmental mechanism of sorafenib resistance
in hepatocellular carcinoma. Cancer Lett. 367:1–11. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nannini M, Pantaleo MA, Maleddu A, Astolfi
A, Formica S and Biasco G: Gene expression profiling in colorectal
cancer using microarray technologies: Results and perspectives.
Cancer Treat Rev. 35:201–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mok SC, Bonome T, Vathipadiekal V, Bell A,
Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al: A
gene signature predictive for outcome in advanced ovarian cancer
identifies a survival factor: Microfibril-associated glycoprotein
2. Cancer Cell. 16:521–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kadioglu O, Saeed M, Kuete V, Greten HJ
and Efferth T: Oridonin targets multiple drug-resistant tumor cells
as determined by in silico and in vitro analyses. Front Pharmacol.
9:3552018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Wang G, Li N, Yu H, Si J and Wang J:
Identification of key genes and pathways associated with obesity in
children. Exp Ther Med. 14:1065–1073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J and
Zhao M: Identification of candidate biomarkers and analysis of
prognostic values in ovarian cancer by integrated bioinformatics
analysis. Med Oncol. 33:1302016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang B, Li C and Zhao J: Identification
of key pathways and genes in colorectal cancer using bioinformatics
analysis. Med Oncol. 33:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carl-McGrath S, Lendeckel U, Ebert M and
Röcken C: Ectopeptidases in tumor biology: A review. Histol
Histopathol. 21:1339–1353. 2006.PubMed/NCBI
|
|
15
|
Cordero OJ, Ayude D, Nogueira M,
Rodriguez-Berrocal FJ and de la Cadena MP: Preoperative serum CD26
levels: Diagnostic efficiency and predictive value for colorectal
cancer. Br J Cancer. 83:1139–1146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ikeda N, Nakajima Y, Tokuhara T, Hattori
N, Sho M, Kanehiro H and Miyake M: Clinical significance of
aminopeptidase N/CD13 expression in human pancreatic carcinoma.
Clin Cancer Res. 9:1503–1508. 2003.PubMed/NCBI
|
|
17
|
Murakami H, Yokoyama A, Kondo K, Nakanishi
S, Kohno N and Miyake M: Circulating aminopeptidase N/CD13 is an
independent prognostic factor in patients with non-small cell lung
cancer. Clin Cancer Res. 11:8674–8679. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Larrinaga G, Blanco L, Sanz B, Perez I,
Gil J, Unda M, Andrés L, Casis L and López JI: The impact of
peptidase activity on clear cell renal cell carcinoma survival. Am
J Physiol Renal Physiol. 303:F1584–F1591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McDonnell DP, Park S, Goulet MT, Jasper J,
Wardell SE, Chang CY, Norris JD, Guyton JR and Nelson ER: Obesity,
cholesterol metabolism, and breast cancer pathogenesis. Cancer Res.
74:4976–4982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin CY, Huo C, Kuo LK, Hiipakka RA, Jones
RB, Lin HP, Hung Y, Su LC, Tseng JC, Kuo YY, et al: Cholestane-3β,
5α, 6β-triol suppresses proliferation, migration, and invasion of
human prostate cancer cells. PLoS One. 8:e657342013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Murtola TJ, Visvanathan K, Artama M,
Vainio H and Pukkala E: Statin use and breast cancer survival: A
nationwide cohort study from finland. PLoS One. 9:e1102312014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cardwell CR, Hicks BM, Hughes C and Murray
LJ: Statin use after colorectal cancer diagnosis and survival: A
population-based cohort study. J Clin Oncol. 32:3177–3183. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jacobs EJ, Newton CC, Thun MJ and Gapstur
SM: Long-term use of cholesterol-lowering drugs and cancer
incidence in a large United States cohort. Cancer Res.
71:1763–1771. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhuo H, Lyu Z, Su J, He J, Pei Y, Cheng X,
Zhou N, Lu X, Zhou S and Zhao Y: Effect of lung squamous cell
carcinoma tumor microenvironment on the CD105+ endothelial cell
proteome. J Proteome Res. 13:4717–4729. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han B, Nakamura M, Mori I, Nakamura Y and
Kakudo K: Urokinase-type plasminogen activator system and breast
cancer (Review). Oncol Rep. 14:105–112. 2005.PubMed/NCBI
|
|
26
|
Tsunoda T, Nakamura T, Ishimoto K, Yamaue
H, Tanimura H, Saijo N and Nishio K: Upregulated expression of
angiogenesis genes and down regulation of cell cycle genes in human
colorectal cancer tissue determined by cDNA macroarray. Anticancer
Res. 21:137–143. 2001.PubMed/NCBI
|
|
27
|
Romero-Cordoba SL, Salido-Guadarrama I,
Rodriguez-Dorantes M and Hidalgo-Miranda A: miRNA biogenesis:
Biological impact in the development of cancer. Cancer Biol Ther.
15:1444–1455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. J Gastrointest Surg. 7:884–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tominaga O, Nita ME, Nagawa H, Fujii S,
Tsuruo T and Muto T: Expressions of cell cycle regulators in human
colorectal cancer cell lines. Jpn J Cancer Res. 88:855–860. 2003.
View Article : Google Scholar
|
|
30
|
van den Berg YW, Osanto S, Reitsma PH and
Versteeg HH: The relationship between tissue factor and cancer
progression: Insights from bench and bedside. Blood. 119:924–932.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pesson M, Volant A, Uguen A, Trillet K, De
La Grange P, Aubry M, Daoulas M, Robaszkiewicz M, Le Gac G, Morel
A, et al: A gene expression and pre-mRNA splicing signature that
marks the adenoma-adenocarcinoma progression in colorectal cancer.
PLoS One. 9:e877612014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsai TH, Song E, Zhu R, Di Poto C, Wang M,
Luo Y, Varghese RS, Tadesse MG, Ziada DH, Desai CS, et al:
LC-MS/MS-based serum proteomics for identification of candidate
biomarkers for hepatocellular carcinoma. Proteomics. 15:2369–2381.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Turati F, Talamini R, Pelucchi C, Polesel
J, Franceschi S, Crispo A, Izzo F, La Vecchia C, Boffetta P and
Montella M: Metabolic syndrome and hepatocellular carcinoma risk.
Br J Cancer. 108:222–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baffy G, Brunt EM and Caldwell SH:
Hepatocellular carcinoma in non-alcoholic fatty liver disease: An
emerging menace. J Hepatol. 56:1384–1391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boroughs LK and DeBerardinis RJ: Metabolic
pathways promoting cancer cell survival and growth. Nat Cell Biol.
17:351–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang W, Huang H, Ding L, Zhu P, Saiyin H,
Ji G, Zuo J, Han D, Pan Y, Ding D, et al: Regulation of cell cycle
of hepatocellular carcinoma by NF90 through modulation of cyclin E1
mRNA stability. Oncogene. 34:4460–4470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Clawson GA, Feldherr CM and Smuckler EA:
Nucleocytoplasmic RNA transport. Mol Cell Biochem. 67:87–99. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Quesada-Calvo F, Massot C, Bertrand V,
Longuespée R, Blétard N, Somja J, Mazzucchelli G, Smargiasso N,
Baiwir D, De Pauw-Gillet MC, et al: OLFM4, KNG1 and Sec24C
identified by proteomics and immunohistochemistry as potential
markers of early colorectal cancer stages. Clin Proteomics.
14:92017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Wang X, Lin S, Chen C, Wang C, Ma
Q and Jiang B: Identification of kininogen-1 as a serum biomarker
for the early detection of advanced colorectal adenoma and
colorectal cancer. PLoS One. 8:e705192013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wahab Abdel AHA, El-Halawany MS, Emam AA,
Elfiky A and Elmageed Abd ZY: Identification of circulating protein
biomarkers in patients with hepatocellular carcinoma concomitantly
infected with chronic hepatitis C virus. Biomarkers. 22:621–628.
2017.PubMed/NCBI
|
|
41
|
He X, Wang Y, Zhang W, Li H, Luo R, Zhou
Y, Liao CL, Huang H, Lv X, Xie Z and He M: Screening differential
expression of serum proteins in AFP-negative HBV-related
hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. Neoplasma.
61:17–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang C, Liu CM, Wei LL, Shi LY, Pan ZF,
Mao LG, Wan XC, Ping ZP, Jiang TT, Chen ZL, et al: A group of novel
serum diagnostic biomarkers for multidrug-resistant tuberculosis by
iTRAQ-2D LC-MS/MS and solexa sequencing. Int J Biol Sci.
12:246–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Koch AE, Halloran MM, Haskell CJ, Shah MR
and Polverini PJ: Angiogenesis mediated by soluble forms of
E-selectin and vascular cell adhesion molecule-1. Nature.
376:517–519. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Byrne GJ, Ghellal A, Iddon J, Blann AD,
Venizelos V, Kumar S, Howell A and Bundred NJ: Serum soluble
vascular cell adhesion molecule-1: Role as a surrogate marker of
angiogenesis. J Natl Cancer Inst. 92:1329–1336. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yoong KF, McNab G, Hubscher SG and Adams
DH: Vascular adhesion protein-1 and ICAM-1 support the adhesion of
tumor-infiltrating lymphocytes to tumor endothelium in human
hepatocellular carcinoma. J Immunol. 160:3978–3988. 1998.PubMed/NCBI
|
|
46
|
Alexiou D, Karayiannakis AJ, Syrigos KN,
Zbar A, Kremmyda A, Bramis I and Tsigris C: Serum levels of
E-selectin, ICAM-1 and VCAM-1 in colorectal cancer patients:
Correlations with clinicopathological features, patient survival
and tumour surgery. Eur J Cancer. 37:2392–2397. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kang X, Wang F, Xie JD, Cao J and Xian PZ:
Clinical evaluation of serum concentrations of intercellular
adhesion molecule-1 in patients with colorectal cancer. World J
Gastroenterol. 11:4250–4253. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
O'Hanlon DM, Fitzsimons H, Lynch J, Tormey
S, Malone C and Given HF: Soluble adhesion molecules (E-selectin,
ICAM-1 and VCAM-1) in breast carcinoma. Eur J Cancer. 38:2252–2257.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ho JW, Poon RT, Tong CS and Fan ST:
Clinical significance of serum vascular cell adhesion molecule-1
levels in patients with hepatocellular carcinoma. World J
Gastroenterol. 10:2014–2018. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iliaz R, Akyuz U, Tekin D, Serilmez M,
Evirgen S, Cavus B, Soydinc H, Duranyildiz D, Karaca C, Demir K, et
al: Role of several cytokines and adhesion molecules in the
diagnosis and prediction of survival of hepatocellular carcinoma.
Arab J Gastroenterol. 17:164–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Diaz-Sanchez A, Matilla A, Nunez O, Rincon
D, Lorente R, Iacono Lo O, Merino B, Hernando A, Campos R, Clemente
G and Bañares R: Serum level of soluble vascular cell adhesion
molecule in patients with hepatocellular carcinoma and its
association with severity of liver disease. Ann Hepatol.
12:236–247. 2013.PubMed/NCBI
|
|
52
|
Scalici JM, Thomas S, Harrer C, Raines TA,
Curran J, Atkins KA, Conaway MR, Duska L, Kelly KA and Slack-Davis
JK: Imaging VCAM-1 as an indicator of treatment efficacy in
metastatic ovarian cancer. J Nucl Med. 54:1883–1889. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ito Y, Azrolan N, O'Connell A, Walsh A and
Breslow JL: Hypertriglyceridemia as a result of human apo CIII gene
expression in transgenic mice. Science. 249:790–793. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chan AW, Wong GL, Chan HY, Tong JH, Yu YH,
Choi PC, Chan HL, To KF and Wong VW: Concurrent fatty liver
increases risk of hepatocellular carcinoma among patients with
chronic hepatitis B. J Gastroenterol Hepatol. 32:667–676. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL,
Hu DF, Luo SY, Huang AC, Li LL and Li GJ: Comparative proteomic
profiles indicating genetic factors may involve in hepatocellular
carcinoma familial aggregation. Cancer Sci. 103:1833–1838. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haukeland JW, Dahl TB, Yndestad A,
Gladhaug IP, Løberg EM, Haaland T, Konopski Z, Wium C, Aasheim ET,
Johansen OE, et al: Fetuin A in nonalcoholic fatty liver disease:
In vivo and in vitro studies. Eur J Endocrinol. 166:503–510. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yilmaz Y, Yonal O, Kurt R, Ari F, Oral AY,
Celikel CA, Korkmaz S, Ulukaya E, Ozdogan O, Imeryuz N, et al:
Serum fetuin A/α2HS-glycoprotein levels in patients with
non-alcoholic fatty liver disease: Relation with liver fibrosis.
Ann Clin Biochem. 47:549–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao ZW, Lin CG, Wu LZ, Luo YK, Fan L,
Dong XF and Zheng H: Serum fetuin-A levels are associated with the
presence and severity of coronary artery disease in patients with
type 2 diabetes. Biomarkers. 18:160–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kaushik SV, Plaisance EP, Kim T, Huang EY,
Mahurin AJ, Grandjean PW and Mathews ST: Extended-release niacin
decreases serum fetuin-A concentrations in individuals with
metabolic syndrome. Diabetes Metab Res Rev. 25:427–434. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kalabay L, Jakab L, Prohaszka Z, Füst G,
Benkö Z, Telegdy L, Lörincz Z, Závodszky P, Arnaud P and Fekete B:
Human fetuin/alpha2HS-glycoprotein level as a novel indicator of
liver cell function and short-term mortality in patients with liver
cirrhosis and liver cancer. Eur J Gastroenterol Hepatol.
14:389–394. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Guillory B, Sakwe AM, Saria M, Thompson P,
Adhiambo C, Koumangoye R, Ballard B, Binhazim A, Cone C,
Jahanen-Dechent W and Ochieng J: Lack of fetuin-A
(alpha2-HS-glycoprotein) reduces mammary tumor incidence and
prolongs tumor latency via the transforming growth factor-beta
signaling pathway in a mouse model of breast cancer. Am J Pathol.
177:2635–2644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Swallow CJ, Partridge EA, Macmillan JC,
Tajirian T, DiGuglielmo GM, Hay K, Szweras M, Jahnen-Dechent W,
Wrana JL, Redston M, et al: alpha2HS-glycoprotein, an antagonist of
transforming growth factor beta in vivo, inhibits intestinal tumor
progression. Cancer Res. 64:6402–6409. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xiang Y, Liu Y, Yang Y, Hu H, Hu P, Ren H
and Zhang D: A secretomic study on human hepatocellular carcinoma
multiple drug-resistant cell lines. Oncol Rep. 34:1249–1260. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hyung SW, Lee MY, Yu JH, Shin B, Jung HJ,
Park JM, Han W, Lee KM, Moon HG, Zhang H, et al: A serum protein
profile predictive of the resistance to neoadjuvant chemotherapy in
advanced breast cancers. Mol Cell Proteomics. 10(M111):
0110232011.PubMed/NCBI
|
|
65
|
Kirschbaum MH and Yarden Y: The ErbB/HER
family of receptor tyrosine kinases: A potential target for
chemoprevention of epithelial neoplasms. J Cell Biochem Suppl.
34:52–60. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Slamon DJ, Godolphin W, Jones LA, Holt JA,
Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J and Ullrich A:
Studies of the HER-2/neu proto-oncogene in human breast and ovarian
cancer. Science. 244:707–712. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Press MF, Pike MC, Chazin VR, Hung G,
Udove JA, Markowicz M, Danyluk J, Godolphin W, Sliwkowski M and
Akita R: Her-2/neu expression in node-negative breast cancer:
Direct tissue quantitation by computerized image analysis and
association of overexpression with increased risk of recurrent
disease. Cancer Res. 53:4960–4970. 1993.PubMed/NCBI
|
|
68
|
Colak D, Chishti MA, Al-Bakheet AB,
Al-Qahtani A, Shoukri MM, Goyns MH, Ozand PT, Quackenbush J, Park
BH and Kaya N: Integrative and comparative genomics analysis of
early hepatocellular carcinoma differentiated from liver
regeneration in young and old. Mol Cancer. 9:1462010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Nakopoulou L, Stefanaki K, Filaktopoulos D
and Giannopoulou I: C-erb-B-2 oncoprotein and epidermal growth
factor receptor in human hepatocellular carcinoma: An
immunohistochemical study. Histol Histopathol. 9:677–682.
1994.PubMed/NCBI
|
|
70
|
Hamazaki K, Yunoki Y, Tagashira H, Mimura
T, Mori M and Orita K: Epidermal growth factor receptor in human
hepatocellular carcinoma. Cancer Detect Prev. 21:355–360.
1997.PubMed/NCBI
|
|
71
|
Prange W and Schirmacher P: Absence of
therapeutically relevant c-erbB-2 expression in human
hepatocellular carcinomas. Oncol Rep. 8:727–730. 2001.PubMed/NCBI
|
|
72
|
Altimari A, Fiorentino M, Gabusi E,
Gruppioni E, Corti B, D'Errico A and Grigioni WF: Investigation of
ErbB1 and ErbB2 expression for therapeutic targeting in primary
liver tumours. Dig Liver Dis. 35:332–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ito Y, Takeda T, Sakon M, Tsujimoto M,
Higashiyama S, Noda K, Miyoshi E, Monden M and Matsuura N:
Expression and clinical significance of erb-B receptor family in
hepatocellular carcinoma. Br J Cancer. 84:1377–1383. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Misale S, Di Nicolantonio F,
Sartore-Bianchi A, Siena S and Bardelli A: Resistance to anti-EGFR
therapy in colorectal cancer: From heterogeneity to convergent
evolution. Cancer Discov. 4:1269–1280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Vaz J, Ansari D, Sasor A and Andersson R:
SPARC: A potential prognostic and therapeutic target in pancreatic
cancer. Pancreas. 44:1024–1035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Nagaraju GP, Dontula R, El-Rayes BF and
Lakka SS: Molecular mechanisms underlying the divergent roles of
SPARC in human carcinogenesis. Carcinogenesis. 35:967–973. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lau CP, Poon RT, Cheung ST, Yu WC and Fan
ST: SPARC and Hevin expression correlate with tumour angiogenesis
in hepatocellular carcinoma. J Pathol. 210:459–468. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Massi D, Franchi A, Borgognoni L, Reali UM
and Santucci M: Osteonectin expression correlates with clinical
outcome in thin cutaneous malignant melanomas. Hum Pathol.
30:339–344. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Golembieski WA, Ge S, Nelson K, Mikkelsen
T and Rempel SA: Increased SPARC expression promotes U87
glioblastoma invasion in vitro. Int J Dev Neurosci. 17:463–472.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Thomas R, True LD, Bassuk JA, Lange PH and
Vessella RL: Differential expression of osteonectin/SPARC during
human prostate cancer progression. Clin Cancer Res. 6:1140–1149.
2000.PubMed/NCBI
|
|
81
|
Wang T, Srivastava S, Hartman M, Buhari
SA, Chan CW, Iau P, Khin LW, Wong A, Tan SH, Goh BC and Lee SC:
High expression of intratumoral stromal proteins is associated with
chemotherapy resistance in breast cancer. Oncotarget.
7:55155–55168. 2016.PubMed/NCBI
|
|
82
|
Seiffert D, Geisterfer M, Gauldie J, Young
E and Podor TJ: IL-6 stimulates vitronectin gene expression in
vivo. J Immunol. 155:3180–3185. 1995.PubMed/NCBI
|
|
83
|
Edwards S, Lalor PF, Tuncer C and Adams
DH: Vitronectin in human hepatic tumours contributes to the
recruitment of lymphocytes in an alpha v beta3-independent manner.
Br J Cancer. 95:1545–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Nejjari M, Hafdi Z, Gouyss G, Fiorentino
M, Béatrix O, Dumortier J, Pourreyron C, Barozzi C, D'errico A,
Grigioni WF and Scoazec JY: Expression, regulation, and function of
alpha V integrins in hepatocellular carcinoma: An in vivo and in
vitro study. Hepatology. 36:418–426. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ferrin G, Ranchal I, Llamoza C,
Rodríguez-Perálvarez ML, Romero-Ruiz A, Aguilar-Melero P,
López-Cillero P, Briceño J, Muntané J, Montero-Álvarez JL and De la
Mata M: Identification of candidate biomarkers for hepatocellular
carcinoma in plasma of HCV-infected cirrhotic patients by 2-D DIGE.
Liver Int. 34:438–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ding Y and Shen Y: Notch increased
vitronection adhesion protects myeloma cells from drug induced
apoptosis. Biochem Biophys Res Commun. 467:717–722. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sun S, Poon RT, Lee NP, Yeung C, Chan KL,
Ng IO, Day PJ and Luk JM: Proteomics of hepatocellular carcinoma:
Serum vimentin as a surrogate marker for small tumors (<or=2
cm). J Proteome Res. 9:1923–1930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lazarova DL and Bordonaro M: Vimentin,
colon cancer progression and resistance to butyrate and other
HDACis. J Cell Mol Med. 20:989–993. 2016. View Article : Google Scholar : PubMed/NCBI
|