Introduction
Osteoarthritis (OA) is a chronic disease
characterized by progressive degradation and loss of articular
cartilage. Although the disease present in various joints, the
weight-bearing joints such as knees and hips are commonly affected.
Furthermore, alterations in other joint tissues such as ligaments,
synovium and subchondral bone are usually noted in OA (1,2).
Generally, the symptoms develop gradually over time and the loss of
cartilage can lead to decreased joint space, stiffness, pain with
increasing loss of function and disability, and finally to the need
for surgical joint replacement (3).
Nowadays, OA is not only viewed as a degenerative disease of
cartilage and different risk factors were associated with OA,
including genetic predisposition, sex, obesity, age, diet,
occupation, metabolic syndromes, injury and mechanical stress
(1,4,5).
Accumulating evidence indicates that inflammation has a critical
role in OA pathogenesis (6), and
recent findings show that the development of OA is in notably
driven by low-grade inflammatory processes and mediated mainly by
the innate immune system (7). The
inflammatory response was identified as the key component, which
promoted synovitis as well as progression of cartilage and bone
destruction in OA via the secretion of chemokines, cytokines and
other molecules, which can be detected in the synovial fluids
(8–10). In addition to their evident role in
inflammation, cytokines can contribute to the pathogenesis of OA
through angiogenesis and chemotaxis (11,12). OA
can take years and even decades to develop and radiography is
routinely used to help in the diagnosis of OA. However, symptoms
often occur before the onset of any radiographic abnormality and
further work should be focused on the identification of new
biological markers as early indicators of OA risk.
Based on this rational, we have genotyped four
single nucleotide polymorphisms (SNPs) located in the promoter
region of the following cytokine genes: IL-4R −3223C>T
(rs2057768), IL-8 −251T>A (rs4073), IL-10 −1082A>G
(rs1800896) and TNF-A −308G>A (rs1800629). This study was
performed in a Romanian population (Eastern European population) in
order to establish whether these cytokine gene SNPs are associated
with OA susceptibility in this region.
Materials and methods
Subjects
In the current hospital-based case-control study we
included 305 Romanian subjects (90 patients diagnosed with OA and
215 controls). All OA cases were diagnosed based on clinical and
radiographic signs, arthroscopic or MRI findings with
Kellgren-Lawrence grade 2 or more. The patients with history of
trauma or skeletal defects were excluded. Matched controls of the
same ethnic and geographical origins were recruited among unrelated
volunteers. The controls were admitted in the Clinical Hospital CF2
from Bucharest and in the Emergency Clinical County Hospital of
Craiova, Romania. The subjects with a positive history of tumor,
autoimmune, other inflammatory or chronic infectious symptoms were
excluded. Written informed consent from all the participants was
obtained and the study was approved by the Ethics Committee of the
University of Medicine and Pharmacy of Craiova (Craiova,
Romania).
SNPs genotyping
Total genomic DNA was isolated from the peripheral
blood leukocytes using the Wizard Genomic DNA Purification kit
(Promega Corporation, Madison, WI, USA) following the
manufacturer's protocol. DNA concentration and purity were
determined with UV spectrophotometry. The identification of SNPs
was performed by TaqMan allelic discrimination real-time PCR.
Validated TaqMan SNP genotyping assays were obtained from Applied
Biosystems (Foster City, CA, USA): IL-4R −3223C>T (rs2057768,
assay C_2769607_10); IL-8 −251T>A (rs4073, assay C_11748116_10);
IL-10 −1082A>G (rs1800896, assay C_1747360_10) and TNF-A
−308G>A (rs1800629, assay C_7514879_10). The procedures for the
PCR reactions and quality control samples have been described
previously (13).
Statistical analysis
The deviation from Hardy-Weinberg equilibrium was
tested among controls by χ2 test. The demographic data
between groups were compared using the χ2 test for sex
and the Student's t-test for age. Using unconditional logistic
regression analysis under codominant and dominant models, we have
calculated the differences in genotype distributions and minor
allele frequencies among OA patients and controls based on Odds
ratios (OR) and 95% confidence intervals (CIs). The reference group
included the subjects that were homozygous for the most common
allele. A stratified analysis by joint location was further
performed aside from the overall association analysis. SPSS
software package (v.17.0; SPSS Inc., Chicago, IL, USA) was used for
data analysis and a P-value of less than 0.05 indicated a
statistically significant difference.
Results
Subjects characteristics
A total of 90 OA patients (63 women and 27 men, mean
age 64.12 years, with a body mass index ≤27) and 215 healthy
controls (150 women and 65 men, mean age 62.69 years) were included
in the present study. In 54 cases the location was within the knee
and in 36 cases within the hip joint. The age and sex distributions
of the groups were comparable (Table
I).
 | Table I.Baseline characteristics of the
patients and controls. |
Table I.
Baseline characteristics of the
patients and controls.
| Characteristic | Osteoarthritis cases
(n=90) | Control (n=215) | P-value |
|---|
| Male/female | 63/27 | 150/65 | 0.97 |
| Age (years) | 64.12 | 62.69 | 0.26 |
| Location |
|
|
|
| Knee | 54 | – | – |
| Hip | 36 | – | – |
Cytokine SNPs and risk of overall
OA
For each polymorphism, the genotype distributions
were consistent with those predicted by the Hardy-Weinberg
equilibrium. A significant association was observed for IL-4R
−3223C>T (rs2057768) SNP. The subjects carrying CT genotype were
at a higher risk for OA (OR 2.03; 95% CI: 1.21–3.42; P=0.007)
compared with the more frequently encountered CC genotype.
Furthermore, the carriers of T allele were at a 1.9 fold elevated
risk for OA in a dominant model (OR 1.92; 95% CI, 1.17–3.17;
P=0.009). The T allele was significantly more frequent in patients
with OA when the allele frequencies were assessed (OR 1.51; 95% CI,
1.03–2.21; P=0.035).
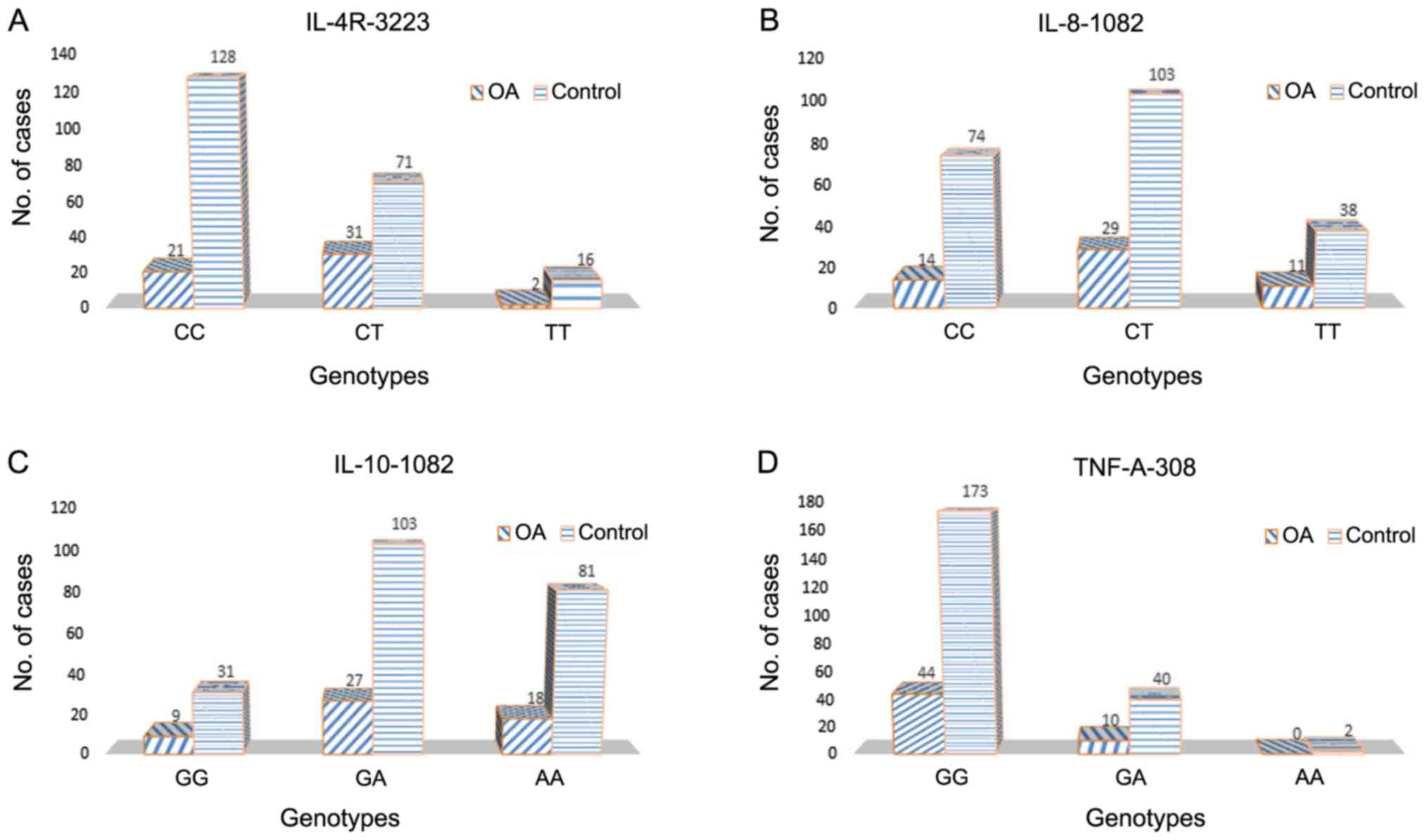
No correlation was noted between OA cases and
controls with regard to IL-8 −251AA, IL-10 −1082GG and TNF-A −308AG
genotypes and overall OA risk (Table
II). In addition, the carriers of IL-10 −1082 G, IL-8 −251A and
TNF-A −308A allele were not associated with an increased risk of OA
in a dominant model.
 | Table II.Genotype frequencies for cytokine
polymorphisms in cases and controls and their association with risk
of osteoarthritis. |
Table II.
Genotype frequencies for cytokine
polymorphisms in cases and controls and their association with risk
of osteoarthritis.
| Polymorphism | Osteoarthritis (n=90)
(%) | Control (n=215)
(%) | OR (95% CI) | P-value |
|---|
| IL-4R
−3223C>T |
|
|
|
|
| CC | 39 (43.33) | 128 (59.54) | Reference | – |
| CT | 44 (48.89) | 71 (33.02) | 2.03 (1.21–3.42) | 0.007 |
| TT | 7 (7.78) | 16 (7.44) | 1.44 (0.55–3.74) | 0.457 |
| T
carriers | 51 (56.67) | 87 (40.46) | 1.92 (1.17–3.17) | 0.009 |
| IL-8 −251T>A |
|
|
|
|
| TT | 22 (24.44) | 74 (34.42) | Reference | – |
| TA | 51 (56.67) | 103 (47.91) | 1.67 (0.93–2.98) | 0.084 |
| AA | 17 (18.89) | 38 (17.67) | 1.51 (0.72–3.17) | 0.280 |
| A
carriers | 68 (75.56) | 141 (65.58) | 1.62 (0.93–2.83) | 0.097 |
| IL-10 −1082
A>G |
|
|
|
|
| AA | 33 (36.67) | 81 (37.67) | Reference | – |
| AG | 43 (47.78) | 103 (47.91) | 1.02
(0.60–1.76) | 0.929 |
| GG | 14 (15.56) | 31 (14.42) | 1.11
(0.52–2.35) | 0.787 |
| G
carriers | 57 (63.33) | 134 (62.33) | 1.04
(0.63–1.74) | 0.868 |
| TNF-A −308
G>A |
|
|
|
|
| GG | 73 (81.11) | 173 (80.47) | Reference | – |
| GA | 17 (18.89) | 40 (18.60) | 1.07
(0.54–1.89) | 0.982 |
| AA | 0 (0.00) | 2 (0.93) | – | – |
| A
carriers | 17 (20.14) | 42 (19.53) | 0.96
(0.51–1.79) | 0.896 |
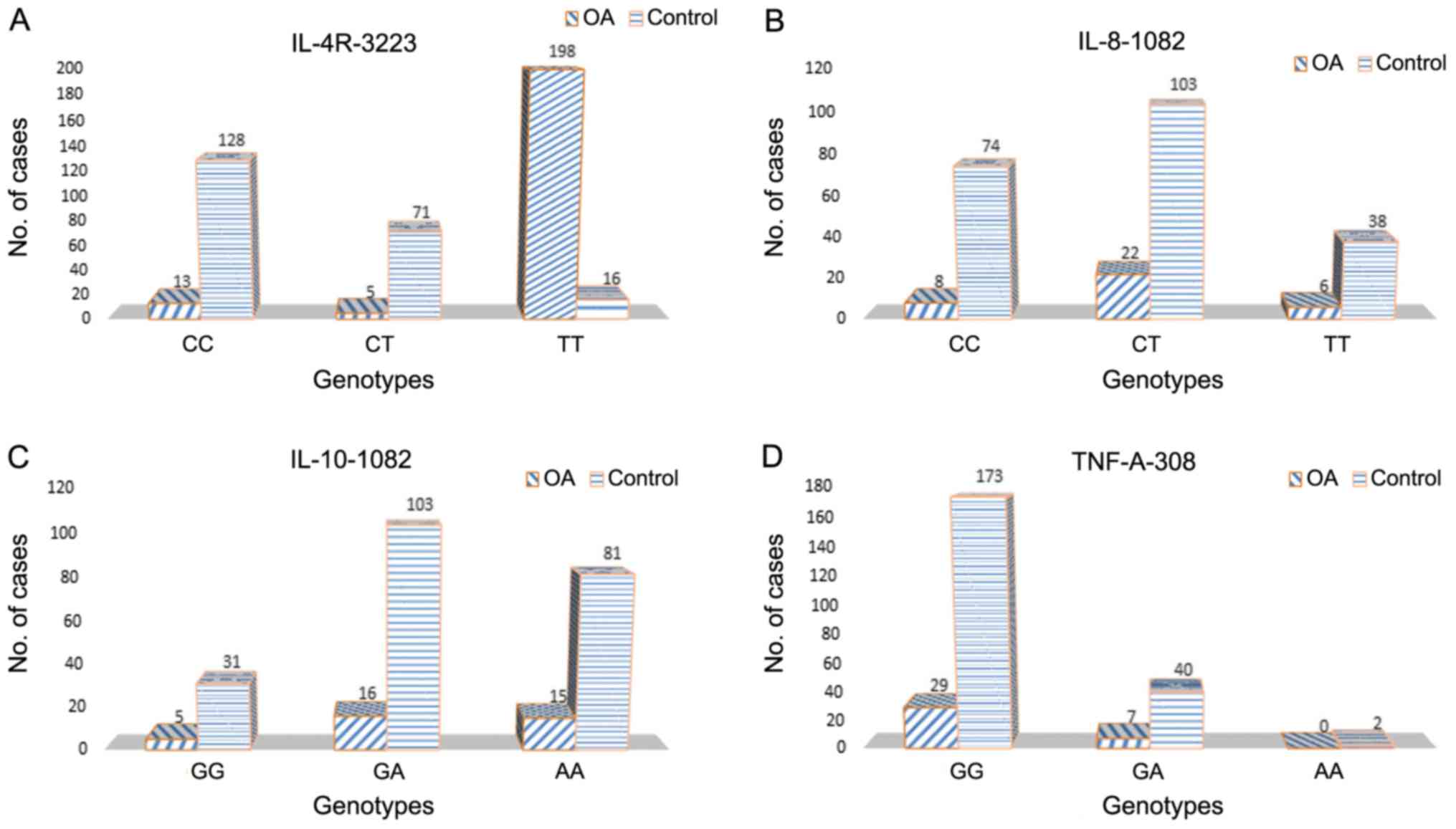
Risk of knee and hip OA by
genotype
In a stratified analysis, the only association
between OA and cytokine polymorphisms was found for IL-4R
−3223C>T polymorphism and was restricted to the knee OA cases.
The T carriers exhibited an increased risk (OR 2.31; 95% CI,
1.26–4.26) (Table III; Fig. 1). No significant differences were
observed between knee or hip OA and controls in the subgroups
analysis for the remaining SNPs (Table
III; Fig. 2).
 | Table III.Comparative analysis between genotype
frequencies and the risk of osteoarthritis in the joint location
subgroups. |
Table III.
Comparative analysis between genotype
frequencies and the risk of osteoarthritis in the joint location
subgroups.
| Polymorphism | Knee, n=54 (%) | OR (95% CI) | P-value | Hip, n=36 (%) | OR (95% CI) | P-value |
|---|
| IL-4R
−3223C>T |
|
|
|
|
|
|
| CC | 21 (38.89) | Reference |
| 18 (50.00) | Reference |
|
| CT | 31 (57.41) | 2.66
(1.42–4.97) | 0.002 | 13 (36.11) | 1.30
(0.60–2.81) | 0.501 |
| TT | 2 (3.70) | 0.76
(0.16–3.56) | 0.728 | 5 (13.89) | 2.22
(0.73–6.80) | 0.153 |
| T
carriers | 33 (61.11) | 2.31
(1.26–4.26) | 0.006 | 18 (50.00) | 1.47
(0.73–2.99) | 0.283 |
| IL-8
−251T>A |
|
|
|
|
|
|
| TT | 14 (25.93) | Reference |
| 8 (22.22) | Reference |
|
| TA | 29 (53.70) | 1.49
(0.74–3.01) | 0.267 | 22 (61.11) | 1.98
(0.83–4.68) | 0.117 |
| AA | 11 (20.37) | 1.53
(0.63–3.69) | 0.342 | 6 (16.67) | 1.46
(0.47–4.51) | 0.509 |
| A
carriers | 40 (74.07) | 1.50
(0.77–2.93) | 0.234 | 28 (77.78) | 1.84
(0.80–4.23) | 0.148 |
| IL-10
−1082A>G |
|
|
|
|
|
|
| AA | 18 (33.33) | Reference |
| 15 (41.67) | Reference |
|
| AG | 27 (50.00) | 1.18
(0.61–2.29) | 0.625 | 16 (44.44) | 0.84
(0.39–1.80) | 0.651 |
| GG | 9 (16.67) | 1.31
(0.53–3.22) | 0.560 | 5 (13.89) | 0.87
(0.29–2.60) | 0.804 |
| G
carriers | 36 (66.67) | 1.20
(0.64–2.27) | 0.554 | 21 (58.33) | 0.85
(0.41–1.73) | 0.648 |
| TNF A −308
G>A |
|
|
|
|
|
|
| GG | 44 (81.48) | Reference |
| 29 (80.56) | Reference |
|
| GA | 10 (18.52) | 0.98
(0.46–2.12) | 0.965 | 7 (19.44) | 1.04
(0.43–2.55) | 0.924 |
| AA | 0 (0.00) | – |
| 0 (0.00) | – |
|
| A
carriers | 10 (18.52) | 0.94
(0.44–2.01) | 0.865 | 7 (19.44) | 0.99
(0.41–2.42) | 0.989 |
Discussion
In the present observational study, we assessed
whether the four promoter SNPs located in the cytokine genes
influence the risk of OA in the Romanian population. The
polymorphisms were selected based on multiple previous reports that
have demonstrated quantitative differences in the transcription
and/or expression, and their involvement in other multifactorial
diseases, where the inflammatory process plays an important role in
pathogenesis.
From the tested SNPs, the only association was
detected for IL-4R rs2057768 C>T SNP. This gene encodes the
alpha chain of the interleukin-4 receptor that can bind the
anti-inflammatory cytokine IL-4. Previous research investigated the
association between IL-4R SNPs and OA susceptibility in different
populations, with inconsistent results. A significant association
was found for both rs1805013 and rs1805016 IL-4R SNPs and hip OA in
a case-control study including UK Caucasians subjects (14). In contrast to Forster et al
(14), another UK study was unable
to replicate the associations of these two SNPs with hip OA and/or
knee OA susceptibility (15).
Furthermore, Vargiolu et al (16) suggested a positive association in an
Italian population between both rs1805013 and rs1805015 located in
the IL-4R gene and overall risk for hand OA, while in the subgroups
analysis only the association between rs1805013 and non-erosive
hand OA was significant. In addition, the possible involvement of
the IL-4/IL-4R axis in the pathogenesis of OA was suggested by an
association of the IL-4 intron 3 VNTR polymorphism and the
incidence of knee OA in a Turkish population (17). No statistically significant
associations were noted between IL-4R SNPs (rs1805015 and
rs1805016) and hand OA in a Finnish study (18).
In the present study no correlation was noted
between the IL-8 −251T>A (rs4073), IL-10 −1082A>G (rs1800896)
and TNF-A −308G>A (rs1800629) SNPs and OA susceptibility. A
statistically significant higher frequency of IL-8 −251TT genotype
and IL-8 −251T allele was noted in patients diagnosed with OA
compared with controls in a Han Chinese population, suggesting that
the TT genotype and the T allele of the IL-8 gene at position-251
confer a high risk in OA. In the same study, the IL-8 SNP located
at position +781C>T influenced the risk of OA (19). Furthermore, IL-10 −1082A>G,
−819T>C and −592A>C gene promoter SNPs were tested in a
Chinese Han population and no significant difference was observed
in the allele and/or haplotype frequencies between end-stage knee
OA and the controls (20). Similar
results were obtained in a Dutch population, where no significant
association was detected between distal interphalangeal OA and
IL-10 SNPs, including IL-10 −1082A>G (21). Moreover, no significant difference in
the genotype distribution between OA individuals and controls was
observed for the IL-10 −1082A>G gene and the hand OA among
Finnish women (18). A positive
correlation between IL-10 G microsatellite SNP and idiopathic knee
OA was noted in a Greek case-control study, and the carriers of the
LL genotype were at 4 times higher risk than the SS genotype
(22).
The association between TNF-A gene and OA
susceptibility has been widely studied, and the published results
remain inconclusive. The TNF-α gene encodes the important
pro-inflammatory cytokine TNF-α and different genetic variants
located in the promoter region have been associated with altered
gene expression, notably at the −308 position (23,24). No
association was observed in the present study between the TNF-α
−308G>A SNP and the risk of OA, which is consistent with the
results from the Turkish Caucasian and Mexican Mestizo population.
Both studies included only knee OA (25,26). On
the contrary, in the Han Chinese population, the TNF-α −308A allele
was found to increase the risk, whereas the TNF-α −238 SNP did not
play a role in OA patients (27).
Similar findings were reported in the other Asian population,
although the risk of OA was significantly higher for carriers of
the TNF-α −308A allele in a Korean Population (28). A recent meta-analysis, which included
983 OA cases and 1355 control subjects indicated a significant
association between TNF-α −308A allele and the risk of OA. An
increased OA risk was found in the recessive genetic model and
higher frequency of both AA genotype and GA genotype was observed
in OA patients compared with the control population. Interestingly,
the association only existed among the Asian population but not
among the Caucasian population (29).
The current study has some limitations. Firstly, our
findings rely on a small sample and the selection bias cannot be
ruled out in this hospital case-control study. OA susceptibility is
influenced by multiple interactions between different other genes,
which were not evaluated and additional data regarding the
biological effects of investigated SNPs are required. Moreover, a
subgroup analysis with regard to additional clinicopathological
parameters could not be performed for this population. In
conclusion, the IL-4R −3223C>T (rs2057768) polymorphism was
correlated with OA susceptibility, mainly for the knee OA, in the
Romanian population. The present results provide new evidence for
the involvement of the IL-4/IL-4R axis in the OA pathogenesis and
additional well-designed large studies are required before final
conclusions can be drawn.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
OCR, DC, MI and EB contributed to the conception of
the study. OCR, AOD and ES collected the clinical data and samples.
MGC, FB, SS and MI performed the experiments. MGC, FB, SS and MI
performed the data analyses. OCR, DC, AOD and EB contributed
significantly to manuscript preparation. OCR, DC, AOD, FB and EB
wrote the manuscript. MI, FB and EB helped perform the analysis
with constructive discussions. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of University of Medicine and Pharmacy of Craiova
approved this study and a written informed consent was provided by
the included participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abramson SB and Attur M: Developments in
the scientific understanding of osteoarthritis. Arthritis Res Ther.
11:2272009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Loeser RF, Goldring SR, Scanzello CR and
Goldring MB: Osteoarthritis: A disease of the joint as an organ.
Arthritis Rheum. 64:1697–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldring MB and Goldring SR:
Osteoarthritis. J Cell Physiol. 213:626–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Felson DT: An update on the pathogenesis
and epidemiology of osteoarthritis. Radiol Clin North Am. 42:1–9,
v. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y and Jordan JM: Epidemiology of
osteoarthritis. Clin Geriatr Med. 26:355–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brooks P: Inflammation as an important
feature of osteoarthritis. Bull World Health Organ. 81:689–690.
2003.PubMed/NCBI
|
|
7
|
Robinson WH, Lepus CM, Wang Q, Raghu H,
Mao R, Lindstrom TM and Sokolove J: Low-grade inflammation as a key
mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol.
12:580–592. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kapoor M, Martel-Pelletier J, Lajeunesse
D, Pelletier JP and Fahmi H: Role of proinflammatory cytokines in
the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 7:33–42.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malemud CJ: Biologic basis of
osteoarthritis: State of the evidence. Curr Opin Rheumatol.
27:289–294. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang F, Hu A, Zhao D, Guo L, Yang L, Wang
B, Tian F, Liu B, Huang S and Xie H: An insertion/deletion
polymorphism at the microRNA-122 binding site in the
interleukin-1alpha 3′-untranslated region is associated with a risk
for osteoarthritis. Mol Med Rep. 12:6199–6206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mapp PI and Walsh DA: Mechanisms and
targets of angiogenesis and nerve growth in osteoarthritis. Nat Rev
Rheumatol. 8:390–398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mabey T and Honsawek S: Cytokines as
biochemical markers for knee osteoarthritis. World J Orthop.
6:95–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Burada F, Dumitrescu T, Nicoli R, Ciurea
ME, Angelescu C, Mixich F and Ioana M: IL-1RN +2018T>C
polymorphism is correlated with colorectal cancer. Mol Biol Rep.
40:2851–2857. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Forster T, Chapman K and Loughlin J:
Common variants within the interleukin 4 receptor alpha gene (IL4R)
are associated with susceptibility to osteoarthritis. Hum Genet.
114:391–395. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Limer KL, Tosh K, Bujac SR, McConnell R,
Doherty S, Nyberg F, Zhang W, Doherty M, Muir KR and Maciewicz RA:
Attempt to replicate published genetic associations in a large,
well-defined osteoarthritis case-control population (the GOAL
study). Osteoarthritis Cartilage. 17:782–789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vargiolu M, Silvestri T, Bonora E, Dolzani
P, Pulsatelli L, Addimanda O, Mancarella L, Punzi L, Fioravanti A,
Facchini A, Romeo G, et al: Interleukin-4/interleukin-4 receptor
gene polymorphisms in hand osteoarthritis. Osteoarthritis
Cartilage. 18:810–816. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yigit S, Inanir A, Tekcan A, Tural E,
Ozturk GT, Kismali G and Karakus N: Significant association of
interleukin-4 gene intron 3 VNTR polymorphism with susceptibility
to knee osteoarthritis. Gene. 537:6–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hämäläinen S, Solovieva S, Vehmas T,
Leino-Arjas P and Hirvonen A: Variations in the TNFalpha gene and
their interactions with the IL4R and IL10 genes in relation to hand
osteoarthritis. BMC Musculoskelet Disord. 15:3112014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He Y, Liang X, Wu X, Meng C, Wu B, Fu D,
Jin S, Yang S and Wang H: Association between interleukin 8 −251
A/T and +781 C/T polymorphisms and osteoarthritis risk. Immunol
Lett. 162:207–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lv C, Xu X, Wang J, Zhang Z, Zhang D, Guo
C, Geng C and Sun Y: Combined effect of cytokine gene polymorphisms
on end-stage knee osteoarthritis from Chinese Han population.
Rheumatol Int. 32:3625–3629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Riyazi N, Kurreeman FA, Huizinga TW,
Dekker FW, Stoeken-Rijsbergen G and Kloppenburg M: The role of
interleukin 10 promoter polymorphisms in the susceptibility of
distal interphalangeal osteoarthritis. J Rheumatol. 32:1571–1575.
2005.PubMed/NCBI
|
|
22
|
Fytili P, Giannatou E, Karachalios T,
Malizos K and Tsezou A: Interleukin-10G and interleukin-10R
microsatellite polymorphisms and osteoarthritis of the knee. Clin
Exp Rheumatol. 23:621–627. 2005.PubMed/NCBI
|
|
23
|
Kroeger KM, Carville KS and Abraham LJ:
The −308 tumor necrosis factor-alpha promoter polymorphism effects
transcription. Mol Immunol. 34:391–399. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uglialoro AM, Turbay D, Pesavento PA,
Delgado JC, McKenzie FE, Gribben JG, Hartl D, Yunis EJ and Goldfeld
AE: Identification of three new single nucleotide polymorphisms in
the human tumor necrosis factor-alpha gene promoter. Tissue
Antigens. 52:359–367. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sezgin M, Barlas IO, Ankarali HC, Altintaş
ZM, Türkmen E, Gökdoğan T, Sahin G and Erdal M: Tumour necrosis
factor alpha −308G/A gene polymorphism: Lack of association with
knee osteoarthritis in a Turkish population. Clin Exp Rheumatol.
26:763–768. 2008.PubMed/NCBI
|
|
26
|
Muñoz-Valle JF, Oregόn-Romero E,
Rangel-Villalobos H, Martínez-Bonilla GE, Castañeda-Saucedo E,
Salgado-Goytia L, Leyva-Vázquez MA, Illades-Aguiar B,
Alarcón-Romero Ldel C, Espinoza-Rojo M and Parra-Rojas I: High
expression of TNF alpha is associated with −308 and −238 TNF alpha
polymorphisms in knee osteoarthritis. Clin Exp Med. 14:61–67. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji B, Shi J, Cheng X, Zhou J, Zhou Q, Cao
C and Pang J: Association analysis of two candidate polymorphisms
in the tumour necrosis factor-alpha gene with osteoarthritis in a
Chinese population. Int Orthop. 37:2061–2063. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han L, Song JH, Yoon JH, Park YG, Lee SW,
Choi YJ, Nam SW, Lee JY and Park WS: TNF-α and TNF-β Polymorphisms
are Associated with Susceptibility to Osteoarthritis in a Korean
Population. Korean J Pathol. 46:30–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kou S and Wu Y: Meta-analysis of tumor
necrosis factor alpha −308 polymorphism and knee osteoarthritis
risk. BMC Musculoskelet Disord. 15:3732014. View Article : Google Scholar : PubMed/NCBI
|