Introduction
Autoimmune inner ear disease (AIED) is one of the
few reversible causes of rapidly progressive, bilateral
sensorineural hearing loss (SNHL) over a period ranging from weeks
to months (1). It has been reported
that AIED has a predilection to occur in young adults, but variable
prevalence is also observed during childhood (4–30%) (2,3). SNHL is
frequently accompanied by tinnitus, persistent vertigo and
increased depression, which disrupt activities and personal
relationships by severely affecting the quality of life (4,5).
Currently, corticosteroids are a cornerstone in the
treatment of AIED; however, a successful reduction of symptoms and
the preservation of hearing can only be achieved in ~60% of
patients following timely corticosteroid administration (6). In addition, prolonged use of high-dose
corticosteroids (>4 weeks) carries risk of serious adverse
events, including hyperglycemia and weight gain (7). In steroid-dependent patients, hearing
immediately worsens if the dose declines (8). The development of novel treatment
strategies is crucial for the effective management of the
disease.
Although the pathogenesis is not fully understood,
cell- and/or humoral-mediated immune injuries serve important roles
in the development of AIED (9). When
a foreign antigen enters the inner ear, it is first processed by
immunocompetent cells present in and around the endolymphatic sac
(ES) (10). These immunocompetent
cells secrete various cytokines, including interleukin (IL)-1β and
tumor necrosis factor (TNF)-α, which recruit inflammatory cells
from the systemic circulation to the cochlea, further amplifying
the immune response and deteriorating inner ear damages (9,11,12).
Several TNF-α (13) or IL-1β
(14) antagonists have been
suggested as potential agents for treating patients with AIED with
no response to corticosteroid treatment. Alternative medicines for
AIED remain rare and further investigations into the crucial genes
for inflammatory response in ES are necessary.
Previously, studies analyzed the global gene
expression profile of the ES in rats (15) and humans (16,17). In
comparison with adjacent dura tissues, several inflammatory
response-associated genes were identified, including macrophage
migration inhibitory factor, small inducible cytokine subfamily e,
member 1, the C-C chemokine ligand (Ccl)21b (serine) and toll-like
receptor (Tlr)7 (15,16). In the current study, the microarray
data from Friis et al (15)
was further analyzed in order to identify inflammatory
response-associated genes in the ES by constructing a
protein-protein interaction (PPI) network, through module analysis
and by common gene screening.
Materials and methods
Microarray data
The microarray data were obtained from the European
Bioinformatics Institute database (ebi.ac.uk) with the accession
number E-MEXP-3022 (15). The
dataset contained three biological replicates of ES plus dura
tissues and three biological replicates of pure dura tissues
obtained from 10-week-old Lewis inbred rats.
Data normalization and identification
of differentially expressed genes (DEG)
Raw data were downloaded from the
A-AFFY-43-Affymetrix GeneChip Rat Genome 230 2.0 (Rat230_2)
platform (ebi.ac.uk/arrayexpress/arrays/A-AFFY-43/?ref=E-MEXP-3022)
and preprocessed to normalize and generate probe-level expression
data using the Robust Multichip Average algorithm (18) as implemented in the ‘affy’ package in
Bioconductor R (http://www.bioconductor.org/packages/release/bioc/html/affy.html).
When multiple probe IDs were matched to the same gene symbol, the
average value of expression was determined and the probe ID closest
to this value was selected to represent the gene symbol.
DEGs between ES plus dura tissues and pure dura
tissues were identified using the Linear Model for Microarray data
t-test method (19) from the
Bioconductor R package (http://www.bioconductor.org/packages/release/bioc/html/limma.html).
Genes were considered differentially expressed at P<0.05 and
|logFC(fold change)|>1.
To determine whether DEGs have the ability to
differentiate between ES plus dura tissues and pure dura tissues,
clustering analysis (20) was
performed to generate a heat map using the ComplexHeatmap R package
(https://github.com/jokergoo/ComplexHeatmap).
PPI network construction
DEGs were mapped into the Search Tool for the
Retrieval of Interacting Genes/Proteins 10.0 database (http://stringdb.org) (21) to obtain PPI pairs. Pairs with PPI
score ≥0.4 were retained to construct the PPI network using
Cytoscape software 2.8 (www.cytoscape.org) (22).
Crucial nodes within the PPI network were identified
by calculating three topological properties using the CytoNCA
plugin (version 2.1.6, parameter: Without weight) in the Cytoscape
software (http://apps.cytoscape.org/apps/cytonca) (23), including the degree [the number of
interactions per node (protein)] (24), the betweenness (the number of
shortest paths that pass through each node) (25) and subgraph centrality (the weighted
sum of all closed walks originating from each node) (26).
Functionally associated and densely interconnected
modules were extracted from the PPI network using Clustering with
Overlapping Neighborhood Expansion (ClusterONE version 1.0;
ftp://ftp.mshri.on.ca/pub/BIND/Tools/MCODE) in
the Cytoscape software (27).
Significant submodules were identified using P<0.01 and nodes
≥6.
Function enrichment analysis
To explain the underlying functions of DEGs, Gene
ontology (GO), including the terms of molecular function (MF),
biological process (BP) and cellular component (CC), and Kyoto
Encyclopedia of Genes and Genomes pathway enrichment analyses were
performed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID) 6.8 online tool (http://david.abcc.ncifcrf.gov) (28) based on hypergeometric tests. Counts
≥2 and P<0.05 were set as cut-off values.
Comparison with previous
literature
To further confirm the key genes in ES for AIED, the
results were also compared with previous studies by Møller et
al (16,17), which investigated the gene expression
profile of ES in humans.
Results
Identification of DEGs
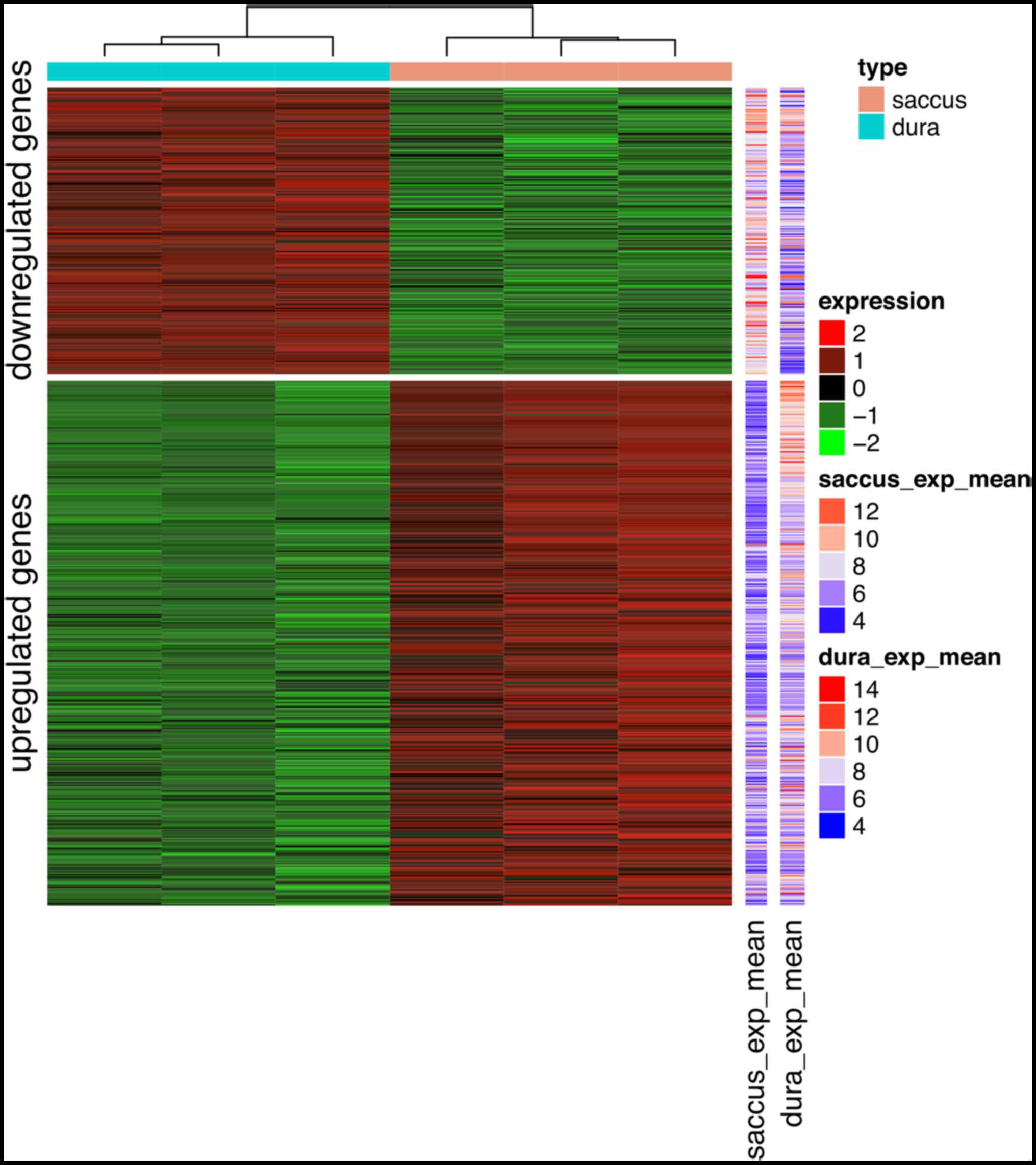
A total of 612 genes were identified as DEGs between
ES plus dura tissues and pure dura tissues based on the set
threshold (P<0.05 and |logFC|>1), including 396 up- and 216
downregulated genes (Table I).
Identified DEGs may allow to distinguish ES plus dura tissues from
pure dura tissues according to the heat map presented in Fig. 1.
 | Table I.Top 20 differentially expressed
genes. |
Table I.
Top 20 differentially expressed
genes.
| Gene | logFC | P-value |
|---|
| Upregulated |
| Ambn | 7.17 | <0.001 |
| Itgb6 | 3.90 | <0.001 |
| Foxi1 | 4.21 | <0.001 |
| Smr3b | 4.89 | <0.001 |
| Atp6v0a4 | 3.87 | <0.001 |
| Ppp1r1b | 3.81 | <0.001 |
| Aldh1a1 | 4.60 | <0.001 |
| Cwh43 | 3.57 | <0.001 |
| Pigr | 4.70 | <0.001 |
| Rab17 | 2.23 | <0.001 |
| Cldn16 | 4.27 | <0.001 |
| Hoxa5 | 4.08 | <0.001 |
| Cldn4 | 3.87 | <0.001 |
| Perp | 4.03 | <0.001 |
| Tbc1d9 | 3.68 | <0.001 |
| Pcdh10 | 4.20 | <0.001 |
| Pinlyp | 3.60 | <0.001 |
| Coch | 3.55 | <0.001 |
| Enpp5 | 3.26 | <0.001 |
| Fbxo2 | 4.00 | <0.001 |
| Downregulated |
| Mcpt1l1 | −3.12 | <0.001 |
| Tpsab1 | −3.64 | <0.001 |
| LOC102555472 | −2.91 | <0.001 |
| Kcne4 | −1.99 | <0.001 |
| Cpxm1 | −3.67 | <0.001 |
| Alx1 | −3.63 | <0.001 |
| Slc5a6 | −1.63 | <0.001 |
| Cpxm2 | −1.88 | <0.001 |
| Fmod | −1.54 | <0.001 |
| Slc6a8 | −1.59 | <0.001 |
| Cdkn1c | −1.48 | <0.001 |
| RGD1305645 | −1.84 | <0.001 |
| Efnb3 | −1.40 | <0.001 |
| Aox1 | −1.76 | <0.001 |
| Olfml3 | −1.39 | <0.001 |
| Krt33b | −1.94 | <0.001 |
| Krt12 | −3.24 | <0.001 |
| Elmod1 | −1.73 | <0.001 |
| Vnn1 | −1.86 | <0.001 |
| Mcpt10 | −2.89 | <0.001 |
Function enrichment analysis of
DEGs
GO enrichment analysis was performed for all DEGs to
reveal underlying functions. As a result, 116 (59 BP, 35 CC and 22
MF) GO terms were enriched for upregulated DEGs, including
GO:0007155-cell adhesion, with α5-integrin (Itga1) and secreted
phosphoprotein 1 (Spp1); GO:0031295-T cell co-stimulation, with
Ccl21 and Ccl19; and GO:0070555-response to IL-1, with Ccl21 and
Ccl19 (Table II). For downregulated
DEGs, 77 (including 48 BP, 11 CC, and 18 MF) GO terms were
enriched. In line with the upregulated DEGs, the majority of
downregulated DEGs was associated with GO:0007155-cell adhesion,
with transforming growth factor (Tgf)-βi; GO:0002224-toll-like
receptor signaling pathway, with Tlr2, Tlr7 and Tlr8; and
GO:0042346-positive regulation of nuclear factor (NF)-κB import
into the nucleus, with Tlr2 and Tlr7 (Table II).
 | Table II.GO enrichment analysis for
differentially expressed genes. |
Table II.
GO enrichment analysis for
differentially expressed genes.
| Regulation | GO term | Specific term | Count | P-value | Genes |
|---|
| Up | BP | GO:0007155-cell
adhesion | 17 | <0.001 | Col18A1, Tnfrsf12A,
Pcdh10, Itga1, Nectin2, Bcam, Adgrg1, Igsf11, Vwf, Ambn, Dsg2,
Cntn2, Lamc2, Emb, Cd24, Dpp4, Spp1 |
|
|
|
GO:0098609-cell-cell adhesion | 12 | 0.003 | Epcam, Cobll1,
Ldha, Krt18, Lad1, Cgn, Baiap2L1, Ppl, Capg, Sptbn2, Perp,
Eps8L1 |
|
|
| GO:0031295-T cell
costimulation | 4 | 0.008 | Ccl21, Ccl19, Cd24,
Dpp4 |
|
|
| GO:0001666-response
to hypoxia | 11 | 0.036 | Muc1, Cd38, Ldha,
Cldn3, Cryab, Arg2, Aldoc, P2Rx2, Cd24, Scnn1B, Dpp4 |
|
|
| GO:0070555-response
to interleukin-1 | 4 | 0.047 | Cd38, Sphk1, Anxa1,
Cited1 |
|
| CC |
GO:0005913-cell-cell adherens
junction | 15 | <0.001 | Cobll1, Ldha, Lad1,
Baiap2L1, Anxa1, Nectin2, Krt18, Ezr, Sorbs1, Cgn, Ppl, Capg,
Sptbn2, Dsc2, Eps8L1 |
|
|
|
GO:0005923-bicellular tight junction | 9 | 0.002 | Epcam, Cldn8,
Cldn16, Pard6B, Igsf5, Cldn4, Cldn3, Cgn, Cldn10 |
|
|
|
GO:0031012-extracellular matrix | 13 | 0.005 | Sbspon, Col18A1,
Coch, Clu, Mmrn1, Vwf, Plscr1, Sbsn, Dsp, Hspb1, Adamts1, Bmp7,
Dpt |
|
|
|
GO:0005578-proteinaceous extracellular
matrix | 12 | 0.012 | Coch, Gpc4, Vwf,
Ambn, Adamts8, Lect1, Col6A6, Egfl6, Gpld1, Lamc2, Adamts1,
Mepe |
|
|
| GO:0030054-cell
junction | 15 | 0.034 | Gabra1, Duox2,
S100A14, Espn, Cadps, Gria2, Pkp2, Loc498368, Pkp4, Sptbn2, Smagp,
Emb, Perp, Htr2B, Dpp4 |
|
| MF | GO:0098641-cadherin
binding involved in cell-cell adhesion | 13 | <0.001 | Cobll1, Ldha, Lad1,
Baiap2L1, Anxa1, Epcam, Krt18, Ezr, Cgn, Ppl, Capg, Sptbn2,
Eps8L1 |
|
|
| GO:0005518-collagen
binding | 6 | 0.005 | Coch, Vwf, Itga1,
Adgrg1, Dpp4, Pcolce2 |
|
|
|
GO:0042379-chemokine receptor binding | 3 | 0.013 | Ccl21, Ccl19,
S100A14 |
|
|
| GO:0031732-CCR7
chemokine receptor binding | 2 | 0.036 | Ccl21, Ccl19 |
|
|
| GO:0050839-cell
adhesion molecule binding | 5 | 0.039 | Ezr, Dsg2, Dsp,
Nectin2, Slc14A2 |
| Down | BP | GO:0007155-cell
adhesion | 13 | <0.001 | Nov, Lama1, Wisp2,
Mybpc1, Fbln5, Comp, Tgfbi, Acan, Thbs2, Lrfn3, Plpp3, Emilin1,
Farp2 |
|
|
| GO:0010811-positive
regulation of cell-substrate adhesion | 6 | <0.001 | Egflam, Smoc1,
Edil3, Vit, Ndnf, Emilin1 |
|
|
|
GO:0002224-toll-like receptor signaling
pathway | 3 | 0.017 | Tlr2, Tlr7,
Tlr8 |
|
|
| GO:0042346-positive
regulation of NF-kappaB import into nucleus | 3 | 0.017 | Tlr2, Grem1,
Tlr7 |
|
|
| GO:0045356-positive
regulation of interferon-alpha biosynthetic process | 2 | 0.042 | Tlr7, Tlr8 |
| Down | CC |
GO:0005578-proteinaceous extracellular
matrix | 26 | <0.001 | Alpl, Fmod, Cthrc1,
Wnt16, Mamdc2, Tgfb3, Mmp16, Vit, Ndnf, Emilin1, Cpz, Nov, Lama1,
Wisp2, Wnt4, Egflam, Sfrp1, Comp, Smoc1, Fbln5, Tgfbi, Acan,
Adamts12, Thbs2, Gpc1, Myoc |
|
|
|
GO:0031012-extracellular matrix | 21 | <0.001 | Fmod, Aebp1, Tgfb3,
Cpxm2, Mmp16, Edil3, Mmp14, Emilin1, Nov, Lama1, Sfrp1, Fbln5,
Comp, Tgfbi, Acan, Cma1, Adamts12, Tpsab1, Pcsk6, Col8A2, Myoc |
|
|
|
GO:0005615-extracellular space | 43 | <0.001 | Alpl, Fmod, Cthrc1,
Rbp4, Wnt16, Aebp1, Prtg, Tgfb3, Grem1, Aldh3A1, Cpz, Rgd1310507,
C1Qtnf5, Wisp2, Wnt4, Comp, Tgfbi, C1Qtnf2, Rgd1305645, Cpa3, Vnn1,
Eno3, Sema3C, Ces1D, Pcsk6, Gpc1, Pcsk5, Myoc, Scg2, Vasn, Igf1,
Cpxm2, Tcn2, Gas6, Clec11A, C8G, Lama1, Sfrp1, Fbln5, Cpxm1, Gdf10,
Tpsab1, Bmp5 |
|
|
|
GO:0005576-extracellular region | 23 | <0.001 | Fcer1A, Fmod, Gas6,
Ndnf, Clec11A, Mcpt1L1, C8G, Nov, Gzmm, Rgd1310507, Olfml3,
C1Qtnf5, Hmcn1, Sfrp1, Comp, Fndc1, Npw, Pdgfrl, Cma1, Tpsb2,
Prss23, Tpsab1, Pla2G2D |
|
|
|
GO:0005614-interstitial matrix | 4 | <0.001 | Egflam, Mamdc2,
Igf1, Vit |
|
| MF | GO:0005178-integrin
binding | 9 | <0.001 | Nov, Wisp2, Fbln5,
Tgfbi, Igf1, Edil3, Mmp14, Plpp3, Thy1 |
|
|
| GO:0008201-heparin
binding | 10 | <0.001 | Nov, Fmod, Wisp2,
Sfrp1, Comp, Pcsk6, Tpsb2, Thbs2, Pla2G2D, Ndnf |
|
|
|
GO:0004181-metallocarboxypeptidase
activity | 5 | <0.001 | Aebp1, Cpxm1, Cpa3,
Cpxm2, Cpz |
|
|
|
GO:0004185-serine-type carboxypeptidase
activity | 4 | <0.001 | Aebp1, Cpxm1,
Cpxm2, Cpz |
|
|
| GO:0005109-frizzled
binding | 5 | <0.001 | Cthrc1, Wnt16,
Wnt4, Sfrp1, Myoc |
Pathway analyses were also performed using DAVID
software. Upregulated DEGs were predicted to participate in six
pathways, including rno04530: Tight junction, with claudin
(Cldn)-4; rno04512: Extracellular matrix (ECM)-receptor
interaction, with Itga1 and Spp1; and rno04670: Leukocyte
transendothelial migration, with Cldn4 (Table III). Downregulated DEGs were
enriched in seven pathways, including inflammation-associated
diseases, rno05144: Malaria, with Tgf-β3 and Tlr2 (Table III).
 | Table III.Kyoto encyclopedia of genes and
genomes pathway enrichment analysis for differentially expressed
genes. |
Table III.
Kyoto encyclopedia of genes and
genomes pathway enrichment analysis for differentially expressed
genes.
| Regulation | Term | Count | P-value | Genes |
|---|
| Up | rno04530:Tight
junction | 11 | <0.001 | Cldn8, Cldn16,
Pard6B, Igsf5, Cldn4, Cldn3, Cgn, Cldn10, Myh14, Myl9, Llgl2 |
|
| rno04966:Collecting
duct acid secretion | 5 | 0.002 | Atp6V1C2, Clcnkb,
Atp6V1G3, Atp6V0A4, Car2 |
|
| rno04974:Protein
digestion and absorption | 8 | 0.002 | Col18A1, Kcnn4,
Col9A3, Col6A6, Mme, Slc1A1, Kcnq1, Dpp4 |
|
|
rno05412:Arrhythmogenic right ventricular
cardiomyopathy | 7 | 0.003 | Dsg2, Pkp2, Itgb6,
Itga1, Dsc2, Dsp, Cacna2D3 |
|
|
rno04512:ECM-receptor interaction | 7 | 0.009 | Vwf, Lamb3, Col6A6,
Itgb6, Itga1, Lamc2, Spp1 |
|
| rno04670:Leukocyte
transendothelial migration | 7 | 0.034 | Cldn8, Cldn16, Ezr,
Cldn4, Cldn3, Cldn10, Myl9 |
| Down |
rno05205:Proteoglycans in cancer | 7 | 0.008 | Wnt16, Wnt4, Tlr2,
Igf1, Fzd2, Gpc1, Twist1 |
|
|
rno04916:Melanogenesis | 5 | 0.011 | Wnt16, Wnt4, Adcy8,
Creb3L1, Fzd2 |
|
|
rno05144:Malaria | 4 | 0.015 | Comp, Tlr2, Tgfb3,
Thbs2 |
|
| rno04977:Vitamin
digestion and absorption | 3 | 0.016 | Slc5A6, Tcn2,
Slc19A2 |
|
| rno05200:Pathways
in cancer | 9 | 0.022 | Lama1, Wnt16, Wnt4,
Adcy8, Rasgrp2, Tgfb3, Skp2, Igf1, Fzd2 |
|
| rno05414:Dilated
cardiomyopathy | 4 | 0.037 | Sgcg, Adcy8, Tgfb3,
Igf1 |
|
| rno04390:Hippo
signaling pathway | 5 | 0.044 | Wnt16, Wnt4, Tgfb3,
Fzd2, Bmp5 |
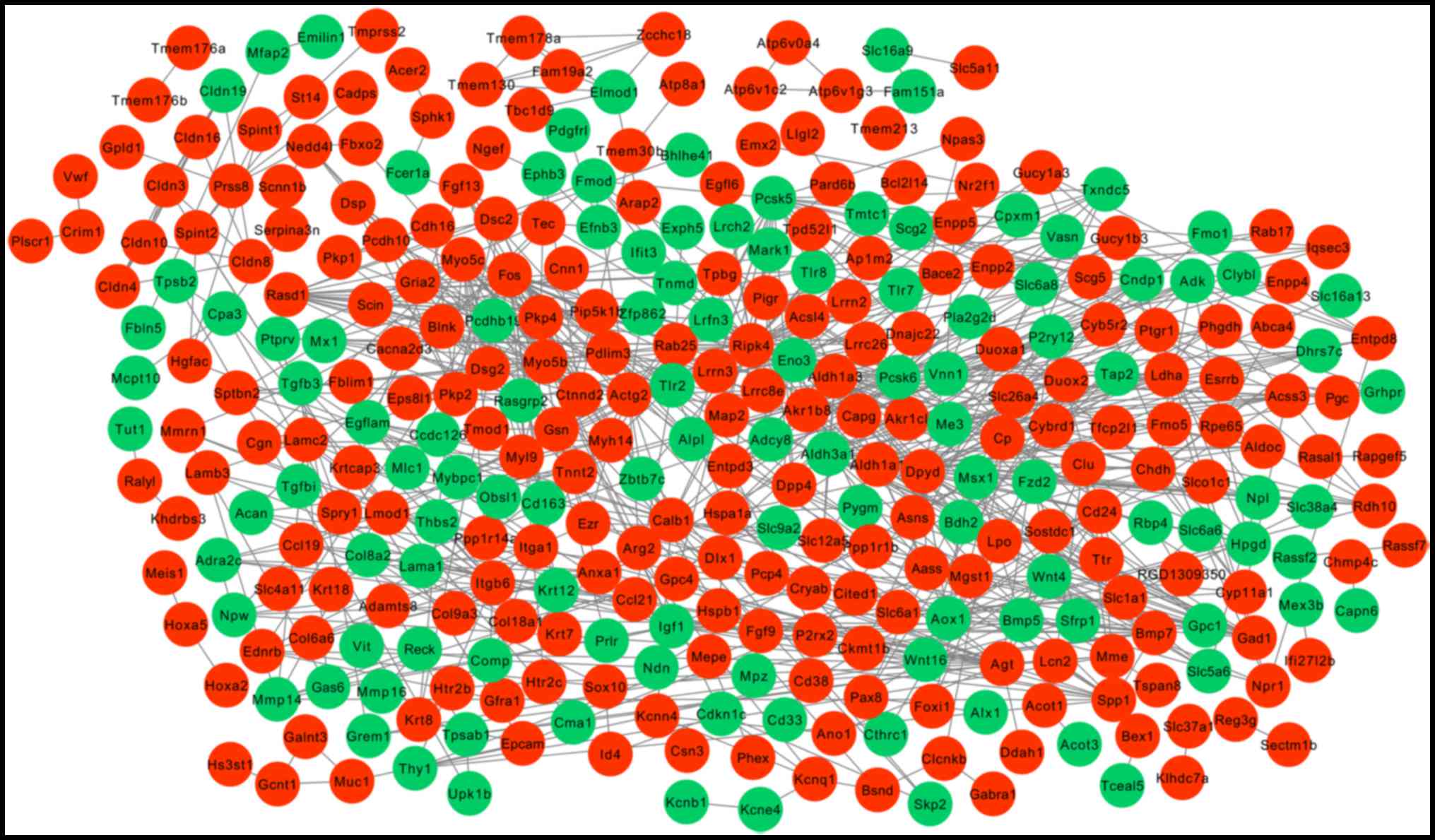
PPI network construction
Following the mapping of DEGs into protein
interactions pairs, a PPI network was constructed, including 338
nodes and 777 edges (interaction relationships; Fig. 2). Key nodes in the PPI network were
screened by calculating the degree, betweenness and subgraph
centrality. Ripk4, Dpyd, Actg2, Fos, Pcsk5, Aldh1a1, Calb1, Pcsk6,
Myo5c, Gad1 and Eno3 were in the top 20 genes of the three
topological characteristics, but were not determined to be enriched
in adhesion or immune pathways. Itga1 and Spp1 were enriched in
adhesion or immune pathways as described above, they were also
found to be crucial according to their degree and/or betweenness
centrality (Table IV).
 | Table IV.Top 20 hub genes presented by
topological property extracted from the protein-protein interaction
network. |
Table IV.
Top 20 hub genes presented by
topological property extracted from the protein-protein interaction
network.
|
| Gene (value) |
|---|
|
|
|
|---|
| Rank | Degreea |
Subgraphb |
Betweennessc |
|---|
| 1 | Ripk4 (47) | Ripk4 (11,351) | Ripk4 (28,108) |
| 2 | Dpyd (31) | Dpyd (8,498) | Fos (15,278) |
| 3 | Actg2 (31) | Actg2 (6,488) | Dpyd (10,661) |
| 4 | Fos (25) | Aldh3a1
(6,317) | Agt (10,644) |
| 5 | Pcsk5 (24) | Aldh1a3
(6,296) | Calb1 (8,620) |
| 6 | Aldh1a1 (23) | Aldh1a1
(6,260) | Spp1 (7,876) |
| 7 | Agt (23) | Pcsk5 (3,030) | Epcam (7,590) |
| 8 | Aldh3a1 (22) | Gad1 (2,611) | Actg2 (6,459) |
| 9 | Aldh1a3 (22) | Calb1 (2,598) | Myo5c (6,141) |
| 10 | Calb1 (22) | Pcsk6 (2,557) | Eno3 (5,163) |
| 11 | Pcsk6 (22) | Rasd1 (2,524) | Txndc5 (4,214) |
| 12 | Myo5c (21) | Myo5c (2,491) | Cd24 (4,135) |
| 13 | Gad1 (16) | Eno3 (2,170) | Krt8 (4,077) |
| 14 | Eno3 (16) | Myo5b (2,134) | Aldh1a1
(3,840) |
| 15 | Myo5b (16) | Rdh10 (1,939) | Pcsk5 (3,802) |
| 16 | Rasd1 (15) | Bdh2 (1,906) | Pcsk6 (3,788) |
| 17 | Itga1 (14) | Hpgd (1,906) | Itga1 (3,674) |
| 18 | Igf1 (14) | Dhrs7c (1,906) | Gad1 (3,617) |
| 19 | Itgb6 (14) | Fos (1,883) | Hspa1a (3,589) |
| 20 | Hspa1a, Spp1
(13) | Cndp1 (1,832) | Itgb6 (3,555) |
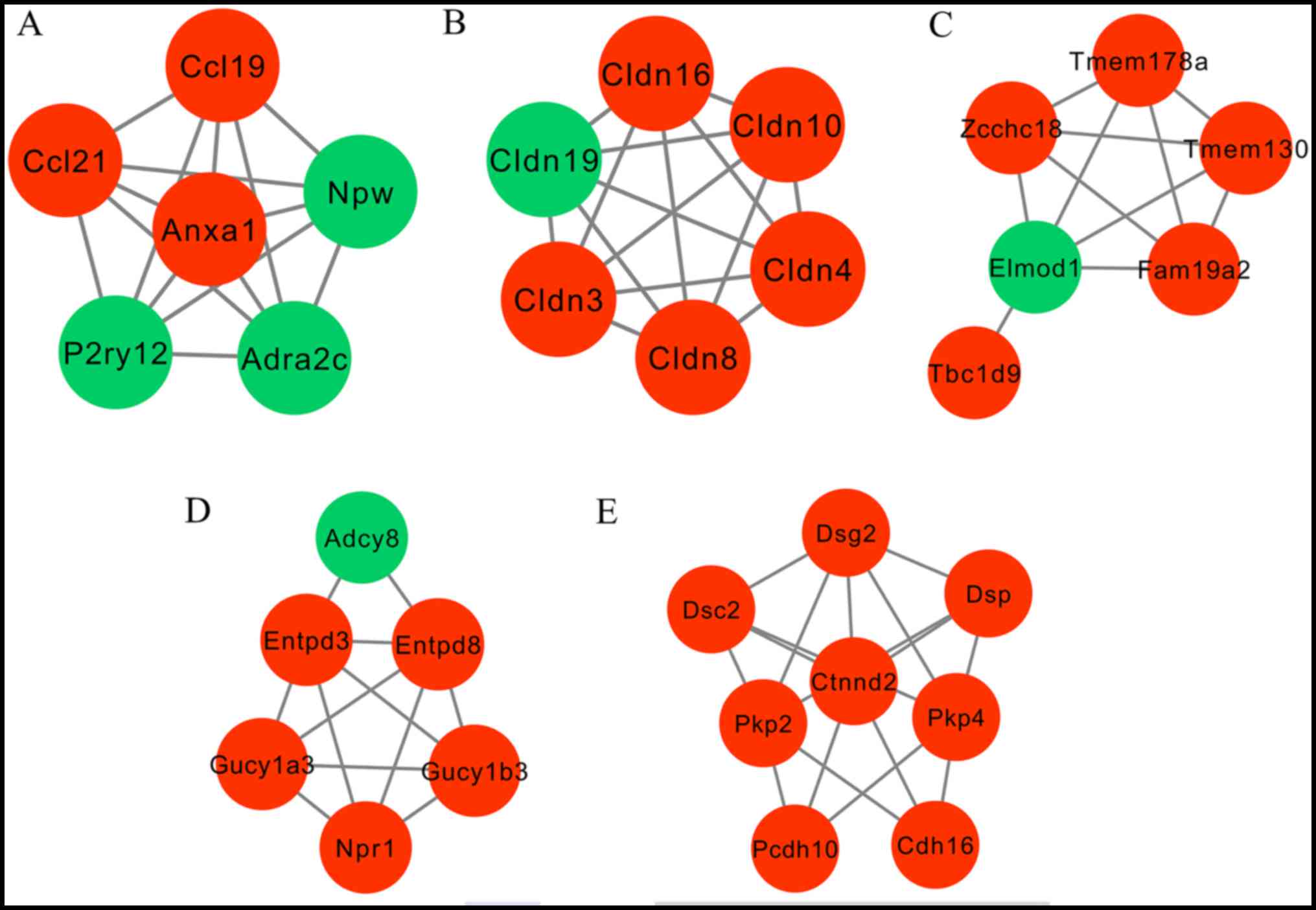
Following ClusterONE analysis, 5 significant
submodules were created (Fig. 3).
Function enrichment analysis revealed that the genes participating
in module 1 were closely associated with inflammation, including
Ccl21 and Ccl19, whereas the genes in module 2 and 5 were
significantly enriched in cell adhesion pathways, including Cldn4
(Fig. 4). In module 3, only one GO
term, GO:0005096-GTPase activator activity, was enriched and module
3 is not presented in Fig. 4.
Identification of common genes in rats
and humans by literature comparison
Møller et al (16) performed a study to investigate gene
expression of the human ES using adjacent dura mater as the
control. The authors identified that ES was a significant
immunological entity within the inner ear. To screen for immune
response-associated genes in the ES, the DEGs from rats from the
current study were compared with human DEGs, including Tlr1, Tlr2,
Tlr3, Tlr4, Tlr7, Fc fragment of IgA and IgM receptor, C-C
chemokine receptor 7, C-X-C chemokine (CXC) ligand 17, CXC receptor
3, α-interferon, IL-17, β-defensins, lactotransferrin precursor,
lactoperoxidase (Lpo), perforin 1, histatin, granzyme (Gzm) and
mucins (Mucs). A total of 6 common genes were identified, including
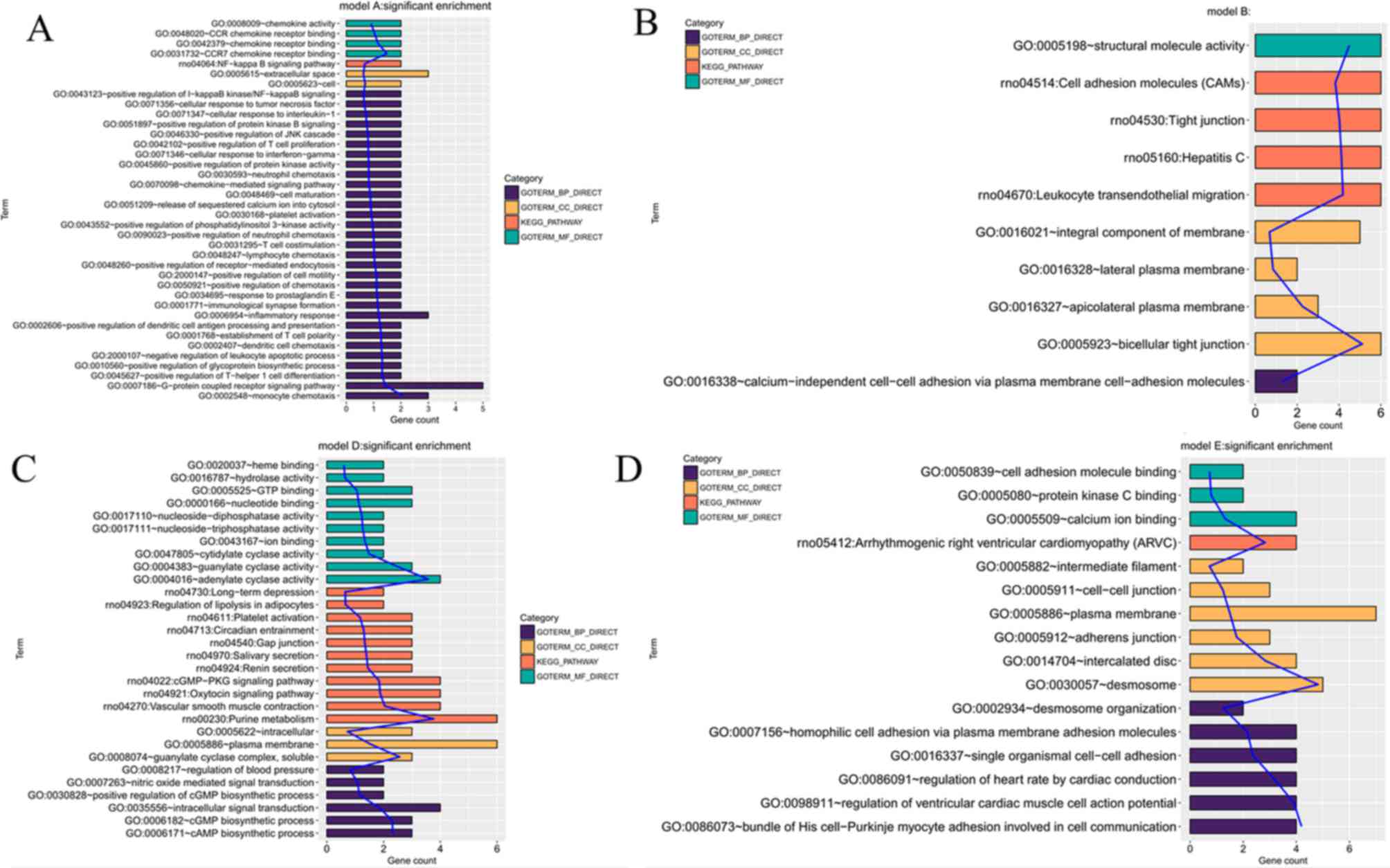
Tlr7, Tlr2, Tlr8, Lpo, Gzmm and Muc1 (Table V). Separate function analysis of
these genes was performed. Tlr7, Tlr2, Tlr8 and Gzmm were enriched
in immune or inflammatory response-associated GO terms or pathways
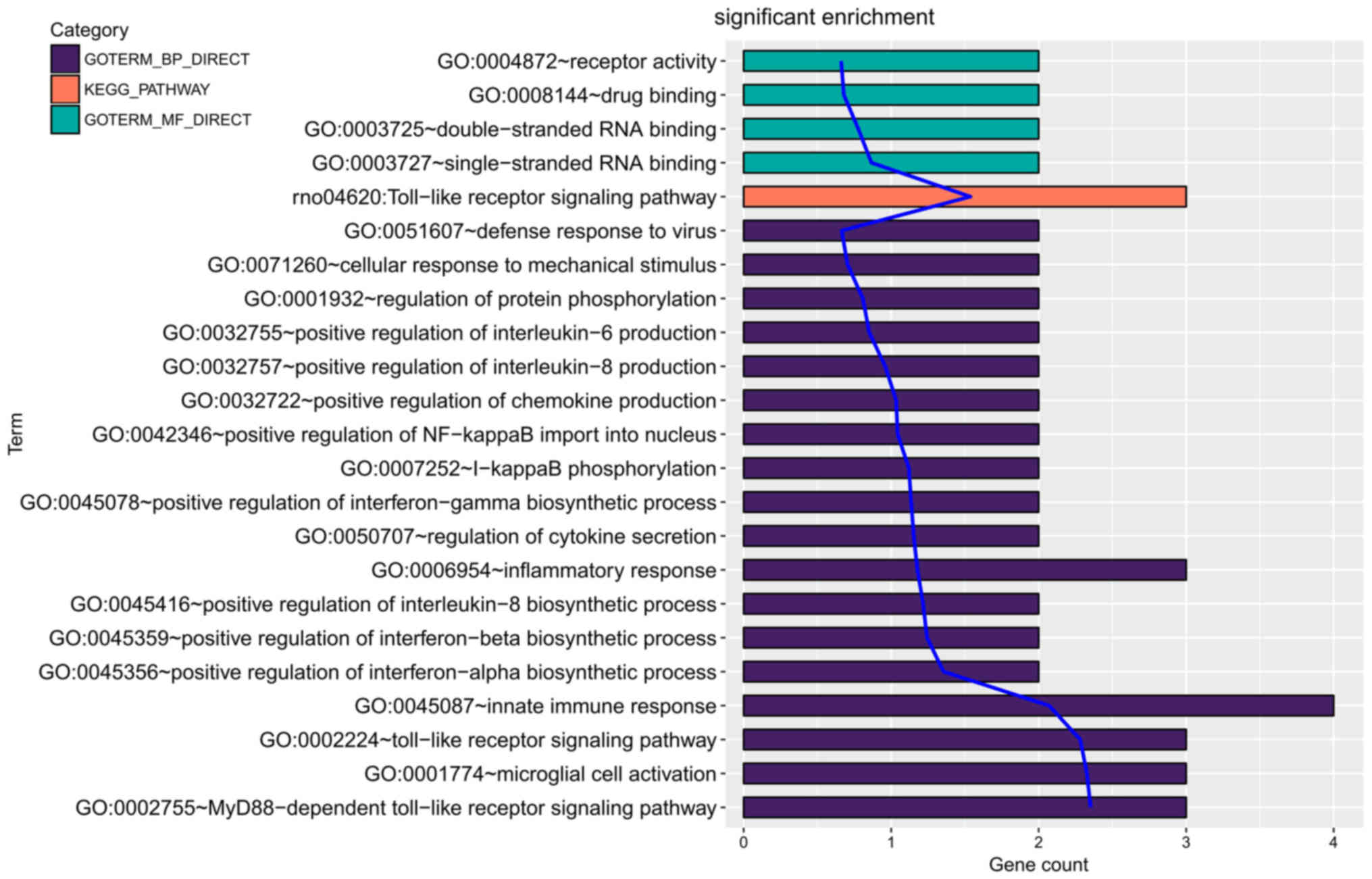
(Fig. 5). No enrichment was observed
for Lpo and Muc1, indicating Tlr7, Tlr2, Tlr8 and Gzmm were of
particular importance.
 | Table V.Genes associated with immune response
identified in rats and humans, obtained from a comparison with a
study by Møller et al (16). |
Table V.
Genes associated with immune response
identified in rats and humans, obtained from a comparison with a
study by Møller et al (16).
| Gene | logFC | P-value |
|---|
| Tlr7 | −1.15 | 0.003 |
| Tlr2 | −1.23 | 0.003 |
| Tlr8 | −1.40 | <0.001 |
| Lpo | 2.02 | 0.004 |
| Gzmm | −1.48 | <0.001 |
| Muc1 | 3.28 | <0.001 |
Discussion
Compared with the previous study from Friis et
al (15), a less restrictive
threshold value [P<0.05 and |logFC(fold change)|>1 vs. false
discovery rate <0.1] was adopted for screening of DEGs in the ES
in the current study. As a result, 619 DEGs were identified
compared with 463 DEGs reported by Friis et al (15). Subsequently, DEGs were subjected to a
PPI network construction, module analysis, function analysis and a
comparison with the results reported by Møller et al
(16), none of which was included in
the work presented by Friis et al (15). The results of the current study may
provide a more solid base compared with previous work and novel
conclusions were obtained. In the present study, a series of
inflammatory response-associated genes were screened. The hub roles
of Itga1, Cd24 and Spp1 were identified by calculating three
topological properties of the PPI network. Ccl21, Ccl19 and Cldn4
were demonstrated to be crucial following significant module
analysis according to the corresponding threshold, which revealed
they were enriched in inflammation pathways. Tlr7, Tlr2, Gzmm and
Tlr8 were identified as common genes associated with inflammatory
responses in rat and human ES (16).
Abnormal expressions of the aforementioned inflammatory-associated
genes may describe the underlying mechanism for the development of
AIED.
Ccl21, Ccl19, Tlr7, Tlr2 and Tlr8 are known
inflammatory-associated genes (29–33,34–36),
making their presence in ES and association with AIED plausible. It
has previously been reported that Ccl19 and Ccl21 were
significantly induced in several autoimmune diseases, including
rheumatoid arthritis (29),
autoimmune encephalomyelitis (30)
and multiple sclerosis (31) which
have been diagnosed in ≤1/3 of patients with AIED (32,33).
High expressions of Ccl19 and Ccl21 were demonstrated to initiate
specific immune responses by co-stimulating the expansion of
CD4+ and CD8+ T cells (32), inducing Th1 (37) and macrophage polarization to secrete
inflammatory cytokines (including IL-8) (29), and mediating antigen presenting cell
trafficking (including dendritic cells) (38). A combination of the continuous local
antigenic stimulation and the maintenance of chronic inflammation
ultimately lead to the development of AIED.
Tlrs are associated with the activation of the
innate immune system by recognizing pathogen-associated molecular
patterns (39). Proinflammatory
effects have been described for Tlrs previously (34), whereas other studies implied
protective effects of activated Tlr2, Tlr7 and Tlr8 in mouse models
of asthma (35,36,40).
Treatment with Tlr7/8 agonist, resiquimod (RSQ), was reported to
reduce allergen-induced airway reactivity and inflammation
(35). The underlying mechanism is
associated with the induction of NF-E2-related factor 2 and
copper/zinc superoxide dismutase (35). The suppressive effect of RSQ in
asthma is further dependent on the induction of IL-27 and is
associated with enhanced expression of the immunomodulatory
molecule B7-homolog 1 on pulmonary antigen presenting cells
(36). Pam3Cys, a Tlr2 agonist
treatment, has been demonstrated to cause significant reductions of
eosinophils and increase numbers of regulatory T cells (Tregs) in
lung infiltrates, resulting in long-term protection against
manifestation of allergic asthma (40). Previous studies revealed that
induction of the generation of antigen specific cluster of
differentiation (CD)4+CD25+forkhead box
P3+Tregs by adipose tissue-derived stem cells
significantly improved hearing function and protected inner hair
cells from loss in previous AIED models (41,42). The
current study speculated that Tlr2 and Tlr7/8 agonist may be
potential agents for the treatment of AIED, which was supported by
the observed downregulation of Tlr7, Tlr2 and Tlr8 in ES.
Gzms comprise a family of serine protease (a, b, h,
k and m) that is constitutively and abundantly expressed in innate
effector natural killer cells to eliminate virus-infected and tumor
cells to prevent inflammation (43,44).
Mice lacking Gzms exhibited a reduced immune tolerance and
increased inflammation levels through a type I interferon-dependent
pathway, leading to the development of autoimmune diseases,
including diabetes (43,45) and ulcerative colitis (46). These two autoimmune diseases were
further associated with SNHL (47,48).
Gzmm was proposed to be downregulated in AIED, which was in
accordance with the results of the current study.
The ECM is a non-cellular component in all tissues
and organs and adherens junctions between cells, and are essential
for the maintenance of the structural and functional integrity of
organs (49). Disruption of ECM and
adherens junctions may contribute to the destruction of the
epithelial barrier and favor the interaction of leukocytes with
endothelial cells, promoting the development of
inflammation-associated diseases (50), including AIED. Cai et al
(51) previously reported that the
expression of ECM gene Itga5 in the basal sections of the sensory
epithelium in the cochlea was positively correlated with greater
hearing loss as a result to exposure to an intense noise for 2 h.
Endogenous overexpression of another ECM gene, SPP1, induces Ccl5
and matrix metalloproteinase-2 activation that contributes to
proinflammatory events (52). Lee
et al (50) recently
demonstrated that the level of Cldn4, coding for a tight junction
protein, was increased in blood and plasma of asthmatic patients
and lung tissues from the model mice, accompanied with significant
increases in tight junction disruptions. A Cldn4 knockdown by small
interfering RNA transfection significantly increased inflammatory
cytokine expression (50). In
accordance with these findings, SPP1, Itga1 and Cldn4 were
upregulated in rats ES, suggesting these genes may be underlying
targets for the treatment of AIED.
In conclusion, the present study revealed a number
of important inflammatory-associated genes, including Tlr2, Tlr7,
Tlr8, Gzmm, Itga1, Spp1 and Cldn4, in ES. Abnormal expression of
these genes may describe the underlying mechanism for the
development of AIED. Further studies with an AIED animal model are
required to confirm these conclusions.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant nos. 81371084 and 81570924).
Availability of data and materials
The E-MEXP-3022 microarray datasets analyzed during
the current study are available in the European Bioinformatics
Institute database repository (ebi.ac.uk).
Author's contributions
JZ and AX participated in the design of the study.
JZ and NW performed the bioinformatics analysis. JZ, NW and AX
contributed to the acquisition and interpretation of the data. JZ
drafted the manuscript. AX revised the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Matsuoka AJ and Harris JP: Autoimmune
inner ear disease: A retrospective review of forty-seven patients.
Audiol Neurootol. 18:228–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang NC and Sataloff RT: Autoimmune inner
ear disease in children. Otol Neurotol. 32:213–216. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dougherty W, Thatayatikom A and Bush ML:
Paediatric autoimmune inner ear disease: A case series. Audiol Med.
13:32–39. 2015.
|
|
4
|
Chen J, Liang J, Ou J and Cai W: Mental
health in adults with sudden sensorineural hearing loss: An
assessment of depressive symptoms and its correlates. J Psychosom
Res. 75:72–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carlsson PI, Hall M, Lind KJ and Danermark
B: Quality of life, psychosocial consequences, and audiological
rehabilitation after sudden sensorineural hearing loss. Int J
Audiol. 50:139–144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niparko JK, Wang NY, Rauch SD, Russell GB,
Espeland MA, Pierce JJ, Bowditch S, Masuda A, Gulya AJ, Gantz BJ,
et al: Serial audiometry in a clinical trial of AIED treatment.
Otol Neurotol. 26:908–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alexander TH, Weisman MH, Derebery JM,
Espeland MA, Gantz BJ, Gulya AJ, Hammerschlag PE, Hannley M, Hughes
GB, Moscicki R, et al: Safety of high-dose corticosteroids for the
treatment of autoimmune inner ear disease. Otol Neurotol.
30:443–448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kanaya T, Kamito T, Shirato M and Unno T:
Steroid dependent sensorineural hearing loss in a patient with
bilateral meniere's disease. Practica Oto-Rhino-Laryngologica.
85:191–196. 1992. View Article : Google Scholar
|
|
9
|
Goodall AF and Siddiq MA: Current
understanding of the pathogenesis of autoimmune inner ear disease:
A review. Clin Otolaryngol. 40:412–419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi M and Harris JP: Anatomic
distribution and localization of immunocompetent cells in normal
mouse endolymphatic sac. Acta Otolaryngol. 106:409–416. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gopen Q, Keithley EM and Harris JP:
Mechanisms underlying autoimmune inner ear disease. Drug Discov
Today Dis Mech. 3:137–142. 2006.https://doi.org/10.1016/j.ddmec.2006.02.006
View Article : Google Scholar
|
|
12
|
Satoh H, Firestein GS, Billings PB, Harris
JP and Keithley EM: Proinflammatory cytokine expression in the
endolymphatic sac during inner ear inflammation. J Assoc Res
Otolaryngol. 4:139–147. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pathak S, Stern C and Vambutas A:
N-Acetylcysteine attenuates tumor necrosis factor alpha levels in
autoimmune inner ear disease patients. Immunol Res. 63:236–245.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vambutas A, Lesser M, Mullooly V, Pathak
S, Zahtz G, Rosen L and Goldofsky E: Early efficacy trial of
anakinra in corticosteroid-resistant autoimmune inner ear disease.
J Clin Invest. 124:4115–4122. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friis M, Martinbertelsen T, Friishansen L,
Winther O, Henao R, Sørensen MS and Qvortrup K: Gene expression of
the endolymphatic sac. Acta Otolaryngol. 131:1257–1263. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Møller MN, Kirkeby S, Vikeså J, Nielsen FC
and Cayé-Thomasen P: Gene expression demonstrates an immunological
capacity of the human endolymphatic sac. Laryngoscope.
125:E269–E275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Møller MN, Kirkeby S, Vikeså J, Nielsen FC
and Cayé-Thomasen P: Neuronal fibers and neurotransmitter receptor
expression in the human endolymphatic sac. Otol Neurotol.
38:765–773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rito T, Deane CM and Reinert G: The
importance of age and high degree, in protein-protein interaction
networks. J Comput Biol. 19:785–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Hernandez JM and Van MP:
Betweenness centrality in a weighted network. Phys Rev E Stat
Nonlin Soft Matter Phys. 77:0461052008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Estrada E and Rodríguez-Velázquez JA:
Subgraph centrality in complex networks. Phys Rev E Stat Nonlin
Soft Matter Phys. 71:0561032005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pickens SR, Chamberlain ND, Volin MV, Pope
RM, Mandelin AM II and Shahrara S: Characterization of CCL19 and
CCL21 in rheumatoid arthritis. Arthritis Rheum. 63:914–922. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Columbacabezas S, Serafini B, Ambrosini E
and Aloisi F: Lymphoid chemokines CCL19 and CCL21 are expressed in
the central nervous system during experimental autoimmune
encephalomyelitis: Implications for the maintenance of chronic
neuroinflammation. Brain Pathol. 13:38–51. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bielecki B, Jatczak-Pawlik I, Wolinski P,
Bednarek A and Glabinski A: Central nervous system and peripheral
expression of CCL19, CCL21 and their receptor CCR7 in experimental
model of multiple sclerosis. Arch Immunol Ther Exp (Warsz).
63:367–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morovic Vergles J, Radic M, Kovacic J and
Salamon L: Successful use of adalimumab for treating rheumatoid
arthritis with autoimmune sensorineural hearing loss: Two birds
with one stone. J Rheumatol. 37:1080–1081. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tanaka M and Tanaka K: Sudden hearing loss
as the initial symptom in Japanese patients with multiple sclerosis
and seropositive neuromyelitis optica spectrum disorders. J
Neuroimmunol. 298:16–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiménezdalmaroni MJ, Gerswhin ME and
Adamopoulos IE: The critical role of toll-like receptors-from
microbial recognition to autoimmunity: A comprehensive review.
Autoimmun Rev. 15:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nadeem A, Siddiqui N, Alharbi NO, Alharbi
MM and Ahmad SF: TLR-7 agonist attenuates airway reactivity and
inflammation through Nrf2-mediated antioxidant protection in a
murine model of allergic asthma. Int J Biochem Cell Biol. 73:53–62.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Daluge K, Jirmo AC, Busse M and Hansen G:
TLR 7/8 agonist resiquimod alleviates murine allergic asthma
through IL-27 production and up regulation of B7-H1 on antigen
presenting cells. Eur Respir J. 46:PA18962015.
|
|
37
|
Flanagan K, Moroziewicz D, Kwak H, Hörig H
and Kaufman HL: The lymphoid chemokine CCL21 costimulates naive T
cell expansion and Th1 polarization of non-regulatory
CD4+ T cells. Cell Immunol. 231:75–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hua J, Stevenson W, Dohlman T, Calcagno N,
Pirmadjid N, Sadrai Z, Chauhan S, Saban D and Dana R: The
CCR7-CCL19/CCL21 axis mediates enhanced antigen-presenting cell
trafficking in high-risk corneal transplantation. Invest Ophthalmol
Vis Sci. 54:12892013.PubMed/NCBI
|
|
39
|
Underhill DM: Collaboration between the
innate immune receptors dectin-1, TLRs, and Nods. Immunol Rev.
219:75–87. 2010. View Article : Google Scholar
|
|
40
|
Nawijn MC, Motta AC, Gras R, Shirinbak S,
Maazi H and van Oosterhout AJ: TLR-2 activation induces regulatory
T cells and long-term suppression of asthma manifestations in mice.
Plos One. 8:e553072013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoo TJ, Du X and Zhou B: The paracrine
effect of mesenchymal human stem cells restored hearing in
β-tubulin induced autoimmune sensorineural hearing loss. Hear Res.
330:57–61. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou Y, Yuan J, Zhou B, Lee AJ, Lee AJ,
Ghawji M Jr and Yoo TJ: The therapeutic efficacy of human adipose
tissue-derived mesenchymal stem cells on experimental autoimmune
hearing loss in mice. Immunology. 133:133–140. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
García-Laorden MI, Stroo I, Blok DC,
Florquin S, Medema JP, de Vos AF and van der Poll T: Granzymes A
and B regulate the local inflammatory response during Klebsiella
pneumoniae Pneumonia. J Innate Immun. 8:258–268. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Walch M, Dotiwala F, Mulik S, Thiery J,
Kirchhausen T, Clayberger C, Krensky AM, Martinvalet D and
Lieberman J: Cytotoxic cells kill intracellular bacteria through
Granulysin-mediated delivery of Granzymes. Cell. 157:1309–1323.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mollah ZU, Quah HS, Graham KL, Jhala G,
Krishnamurthy B, Francisca MDJ, Chee J, Trivedi PM, Pappas EG,
Mackin L, et al: Granzyme A-deficiency breaks immune tolerance and
promotes autoimmune diabetes through a type I interferon-dependent
pathway. Diabetes. 66:3041–3050. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Souza-Fonseca-Guimaraes F, Krasnova Y,
Putoczki T, Miles K, Macdonald KP, Town L, Shi W, Gobe GC, Mcdade
L, Mielke LA, et al: Granzyme M has a critical role in providing
innate immune protection in ulcerative colitis. Cell Death Dis.
7:e23022016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Casella G, Corbetta D, Zolezzi M, Di Bella
C, Villanacci V, Salemme M, Milanesi U, Antonelli E, Baldini V and
Bassotti G: Symptomatic sensorineural hearing loss in patients with
ulcerative colitis. Tech Coloproctol. 19:729–731. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lin CF, Lee KJ, Yu SS and Lin YS: Effect
of comorbid diabetes and hypercholesterolemia on the prognosis of
idiopathic sudden sensorineural hearing loss. Laryngoscope.
126:142–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burridge K, Fath K, Kelly T, Nuckolls G
and Turner C: Focal Adhesions: Transmembrane junctions between the
extracellular matrix and the cytoskeleton. Annu Rev Cell Biol.
4:487–525. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lee PH, Kim BG, Lee SH, Lee JH, Park SW,
Kim DJ, Park CS, Leikauf GD and Jang AS: Alteration in Claudin-4
contributes to airway inflammation and responsiveness in asthma.
Allergy Asthma Immunol Res. 10:25–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cai Q, Patel M, Coling D and Hu BH:
Transcriptional changes in adhesion-related genes are site-specific
during noise-induced cochlear pathogenesis. Neurobiol Dis.
45:723–732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Caballero EP, Santamaría MH and Corral RS:
Endogenous osteopontin induces myocardial CCL5 and MMP-2 activation
that contributes to inflammation and cardiac remodeling in a mouse
model of chronic Chagas heart disease. Biochim Biophys Acta.
1864:11–23. 2017. View Article : Google Scholar : PubMed/NCBI
|